Midkine and NANOG Have Similar Immunohistochemical Expression Patterns and Contribute Equally to an Adverse Prognosis of Oral Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Selected Patient Information
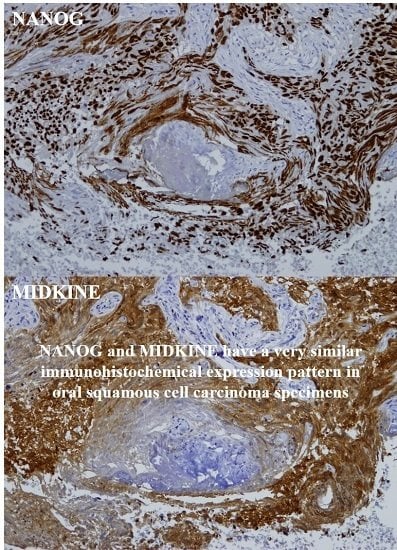
2.2. Expression Patterns of NANOG and MK in OSCC Pretreatment Biopsy Specimens
2.3. Association Between Clinicopathological Features and the Expression Patterns of NANOG and MK
2.4. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Immunohistochemical Analysis
4.3. Statistical Analysis and Overall Survival Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| OSCC | Oral squamous cell carcinoma |
| MK | Midkine |
| IHC | Immunohistochemistry or immunohistochemical |
| P53 | Tumor suppress gene p53 |
| OCT4 | Octamer-binding transcription factor 4 |
| SOX2 | Sex determining region Y-box 2 |
| STAT3 | Signal transducer and activator of transcription 3 |
| CSCs | Cancer stem cells |
| ESCs | Embryonic stem cells |
| Surg | Surgery-only group |
| Surg + RT | Surgery combined with adjuvant radiotherapy or concurrent chemoradiotherapy group |
| PaRT | Palliative radiotherapy or no treatment group |
| NF-κB | Nuclear factor-κB |
| DAZL | Deleted in azoospermia-like gene |
| TTF-1 | Thyroid transcription factor-1 |
| VEGF | Vascular endothelial growth factor |
References
- Russo, D.; Merolla, F.; Mascolo, M.; Ilardi, G.; Romano, S.; Varricchio, S.; Napolitano, V.; Celetti, A.; Postiglione, L.; Di Lorenzo, P.P.; et al. FKBP51 immunohistochemical expression: A new prognostic biomarker for OSCC? Int. J. Mol. Sci. 2017, 18, 443. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Fujimori, H.; Ueda, M.; Jono, H.; Shinriki, S.; Ota, T.; Sueyoshi, T.; Taura, M.; Taguma, A.; Kai, H.; et al. Midkine expression is correlated with an adverse prognosis and is down-regulated by p53 in oral squamous cell carcinoma. Int. J. Oncol. 2010, 37, 797–804. [Google Scholar] [PubMed]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Baillie, R.; Tan, S.T.; Itingeang, T. Cancer stem cells in oral cavity squamous cell carcinoma: A review. Front. Oncol. 2017, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Søland, T.M.; Brusevold, I.J. Prognostic molecular markers in cancer—Quo vadis? Histopathology 2013, 63, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Mallett, S.; Timmer, A.; Sauerbrei, W.; Altman, D.G. Reporting of prognostic studies of tumor markers: A review of published articles in relation to REMARK guidelines. Br. J. Cancer 2010, 102, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kang, Y.H.; Lee, J.S.; Byun, J.H.; Kim, U.K.; Jang, S.J.; Rho, G.J.; Park, B.W. Positive expression of NANOG, mutant p53, and CD44 is directly associated with clinicopathological features and poor prognosis of oral squamous cell carcinoma. BMC Oral Health 2015, 15, 153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Filho, M.S.; Nör, J.E. The biology of head and neck cancer stem cells. Oral Oncol. 2012, 48, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, G.; Sawant, S.S.; Hague, A.; Kingsley, K.; Devaraj, H. Association of differential β-catenin expression with Oct-4 and Nanog in oral squamous cell carcinoma and their correlation with clinicopathological factors and prognosis. Head Neck 2015, 37, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Chiou, S.H.; Yu, C.C.; Huang, C.Y.; Lin, S.C.; Liu, C.J.; Tsai, T.H.; Chou, S.H.; Chien, C.S.; Ku, H.H.; Lo, J.F. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin. Cancer Res. 2008, 14, 4085–4095. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Ohnishi, Y.; Inoue, H.; Wato, M.; Tanaka, A.; Kakudo, K.; Nozaki, M. NANOG expression correlates with differentiation, metastasis and resistance to preoperative adjuvant therapy in oral squamous cell carcinoma. Oncol. Lett. 2014, 7, 35–40. [Google Scholar] [PubMed]
- Tsai, L.L.; Yu, C.C.; Chang, Y.C.; Yu, C.H.; Chou, M.Y. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Jono, H.; Ando, Y. Midkine: A novel prognostic biomarker for cancer. Cancers 2010, 2, 624–641. [Google Scholar] [CrossRef] [PubMed]
- Ota, K.; Fujimori, H.; Ueda, M.; Shiniriki, S.; Kudo, M.; Jono, H.; Fukuyoshi, Y.; Suqiuchi, H.; Iwase, H.; Shinohara, M.; et al. Midkine as a prognostic biomarker in oral squamous cell carcinoma. Br. J. Cancer 2008, 99, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Ruan, M.; Ji, T.; Wu, Z.; Zhou, J.; Zhang, C. Evaluation of expression of midkine in oral squamous cell carcinoma and its correlation with tumor angiogenesis. Int. J. Oral Maxillofac. Surg. 2007, 36, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, S.; Take, M.; Pedraza, C.; Muramatsu, T. Mapping and characterization of a retinoic acid-responsive enhancer of midkine, a novel heparin-binding growth/differentiation factor with neurotrophic activity. J. Biochem. 1994, 115, 1088–1096. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Park, B.W.; Toms, D.; Li, J. Midkine promotes proliferation of primordial germ cells by inhibiting the expression of the Deleted in Azoospermia-Like gene. Endocrinology 2012, 153, 3482–3492. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T. Midkine, a heparin-binding cytokine with multiple roles in development, repair and diseases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010, 86, 410–425. [Google Scholar] [CrossRef] [PubMed]
- Jham, B.C.; Costa, N.L.; Silva, J.M.; de Miranda, A.C.; Oliveira, J.C.; Silva, T.A.; Batista, A.C. Midkine expression in oral squamous cell carcinoma and leukoplakia. J. Oral Pathol. Med. 2012, 41, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Park, B.W.; Jang, S.J.; Byun, J.H.; Kang, Y.H.; Choi, M.J.; Park, W.U.; Lee, W.J.; Rho, G.J. Cryopreservation of human dental follicle tissue for use as a resource of autologous mesenchymal stem cells. J. Tissue Eng. Reg. Med. 2017, 11, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Shinchi, H.; Kurahara, H.; Mataki, Y.; Noma, H.; Maemura, K.; Aridome, K.; Yokomine, T.; Natsugoe, S.; Aikou, T.; et al. Clinical significance of midkine expression in pancreatic head carcinoma. Br. J. Cancer 2007, 97, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Krzystek-Korpacka, M.; Matusiewicz, M.; Diakowska, D.; Grabowski, K.; Blachut, K.; Kustrzeba-Wojcicka, I.; Bana, T. Serum midkine depends on lymph node involvement and correlates with circulating VEGF-C in oesophageal squamous cell carcinoma. Biomarkers 2007, 12, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Ota, T.; Jono, H.; Ota, K.; Shinriki, S.; Ueda, M.; Sueyoshi, T.; Nakatani, K.; Hiraishi, Y.; Wada, T.; Fujita, S.; et al. Downregulation of midkine induces cisplatin resistance in human oral squamous cell carcinoma. Oncol. Rep. 2012, 27, 1674–1680. [Google Scholar] [PubMed]
- Mirkin, B.L.; Clark, S.; Zheng, X.; Chu, F.; White, B.D.; Greene, M.; Rebbaa, A. Identification of midkine as a mediator for intercellular transfer of drug resistance. Oncogene 2005, 24, 4965–4974. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Kim, I.J.; Park, J.H.; Shin, Y.; Ku, J.L.; Jung, M.S.; Yoo, B.C.; Kim, H.K.; Park, J.G. Identification of genes with differential expression in acquired drug-resistant gastric cancer cells using high-density oligonucleotide microarrays. Clin. Cancer Res. 2004, 10, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, P.R.; Mucenski, M.L.; Whitsett, J.A. Thyroid transcription factor (TTF)-1 regulates the expression of midkine (MK) during lung morphogenesis. Dev. Dyn. 2003, 227, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Ugai, S.; O-Wang, J.; Namba, M.; Kadomatsu, K.; Muramatsu, T.; Matsubara, S.; Sakiyama, S.; Tagawa, M. Cell growth- and P53-dependent transcriptional activity of the midkine promoter confers suicide gene expression in tumor cells. Oncol. Rep. 2003, 10, 1301–1305. [Google Scholar] [CrossRef] [PubMed]
- Ota, T.; Ota, K.; Jono, H.; Fujimori, H.; Ueda, M.; Shinriki, S.; Sueyoshi, T.; Shinohara, M.; Ando, Y. Midkine expression in malignant salivary gland tumors and its role in tumor angiogenesis. Oral Oncol. 2010, 46, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, E.H.; Frank, B.T.; Chinn, L.; Coxon, A.; Li, S.; Polesso, F.; Slavin, A.; Ruefli-Brasse, A.; Wesche, H. The growth factor Midkine antagonizes VEGF signaling in vitro and in vivo. Neoplasia 2008, 10, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.T.; Liu, S.Y.; Zheng, P.S. Cytoplasmic NANOG-positive stromal cells promote human cervical cancer progression. Am. J. Pathol. 2012, 181, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Li, S.; Peng, B.; Ye, Y.; Deng, X.; Yao, K. Embryonic stem cellsmarkers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS ONE 2013, 8, e56324. [Google Scholar]
- Shibata, Y.; Muramatsu, T.; Hirai, M.; Inui, T.; Kimura, T.; Saito, H.; McCormick, L.M.; Bu, G.; Kadomatsu, K. Nuclear targeting by growth factor midkine. Mol. Cell. Biol. 2002, 22, 6788–6796. [Google Scholar] [CrossRef] [PubMed]
- Barnes, L.; Eveson, J.W.; Reichart, P.; Sidransky, D. WHO Classification of Tumours, Pathology and Genetics of Head and Neck Tumours; IARC Press: Lyon, France, 2005. [Google Scholar]






| NANOG | MIDKINE | ||||||
|---|---|---|---|---|---|---|---|
| Total No. | +++ | ++ | +&− | +++ | ++ | +&− | |
| No. of Cases | 62 | 21 | 22 | 19 | 37 | 14 | 11 |
| Tumor sites | |||||||
| Gingiva | 28 | 11 | 8 | 9 | 17 | 5 | 6 |
| Cheek | 6 | 1 | 3 | 2 | 3 | 2 | 1 |
| Palate | 11 | 3 | 6 | 2 | 6 | 5 | 0 |
| Tongue | 10 | 2 | 3 | 5 | 5 | 2 | 3 |
| FOM | 7 | 4 | 2 | 1 | 6 | 0 | 1 |
| Tumor stage | |||||||
| I+II | 27 | 6 | 9 | 12 | 14 | 4 | 9 |
| III+IV | 35 | 15 | 13 | 7 | 23 | 10 | 2 |
| Neck node | |||||||
| Node (−) | 39 | 9 | 14 | 16 | 20 | 8 | 11 |
| Node (+) | 23 | 12 | 8 | 3 | 17 | 6 | 0 |
| Histo grade | |||||||
| Well | 23 | 4 | 4 | 15 | 8 | 5 | 10 |
| Mod + Poor | 39 | 17 | 18 | 4 | 29 | 9 | 1 |
| Variable | Overall Survival | |
|---|---|---|
| p Value | 95% CI | |
| MIDKINE | ||
| MK(+/−) | 0.031 * | 105.575–150.425 |
| MK(++/+++) | 39.938–70.398 | |
| NANOG | ||
| NANOG(+/−) | 0.022 * | 62.674–134.938 |
| NANOG(++/+++) | 35.759–68.475 | |
| MK + NANOG | ||
| MK(+/−)/NA(+/−) | 0.015 * | 93.569–137.210 |
| MK(++)/NA(+/−) | 34.481–84.387 | |
| MK(++)/NA(++) | 9.155–20.009 | |
| Neck node | ||
| Node (−) | 0.001 ** | 72.644–114.863 |
| Node (+) | 14.640–32.900 | |
| Histological grade | ||
| Well | 0.008 ** | 75.600–131.275 |
| Moderately | 26.664–62.813 | |
| Poorly | 9.688–48.732 | |
| Tumor stage | ||
| Stage I+II | 0.023 * | 63.238–120.872 |
| Stage III+IV | 31.809–67.282 | |
| Treatment | ||
| Surg | 0.001 ** | 63.307–115.683 |
| Surg + RT | 83.268–137.232 | |
| PaRT | 9.716–16.344 | |
| Variable | Overall Survival | ||
|---|---|---|---|
| p Value | Hazard Ratio | 95% Cl | |
| Neck node | 0.004 * | 2.091 | 0.859–5.093 |
| Histological grade | 0.003 * | 3.570 | 1.221–10.435 |
| Tumor stage | 0.130 | 2.195 | 0.057–7.598 |
| NANOG | 0.050 * | 4.135 | 2.018–25.037 |
| MIDKINE | 0.045 * | 3.331 | 1.079–27.398 |
| MK + NANOG | 0.049 * | 4.081 | 2.017–22.266 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-M.; Kang, Y.-H.; Byun, J.-H.; Jang, S.-J.; Rho, G.-J.; Lee, J.-S.; Park, B.-W. Midkine and NANOG Have Similar Immunohistochemical Expression Patterns and Contribute Equally to an Adverse Prognosis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 2339. https://doi.org/10.3390/ijms18112339
Kim H-M, Kang Y-H, Byun J-H, Jang S-J, Rho G-J, Lee J-S, Park B-W. Midkine and NANOG Have Similar Immunohistochemical Expression Patterns and Contribute Equally to an Adverse Prognosis of Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2017; 18(11):2339. https://doi.org/10.3390/ijms18112339
Chicago/Turabian StyleKim, Hyun-Min, Young-Hoon Kang, June-Ho Byun, Si-Jung Jang, Gyu-Jin Rho, Jong-Sil Lee, and Bong-Wook Park. 2017. "Midkine and NANOG Have Similar Immunohistochemical Expression Patterns and Contribute Equally to an Adverse Prognosis of Oral Squamous Cell Carcinoma" International Journal of Molecular Sciences 18, no. 11: 2339. https://doi.org/10.3390/ijms18112339
APA StyleKim, H.-M., Kang, Y.-H., Byun, J.-H., Jang, S.-J., Rho, G.-J., Lee, J.-S., & Park, B.-W. (2017). Midkine and NANOG Have Similar Immunohistochemical Expression Patterns and Contribute Equally to an Adverse Prognosis of Oral Squamous Cell Carcinoma. International Journal of Molecular Sciences, 18(11), 2339. https://doi.org/10.3390/ijms18112339