Buprofezin Is Metabolized by CYP353D1v2, a Cytochrome P450 Associated with Imidacloprid Resistance in Laodelphax striatellus
Abstract
:1. Introduction
2. Results
2.1. Functional Expression of CYP353D1v2 in Sf9 Cells
2.2. Enzyme Kinetics
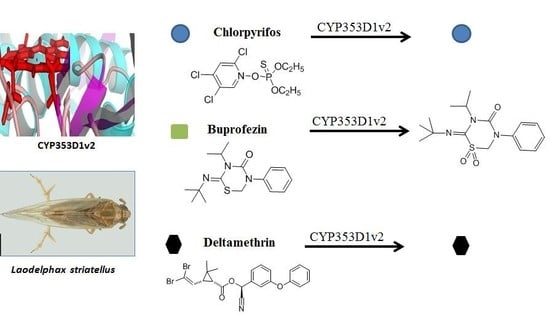
2.3. Insecticides Metabolism Assay
2.4. Identification of Buprofezin Metabolite
3. Discussion
4. Materials and Methods
4.1. Insecticide and Chemical
4.2. Cloning and Functional Expression of CYP353D1v2 and Preparation of Microsomes
4.3. Enzyme Kinetics with p-Nitroanisole
4.4. Insecticide Metabolism Assay
4.5. UPLC-MS/MS Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Konno, T. Buprofezin; A reliable IGR for the control of rice pests. In Pest Management Rice; Springer: Berlin, Germany, 1990; pp. 210–222. [Google Scholar]
- De Cock, A.; Degheele, D. Buprofezin: A novel chitin synthesis inhibitor affecting specifically planthoppers, whiteflies and scale insects. In Insecticides with Novel Modes of Action; Springer: Berlin/Heidelberg, Germany, 1998; pp. 74–91. [Google Scholar]
- Nagata, T.; Masda, T.; Moriya, S. Development of insecticide resistance in the brown planthopper: Nilaparvata lugens STAL (Hemiptera: Delphacidae). Appl. Entomol. Zool. 1979, 14, 264–269. [Google Scholar] [CrossRef]
- Nagata, T.; Ohira, Y. Insecticide resistance of the small brown planthopper, Laodelphax striatellus Fallén (Hemiptera: Delphacidae), collected in Kyushu and on the East China Sea. Appl. Entomol. Zool. 1986, 21, 216–219. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Han, Z.; Liu, Y.; Fang, J. Cross-resistance and possible mechanisms of chlorpyrifos resistance in Laodelphax striatellus (Fallén). Pest Manag. Sci. 2010, 66, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Wu, J.; Huang, S.; Mu, L.; Han, Z. Insecticide resistance in field populations of Laodelphax striatellus Fallén (Homoptera: Delphacidae) in China and its possible mechanisms. Int. J. Pest Manag. 2008, 54, 13–19. [Google Scholar] [CrossRef]
- Yanhua, W.; Changxing, W.; Xueping, Z. Advances in the research of insecticide resistance of the small brown planthopper, Laodelphax striatellus. Plant Prot. 2011, 4, 29–35. (In Chinese) [Google Scholar]
- Zhang, Y.; Guo, H.; Yang, Q.; Li, S.; Wang, L.; Zhang, G.; Fang, J. Overexpression of a P450 gene (CYP6CW1) in buprofezin-resistant Laodelphax striatellus (Fallén). Pestic. Biochem. Physiol. 2012, 104, 277–282. [Google Scholar] [CrossRef]
- Feyereisen, R. Insect P450 enzymes. Ann. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, M.; Han, Z. Overexpression of multiple detoxification genes in deltamethrin resistant Laodelphax striatellus (Hemiptera: Delphacidae) in China. PLoS ONE 2013, 8, e79443. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, M.; Han, Z. Biochemical and molecular characterisation and cross-resistance in field and laboratory chlorpyrifos-resistant strains of Laodelphax striatellus (Hemiptera: Delphacidae) from eastern China. Pest Manag. Sci. 2014, 70, 1118–1129. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.E.A.; Zhang, W.; Feng, A.; Qiou, X.; Zhao, W.; Han, Z. Constitutive overexpression of cytochrome P450 associated with imidacloprid resistance in Laodelphax striatellus (Fallén). Pest Manag. Sci. 2016, 72, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.; Zhang, W.; Han, Z. Cytochrome P450 CYP4DE1 and CYP6CW3v2 contribute to ethiprole resistance in Laodelphax striatellus (Fallén). Insect Mol. Biol. 2015, 24, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Parthasarathy, R.; Bai, H.; Woithe, K.; Kaussmann, M.; Nauen, R.; Harrison, D.A.; Palli, S.R. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2010, 107, 8557–8562. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wen, Y.; Yang, B.; Zhang, Y.; Liu, S.; Liu, Z.; Han, Z. Biochemical mechanisms of imidacloprid resistance in Nilaparvata lugens: Over-expression of cytochrome P450 CYP6AY1. Insect Biochem. Mol. Biol. 2013, 43, 1021–1027. [Google Scholar] [CrossRef] [PubMed]
- Joußen, N.; Agnolet, S.; Lorenz, S.; Schöne, S.E.; Ellinger, R.; Schneider, B.; Heckel, D.G. Resistance of Australian Helicoverpa armigera to fenvalerate is due to the chimeric P450 enzyme CYP337B3. Proc. Natl. Acad. Sci. USA 2012, 109, 15206–15211. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.J.; Bibby, J.; Pignatelli, P.; Muangnoicharoen, S.; O’Neill, P.M.; Lian, L.-Y.; Müller, P.; Nikou, D.; Steven, A.; Hemingway, J. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: Sequential metabolism of deltamethrin revealed. Insect Biochem. Mol. Biol. 2011, 41, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.E.A.; Miah, M.A.; Wu, M.; Zhang, H.J.; Jiang, L.; Han, Z. Imidacloprid is degraded by CYP353D1v2, a cytochrome P450 overexpressed in a resistant strain of Laodelphax striatellus. Pest Manag. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Liu, J.; Li, X.; Chi, J.; Liu, Y. Resistance risk, cross-resistance and biochemical resistance mechanism of Laodelphax striatellus to buprofezin. J. Appl. Ecol. 2016, 27, 255–262. [Google Scholar]
- Scott, J.G. Cytochromes P450 and insecticide resistance. Insect Biochem. Mol. Biol. 1999, 29, 757–777. [Google Scholar] [CrossRef]
- Nauen, R.; Vontas, J.; Kaussmann, M.; Wölfel, K. Pymetrozine is hydroxylated by CYP6CM1, a cytochrome P450 conferring neonicotinoid resistance in Bemisia tabaci. Pest Manag. Sci. 2013, 69, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Roditakis, E.; Morou, E.; Tsagkarakou, A.; Riga, M.; Nauen, R.; Paine, M.; Morin, S.; Vontas, J. Assessment of the Bemisia tabaci CYP6CM1vQ transcript and protein levels in laboratory and field-derived imidacloprid-resistant insects and cross-metabolism potential of the recombinant enzyme. Insect Sci. 2011, 18, 23–29. [Google Scholar] [CrossRef]
- Joußen, N.; Heckel, D.G.; Haas, M.; Schuphan, I.; Schmidt, B. Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1-overexpressing strains of Drosophila melanogaster, leading to resistance. Pest Manag. Sci. 2008, 64, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.N.; Stevenson, B.J.; Müller, P.; Wilding, C.S.; Egyir-Yawson, A.; Field, S.G.; Hemingway, J.; Paine, M.J.; Ranson, H.; Donnelly, M.J. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc. Natl. Acad. Sci. USA 2012, 109, 6147–6152. [Google Scholar] [CrossRef] [PubMed]
- Yunta, C.; Grisales, N.; Nász, S.; Hemmings, K.; Pignatelli, P.; Voice, M.; Ranson, H.; Paine, M.J. Pyriproxyfen is metabolized by P450s associated with pyrethroid resistance in An. gambiae. Insect Biochem. Mol. Biol. 2016, 78, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Elzaki, M.E.A.; Pu, J.; Zhu, Y.; Zhang, W.; Sun, H.; Wu, M.; Han, Z. Cross-resistance among common insecticides and its possible mechanism in Laodelphax striatellus Fallén (Hemiptera: Delphacidae). Orient. Insects 2017, 1–14. [Google Scholar] [CrossRef]
- Chandor-Proust, A.; Bibby, J.; Régent-Kloeckner, M.; Roux, J.; Guittard-Crilat, E.; Poupardin, R.; Riaz, M.A.; Paine, M.; Dauphin-Villemant, C.; Reynaud, S. The central role of mosquito cytochrome P450 CYP6Zs in insecticide detoxification revealed by functional expression and structural modelling. Biochem. J. 2013, 455, 75–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, I.R.; Shephard, E.A.; de Montellano, P.R.O. Cytochrome P450 Protocols; Springer: New York, NY, USA, 1998; Volume 107. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Omura, T. The carbon monoxide-binding pigment of liver microsomes. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [PubMed]
- Pritchard, M.P.; McLaughlin, L.; Friedberg, T. Establishment of functional human cytochrome P450 monooxygenase systems in Escherichia coli. In Cytochrome P450 Protocols; Humana Press: Totowa, NJ, USA, 2006; pp. 19–29. [Google Scholar]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elzaki, M.E.A.; Miah, M.A.; Han, Z. Buprofezin Is Metabolized by CYP353D1v2, a Cytochrome P450 Associated with Imidacloprid Resistance in Laodelphax striatellus. Int. J. Mol. Sci. 2017, 18, 2564. https://doi.org/10.3390/ijms18122564
Elzaki MEA, Miah MA, Han Z. Buprofezin Is Metabolized by CYP353D1v2, a Cytochrome P450 Associated with Imidacloprid Resistance in Laodelphax striatellus. International Journal of Molecular Sciences. 2017; 18(12):2564. https://doi.org/10.3390/ijms18122564
Chicago/Turabian StyleElzaki, Mohammed Esmail Abdalla, Mohammad Asaduzzaman Miah, and Zhaojun Han. 2017. "Buprofezin Is Metabolized by CYP353D1v2, a Cytochrome P450 Associated with Imidacloprid Resistance in Laodelphax striatellus" International Journal of Molecular Sciences 18, no. 12: 2564. https://doi.org/10.3390/ijms18122564
APA StyleElzaki, M. E. A., Miah, M. A., & Han, Z. (2017). Buprofezin Is Metabolized by CYP353D1v2, a Cytochrome P450 Associated with Imidacloprid Resistance in Laodelphax striatellus. International Journal of Molecular Sciences, 18(12), 2564. https://doi.org/10.3390/ijms18122564