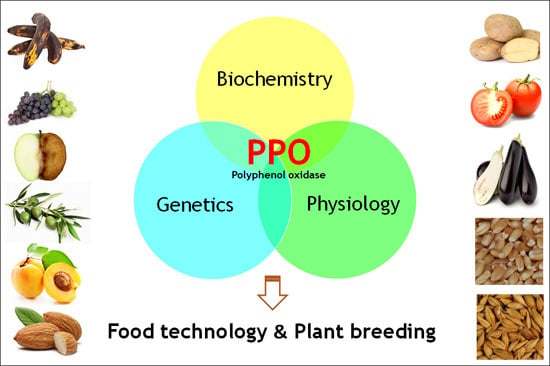
Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects
Abstract
:1. Introduction
2. Polyphenol Oxidases (PPO) and Browning
2.1. PPO Biochemical Properties
2.2. Enzymatic Browning in Crops and Food Processing
2.3. Technological Strategies for Browning Prevention
3. Physiological Roles of PPOs
4. Genomic and Genetic Aspects
4.1. Solanaceae
4.2. Poaceae
4.3. Other Species
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Oliveira, C.M.; Ferreira, A.C.S.; de Freitas, V.; Silva, A.M. Oxidation mechanisms occurring in wines. Food Res. Int. 2011, 44, 1115–1126. [Google Scholar] [CrossRef]
- Van Gelder, C.W.G.; Flurkey, W.H.; Wichers, H.J. Sequence and structural features of plant and fungal tyrosinases. Phytochemistry 1997, 45, 1309–1323. [Google Scholar] [CrossRef]
- Golan-Goldhirsh, A.; Whitaker, J.R.; Kahn, V. Relation structure of polyphenol oxidase and prevention of browning. In Nutritional and Toxicological Aspects of Food Safety; Friedman, M., Ed.; Plenum: New York, NY, USA, 1984; pp. 457–495. [Google Scholar]
- Wong, D.W.S. Mechanism and Theory in Food Chemistry, 1st ed.; Springer: New York, NY, USA, 1989. [Google Scholar]
- Rolff, M.; Schottenheim, J.; Decker, H.; Tuczek, F. Copper-O2 reactivity of tyrosinase models towards external monophenolic substrates: Molecular mechanism and comparison with the enzyme. Chem. Soc. Rev. 2011, 40, 4077–4098. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Food browning and its prevention: An overview. J. Agric. Food Chem. 1996, 44, 631–653. [Google Scholar] [CrossRef]
- Mayer, A.M. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry 2006, 67, 2318–2331. [Google Scholar] [CrossRef] [PubMed]
- Hurrell, R.F.; Finot, P.A. Nutritional consequences of the reactions between proteins and oxidized polyphenols. In Nutritional and Toxicological Aspects of Food Safety; Friedman, M., Ed.; Plenum: New York, NY, USA, 1984; pp. 423–435. [Google Scholar]
- Vamos-Vigyazo, L. Polyphenol oxidase and peroxidase in fruits and vegetables. Crit. Rev. Food Sci. 1981, 15, 49–127. [Google Scholar] [CrossRef] [PubMed]
- Yoruk, R.; Marshall, M.R. Physicochemical properties and function of plant polyphenol oxidase: A review. J. Food Biochem. 2003, 27, 361–422. [Google Scholar] [CrossRef]
- Sánchez-Ferrer, A.; Rodríguez-López, J.N.; García-Cánovas, F.; García-Carmona, F. Tyrosinase: A comprehensive review of its mechanism. Biochim. Biophys. Acta 1995, 1247, 1–11. [Google Scholar] [CrossRef]
- Fronk, P.; Hartmann, H.; Bauer, M.; Solem, E.; Jaenicke, E.; Tenzer, S.; Decker, H. Polyphenoloxidase from Riesling and Dornfelder wine grapes (Vitis vinifera) is a tyrosinase. Food Chem. 2015, 183, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, K.C.; Duke, S.O. Function of polyphenol oxidase in higher plants. Plant Physiol. 1984, 60, 106–112. [Google Scholar] [CrossRef]
- Lerner, H.R.; Mayer, A.M.; Harel, E. Evidence for conformational changes in grape catechol oxidase. Phytochemistry 1972, 11, 2415–2421. [Google Scholar] [CrossRef]
- Jolley, R.L.; Robb, D.A.; Mason, H.S. The multiple forms of mushroom tyrosinase. J. Biol. Chem. 1969, 244, 1593–1599. [Google Scholar] [PubMed]
- Gregory, R.P.F.; Bendall, D.S. The purification and some properties of the polyphenoloxidase from tea (Camellia sinensis L.). Biochem. J. 1966, 101, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Flurkey, W.H.; Jen, J.J. Purification of peach polyphenoloxidase in the presence of added protease inhibitors. J. Food Biochem. 1980, 4, 29–41. [Google Scholar] [CrossRef]
- Whitaker, J.R. Principles of Enzymology for the Food Sciences, 2nd ed.; CRC Press: New York, NY, USA, 1994; pp. 271–556. [Google Scholar]
- Janovitz-Klapp, A.; Richard, F.; Nicolas, J. Polyphenoloxidase from apple, partial purification and some properties. Phytochemistry 1989, 28, 2903–2907. [Google Scholar] [CrossRef]
- Valero, E.; Garcia-Carmona, F. pH-dependent effect of sodium chloride on latent grape polyphenol oxidase. J. Agric. Food Chem. 1998, 46, 2447–2451. [Google Scholar] [CrossRef]
- Fraignier, M.; Marques, L.; Fleuriet, A.; Macheix, J. Biochemical and immunochemical characteristics of polyphenol oxidase from different fruits of Prunus. J. Agric. Food Chem. 1995, 43, 2375–2380. [Google Scholar] [CrossRef]
- Zhou, P.; Smith, N.L.; Lee, C.Y. Potential purification and some properties of Monroe apple peel polyphenol oxidase. J. Agric. Food Chem. 1993, 41, 532–536. [Google Scholar] [CrossRef]
- Wesche-Ebeling, P.; Montgomery, M.W. Strawberry polyphenoloxidase: Extraction and partial characterization. J. Food Sci. 1990, 55, 1320–1324. [Google Scholar] [CrossRef]
- Park, E.Y.; Luh, B.S. Polyphenol oxidase of kiwifruit. J. Food Sci. 1985, 50, 678–684. [Google Scholar] [CrossRef]
- Ünal, M.Ü.; Taş, C.; Şener, A. Determination of biochemical properties of polyphenol oxidase from domat olives. GIDA 2011, 36, 185–192. [Google Scholar]
- Gençer, N.; Sinan, S.; Arslan, O. Kinetic properties of polyphenol oxidase obtained from various olives (Olea europea L.). Asian J. Chem. 2012, 24, 2159–2161. [Google Scholar]
- Segovia-Bravo, K.A.; Jarén-Galán, M.; García-García, P.; Garrido-Fernández, A. Characterization of polyphenol oxidase from the Manzanilla cultivar (Olea europaea pomiformis) and prevention of browning reactions in bruised olive fruits. J. Agric. Food Chem. 2007, 55, 6515–6520. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Ferrer, A.; Laveda, F.; Garcia-Carmona, F. Partial purification of soluble potato polyphenol oxidase by partitioning in aqueous two-phase system. J. Agric. Food Chem. 1993, 41, 1219–1224. [Google Scholar] [CrossRef]
- Interesse, F.S.; Ruggiero, P.; D’Avella, G.; Lamparelli, F. Partial purification of some properties of wheat (Triticum aestivum) o-diphenolase. J. Sci. Food Agric. 1980, 31, 459–466. [Google Scholar] [CrossRef]
- Nozue, M.; Souri, M.; Arakawa, D.; Kojima, M. Purification and characterization of two isoforms of chlorogenic acid oxidase from sweet potato cells in suspension culture. J. Plant Physiol. 1998, 153, 552–557. [Google Scholar] [CrossRef]
- Heimdal, H.; Larsen, L.M.; Poll, L. Characterization of polyphenol oxidase from photosynthetic and vascular lettuce tissues (Lactuca sativa). J. Agric. Food Chem. 1994, 42, 1428–1433. [Google Scholar] [CrossRef]
- Sheptovitsky, Y.G.; Brudvig, G.W. Isolation and characterization of spinach photosystem I1 membrane-associated catalase and polyphenol oxidase. Biochemistry 1996, 35, 16255–16263. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gilabert, M.; Carmona, F.G. Characterization of catecholase and cresolase activities of eggplant polyphenol oxidase. J. Agric. Food Chem. 2000, 48, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Taticchi, A.; Esposto, S.; Veneziani, G.; Urbani, S.; Selvaggini, R.; Servili, M. The influence of the malaxation temperature on the activity of polyphenoloxidase and peroxidase and on the phenolic composition of virgin olive oil. Food Chem. 2013, 136, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Serradell, M.D.L.A.; Rozenfeld, P.A.; Martinez, G.A.; Civello, P.M.; Chaves, A.R.; Anon, M.C. Polyphenoloxidase activity from strawberry fruit (Fragaria × ananassa, Duch., cv. Selva) characterization and partial purification. J. Sci. Food Agric. 2000, 80, 1421–1427. [Google Scholar]
- Miller, A.R.; Kelley, T.J.; Mujer, C.V. Anodic peroxidase isoenzymes and polyphenol oxidase activity from cucumber fruit: Tissue and substrate specificity. Phytochemistry 1990, 29, 705–709. [Google Scholar] [CrossRef]
- Robinson, S.P.; Loveys, B.R.; Chacko, E.K. Polyphenoloxidase enzymes in the sap and skin of mango fruit. Aust. J. Plant Physiol. 1993, 20, 99–107. [Google Scholar]
- Lee, P.M.; Lee, K.; Karim, M.I.A. Biochemical studies of cocoa bean polyphenol oxidase. J. Sci. Food Agric. 1991, 55, 251–260. [Google Scholar] [CrossRef]
- Lourenco, E.J.; Neves, V.A.; Silva, M.A.D. Polyphenol oxidase from sweet potato: Purification and properties. J. Agric. Food Chem. 1992, 40, 2369–2373. [Google Scholar] [CrossRef]
- Reinkensmeier, A.; Steinbrenner, K.; Homann, T.; Bußler, S.; Rohn, S.; Rawel, H.M. Monitoring the apple polyphenol oxidase-modulated adduct formation of phenolic and amino compounds. Food Chem. 2016, 194, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Gooding, P.S.; Bird, C.; Robinson, S.P. Molecular cloning and characterization of banana fruit polyphenol oxidase. Planta 2001, 213, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Rathjen, A.H.; Robinson, S.P. Aberrant processing of polyphenol oxidase in a variegated grapevine mutant. Plant Physiol. 1992, 99, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Saba, M.K.; Moradi, S. Internal browning disorder of eight pear cultivars affected by bioactive constituents and enzyme activity. Food Chem. 2016, 205, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Docimo, T.; Francese, G.; de Palma, M.; Mennella, D.; Toppino, L.; Lo Scalzo, R.; Mennella, G.; Tucci, M. Insights in the fruit flesh browning mechanisms in Solanum melongena genetic lines with opposite post-cut behavior. J. Agric. Food Chem. 2016, 64, 4675–4685. [Google Scholar] [CrossRef] [PubMed]
- Taranto, F.; Delvecchio, L.N.; Mangini, G.; Del Faro, L.; Blanco, A.; Pasqualone, A. Molecular and physico-chemical evaluation of enzymatic browning of whole meal and dough in a collection of tetraploid wheats. J. Cereal Sci. 2012, 55, 405–414. [Google Scholar] [CrossRef]
- Pasqualone, A.; Delvecchio, L.N.; Mangini, G.; Taranto, F.; Blanco, A. Variability of total soluble phenolic compounds and antioxidant activity in a collection of tetraploid wheat. Agric. Food Sci. 2014, 23, 307–316. [Google Scholar]
- Ullah Tea, M.R. Food Enzymotogy; Fox, P.F., Ed.; Elsevier Science: New York, NY, USA, 1991; pp. 163–187. [Google Scholar]
- Amorim, H.W.; Melo, M. Significance of enzymes in nonalcoholic coffee beverage. In Food Enzymlogy; Fox, P.F., Ed.; Elsevier Science: New York, NY, USA, 1991; pp. 189–209. [Google Scholar]
- Jaworski, A.W.; Lee, C.Y. Fractionation of HPLC determination of grape phenolics. J. Agric. Food Chem. 1987, 35, 257–259. [Google Scholar] [CrossRef]
- Murata, M.; Tsurutani, M.; Tomita, M.; Homma, S.; Kaneko, K. Relationship between apple ripening and browning: Changes in polyphenol content and polyphenol oxidase. J. Agric. Food Chem. 1995, 43, 1115–1121. [Google Scholar] [CrossRef]
- Raymond, J.; Rakariyatham, N.; Azanza, J.L. Purification and some properties of polyphenoloxidase from sunflower seeds. Phytochemistry 1993, 34, 927–931. [Google Scholar] [CrossRef]
- Stommel, J.R.; Whitaker, B.D. Phenolic acid content and composition of eggplant fruit in a germplasm core subset. J. Am. Soc. Hortic. Sci. 2003, 128, 704–710. [Google Scholar]
- Laddomada, B.; Durante, M.; Mangini, G.; D’Amico, L.; Lenucci, M.S.; Simeone, R.; Piarulli, L.; Mita, G.; Blanco, A. Genetic variation for phenolic acids concentration and composition in a tetraploid wheat (Triticum turgidum L.) collection. Genet. Resour. Crop Evol. 2016. [Google Scholar] [CrossRef]
- Macheix, J.; Fleuriet, A.; Billot, J. Phenolic compounds in fruit processing. In Fruit Phenolics; Macheix, J., Fleuriet, A., Billot, J., Eds.; CRC Press: Boca Raton, FL, USA, 1990; pp. 239–312. [Google Scholar]
- Lee, C.Y.; Jaworski, A.W. Phenolics and browning potential of white grapes grown in New York. Am. J. Enol. Vitic. 1988, 39, 337–340. [Google Scholar]
- Li, H.; Guo, A.; Wang, H. Mechanisms of oxidative browning of wine. Food Chem. 2008, 108, 1–13. [Google Scholar] [CrossRef]
- Ortega-García, F.; Blanco, S.; Peinado, M.A.; Peragón, J. Polyphenol oxidase and its relationship with oleuropein concentration in fruits and leaves of olive (Olea europaea) cv. Picual trees during fruit ripening. Tree Physiol. 2008, 28, 45–54. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, R.; Romero-Segura, C.; Sanz, C.; Sánchez-Ortiz, A.; Pérez, A.G. Role of polyphenol oxidase and peroxidase in shaping the phenolic profile of virgin olive oil. Food Res. Int. 2011, 44, 629–635. [Google Scholar] [CrossRef]
- Ortega-García, F.; Peragón, J. The response of phenylalanine ammonia lyase, polyphenol oxidase and phenols to cold stress in the olive tree (Olea europaea L. cv. Picual). J. Sci. Food Agric. 2009, 89, 1565–1573. [Google Scholar] [CrossRef]
- Servili, M.; Taticchi, A.; Esposto, S.; Urbani, S.; Selvaggini, R.; Montedoro, G. Influence of the decrease in oxygen during malaxation of olive paste on the composition of volatiles and phenolic compounds in virgin olive oil. J. Agric. Food Chem. 2008, 56, 10048–10055. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Sheoran, I.S. Enzymic browning of whole wheat meal flour. J. Sci. Food Agric. 1972, 23, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Baik, B.K.; Czuchajowska, Z.U.; Pomeranz, Y.E. Discoloration of dough for oriental noodles. Cereal Chem. 1995, 72, 198–204. [Google Scholar]
- Dexter, J.E.; Preston, K.R.; Matsuo, R.R.; Tipples, K.H. Development of a high extraction flow for the GRL pilot mill to evaluate Canadian wheat potential for the Chinese market. Can. Inst. Food Sci. Technol. J. 1984, 14, 253–259. [Google Scholar] [CrossRef]
- Faridi, H. Flat breads. In Wheat: Chemistry and Technology, 3rd ed.; Pomeranz, Y., Ed.; AACC International: St. Paul, MA, USA, 1988; Volume 2, pp. 457–506. [Google Scholar]
- Miskelly, D.M. Flour components affecting pasta and noodle color. J. Sci. Food Agric. 1984, 35, 463–471. [Google Scholar] [CrossRef]
- Morris, C.F.; Jeffers, H.C.; Engle, D.A. Effect of processing, formula and measurement variables on alkaline noodle color-toward an optimized laboratory system. Cereal Chem. 2000, 77, 77–85. [Google Scholar] [CrossRef]
- Pasqualone, A. Italian durum wheat breads. In Bread Consumption and Health; Clerici, P.S.M.T., Ed.; Nova Science Publisher Inc.: Hauppauge, NY, USA, 2012; pp. 57–80. [Google Scholar]
- Hatcher, D.W.; Kruger, J.E. Distribution of polyphenol oxidase in flour millstreams of Canadian common wheat classes milled to three extraction rates. Cereal Chem. 1993, 70, 51–55. [Google Scholar]
- Demeke, T.; Morris, C. Molecular characterization of wheat polyphenol oxidase (PPO). Theor. Appl. Genet. 2002, 104, 813–818. [Google Scholar] [PubMed]
- Feillet, P.; Autran, J.C.; Icard-Verniere, C. Pasta brownness: An assessment. J. Cereal Sci. 2000, 32, 215–233. [Google Scholar] [CrossRef]
- Pasqualone, A.; Delvecchio, L.N.; Gambacorta, G.; Laddomada, B.; Urso, V.; Mazzaglia, A.; Ruisi, P.; di Miceli, G. Effect of supplementation with wheat bran aqueous extracts obtained by ultrasound-assisted technologies on the sensory properties and the antioxidant activity of dry pasta. Nat. Prod. Commun. 2015, 10, 1739–1742. [Google Scholar] [PubMed]
- Pasqualone, A.; Gambacorta, G.; Summo, C.; Caponio, F.; di Miceli, G.; Flagella, Z.; Marrese, P.P.; Piro, G.; Perrotta, C.; de Bellis, L.; et al. Functional, textural and sensory properties of dry pasta supplemented with lyophilized tomato matrix or with durum wheat bran extracts produced by supercritical carbon dioxide or ultrasound. Food Chem. 2016, 213, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.; Degl’Innocenti, E.; Guglielminetti, L.; Guidi, L. Role of ascorbic acid in the inhibition of polyphenol oxidase and the prevention of browning in different browning-sensitive Lactuca sativa var. capitata (L.) and Eruca sativa (Mill.) stored as fresh-cut produce. J. Sci. Food Agric. 2013, 93, 1814–1819. [Google Scholar] [PubMed]
- Soares, J.M.; Fonseca, G.G. Effect of L-ascorbic acid and sodium metabisulfite in the inhibition of the enzymatic browning of minimally processed apple. Int. J. Agric. Res. 2008, 3, 196–201. [Google Scholar]
- Richard-Forget, F.C.; Goupy, P.M.; Nicolas, J.J. Cysteine as an inhibitor of enzymic browning. 2. Kinetic studies. J. Agric. Food Chem. 1992, 40, 2108–2113. [Google Scholar] [CrossRef]
- Gacchea, R.N.; Shete, A.M.; Dhole, N.A.; Ghole, V.S. Reversible inhibition of polyphenol oxidase from apple using l-cysteine. Indian J. Chem. Technol. 2006, 13, 459–463. [Google Scholar]
- Ashie, I.N.A.; Simpson, B.K.; Smith, J.P. Mechanisms for controlling enzymatic reactions in foods. Crit. Rev. Food Sci. Nutr. 1996, 36, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Sapers, G.M. Browning of foods: Control by sulfites, antioxidants, and other means. Food Technol. 1993, 47, 75–84. [Google Scholar]
- Nicolas, J.J.; Richard-Forget, F.C.; Goupy, P.M.; Amiot, M.; Aubert, S.Y. Enzymatic browning reactions in apple and apple products. Crit. Rev. Food Sci. 1994, 34, 109–157. [Google Scholar] [CrossRef] [PubMed]
- Eskin, N.A.M. Biochemistry of food spoilage: Enzymatic browning. In Biochemistry of Foods; Eskin, N.A.M., Ed.; Academic Press: New York, NY, USA, 1990; pp. 401–432. [Google Scholar]
- Martinez, M.V.; Whitaker, J.R. The biochemistry and control of enzymatic browning. Trends Food Sci. Technol. 1995, 6, 195–200. [Google Scholar] [CrossRef]
- Gultekin, F.; Doguc, D.K. Allergic and immunologic reactions to food additives. Clin. Rev. Allerg. Immunol. 2013, 45, 6–29. [Google Scholar] [CrossRef] [PubMed]
- Fayad, N.; Marchal, L.; Billaud, C.; Nicolas, J. Comparison of β-cyclodextrin effect on polyphenol oxidation catalyzed by purified polyphenol oxidase from different sources. J. Agric. Food Chem. 1997, 45, 2442–2446. [Google Scholar] [CrossRef]
- Hicks, K.B.; Haines, R.M.; Tong, C.B.; Sapers, G.M.; El-Atawy, Y.; Irwin, P.L.; Seib, P.A. Inhibition of enzymatic browning in fresh fruit and vegetable juices by soluble and insoluble forms of β-cyclodextrin alone or in combination with phosphates. J. Agric. Food Chem. 1996, 44, 2591–2594. [Google Scholar] [CrossRef]
- Singh, V.; Jadhav, S.B.; Singhal, R.S. Interaction of polyphenol oxidase of Solanum tuberosum with β-cyclodextrin: Process details and applications. Int. J. Biol. Macromol. 2015, 80, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Fesler, M.; Salvatori, D.; Gomez, P.; Alzamora, S.M. Convective air drying of apples as affected by blanching and calcium impregnation. J. Food Eng. 2008, 87, 323–332. [Google Scholar] [CrossRef]
- Severini, C.; Baiano, A.; de Pilli, T.; Carbone, B.F.; Derossi, A. Combined treatments of blanching and dehydration: Study on potato cubes. J. Food Eng. 2005, 68, 289–296. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, M.; Arun, S.; Mujumdar, A.S. Effects of vacuum and microwave freeze drying on microstructure and quality of potato slices. J. Food Eng. 2010, 101, 131–139. [Google Scholar] [CrossRef]
- Kidmose, U.; Martens, H.J. Changes in texture, microstructure and nutritional quality of carrot slices during blanching and freezing. J. Sci. Food Agric. 1999, 79, 1747–1753. [Google Scholar] [CrossRef]
- Mukherjee, S.; Chattopadhyay, P.K. Whirling bed blanching of potato cubes and its effects on product quality. J. Food Eng. 2007, 78, 52–60. [Google Scholar] [CrossRef]
- Sapers, G.M.; Miller, R.C. Heated ascorbic/citric acid solution as browning inhibitor for pre-peeled potatoes. J. Food Sci. 1995, 60, 762–766. [Google Scholar] [CrossRef]
- Severini, C.; Baiano, A.; de Pilli, T.; Romaniello, R.; Derossi, A. Prevention of enzymatic browning in sliced potatoes by blanching in boiling saline solutions. LWT Food Sci. Technol. 2003, 36, 657–665. [Google Scholar] [CrossRef]
- Severini, C.; de Pilli, T.; Baiano, A.; Mastrocola, D.; Massini, R. Study on preventing enzymatic browning of potato by microwave blanching. Sci. Aliment. 2001, 21, 149–160. [Google Scholar] [CrossRef]
- De Leonardis, A.; Lustrato, G.; Macciola, V.; Ranalli, G. Application of chemical and physical agents in model systems to controlling phenoloxidase enzymes. Eur. Food Res. Technol. 2010, 231, 603–610. [Google Scholar] [CrossRef]
- Redondo, D.; Venturini, M.E.; Oria, R.; Arias, E. Inhibitory effect of microwaved thinned nectarine extracts on polyphenol oxidase activity. Food Chem. 2016, 197, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Ngazee, E.; Icishahayo, D.; Coutinho, T.A.; Van der Waals, J.E. Role of polyphenol oxidase, peroxidase, phenylalanine ammonia lyase, chlorogenic acid, and total soluble phenols in resistance of potatoes to soft rot. Plant Dis. 2012, 96, 186–192. [Google Scholar]
- Vanitha, S.C.; Niranjana, S.R.; Umesha, S. Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. J. Phytopathol. 2009, 157, 552–557. [Google Scholar] [CrossRef]
- Kavitha, R.; Umesha, S. Regulation of defense-related enzymes associated with bacterial spot resistance in tomato. Phytoparasitica 2008, 36, 144–159. [Google Scholar] [CrossRef]
- Niranjan Ray, S.; Sarosh, B.R.; Shetty, H.S. Induction and accumulation of polyphenol oxidase activities as implicated in development of resistance against pearl millet downy mildew disease. Funct. Plant Biol. 2006, 33, 563–571. [Google Scholar]
- Mohammadi, M.; Kazemi, H. Changes in peroxidase and polyphenol oxidase activities in susceptible and resistant wheat heads inoculated with Fusarium graminearum and induced resistance. Plant Sci. 2002, 162, 491–498. [Google Scholar] [CrossRef]
- Raju, S.; Jayalakshmi, S.K.; Sreeramulu, K. Comparative study on the induction of defense related enzymes in two different cultivars of chickpea (Cicer arietinum L.) genotypes by salicylic acid, spermin and Fusarium oxysporum f. sp. ciceri. Aust. J. Crop Sci. 2008, 2, 121–140. [Google Scholar]
- Jayalakshmi, S.K.; Raju, S.; Rani, S.U.; Benagi, V.I.; Sreeramulu, K. Trichoderma harzianum L1 as a potential source for lytic enzymes and elicitor of defense responses in chickpea (Cicer arietinum L.) against wilt disease caused by Fusarium oxysporum f. sp. ciceri. Aust. J. Crop Sci. 2009, 3, 44–52. [Google Scholar]
- Castañera, P.; Steffens, J.C.; Tingey, W.M. Biological performance of Colorado potato beetle larvae on potato genotypes with differing levels of polyphenol oxidase. J. Chem. Ecol. 1996, 22, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Steffens, J.C. Overexpression of polyphenol oxidase in transgenic tomato plants results in enhanced bacterial disease resistance. Planta 2002, 2, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Melkonian, J.; Wolfe, D.W.; Steffens, J.C. Suppression of polyphenol oxidases increases stress tolerance in tomato. Plant Sci. 2004, 167, 693–703. [Google Scholar] [CrossRef]
- Wang, J.; Constabel, C.P. Polyphenol oxidase overexpression in transgenic Populus enhances resistance to herbivory by forest tent caterpillar (Malacosoma disstria). Planta 2004, 220, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.T.; Taylor, J.S.; Constabel, C.P. The polyphenol oxidase gene family in plants: Lineage-specific duplication and gene expansion. BMC Genom. 2012, 13, 395. [Google Scholar] [CrossRef] [PubMed]
- Araji, S.; Grammer, T.A.; Gertzen, R.; Anderson, S.D.; Mikulic-Petkovsek, M.; Veberic, R.; Phu, M.L.; Solar, A.; Leslie, C.A.; Dandekar, A.M.; et al. Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 2014, 164, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Boeck, T.; Winters, A.L.; Webb, K.J.; Kingston-Smith, A.H. Polyphenol oxidase in leaves; is there any significance to the chloroplastic localization? J. Exp. Bot. 2015, 66, 3571–3579. [Google Scholar] [CrossRef] [PubMed]
- Boeckx, T.; Webster, R.; Winters, A.L.; Webb, K.J.; Gay, A.; Kingston-Smith, A.H. Polyphenol oxidase-mediated protection against oxidative stress is not associated with enhanced photosynthetic efficiency. Ann. Bot. 2015, 116, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Hunt, M.D.; Steffens, J.C. Antisense down regulation of poly-phenol oxidase results in enhanced disease susceptibility. Planta 2004, 220, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Shetty, SM.; Chandrashekar, A.; Venkatesh, Y.P. Eggplant polyphenol oxidase multigene family: Cloning, phylogeny, expression analyses and immunolocalization in response to wounding. Phytochemistry 2011, 72, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Tantasawat, P.; Steffens, J.C. Tomato polyphenol oxidase B is spatially and temporally regulated during development and in response to ethylene. Molecules 2011, 16, 493–517. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Bhagwat, B.; Lane, D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Newman, S.M.; Eannetta, N.T.; Yu, H.; Prince, J.P.; de Vicente, M.C.; Tanksley, S.D.; Steffens, J.C. Organization of the tomato polyphenol oxidase gene family. Plant Mol. Biol. 1993, 21, 1035–1051. [Google Scholar] [CrossRef] [PubMed]
- Thipyapong, P.; Steffens, J.C. Tomato polyphenol oxidase: Differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol. 1997, 115, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Thygesen, P.W.; Dry, I.B.; Robinson, S.P. Polyphenol oxidase in potato. Plant Physiol. 1995, 109, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Liu, C.; Su, Y.; Tong, Y.; Liu, H. Bioinformatic prediction of upstream microRNAs of PPO and novel microRNAs in potato. Can. J. Plant Sci. 2015, 95, 871–877. [Google Scholar] [CrossRef]
- Gramazio, P.; Prohens, J.; Plazas, M.; Andújar, I.; Castillo, E.; Herraiz, F.J.; Knapp, S.; Meyer, S.R.; Vilanova, S. Location of the chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map of eggplant. BMC Plant Biol. 2014, 14, 350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiménez, M.; Dubcovsky, J. Chromosome location of genes affecting polyphenol oxidase activity in seeds of common and durum wheat. Plant Breed. 1999, 118, 395–398. [Google Scholar] [CrossRef]
- Zhang, L.P.; Ge, X.X.; He, Z.H.; Wang, D.S.; Yan, J.; Xia, X.C.; Sutherland, M.W. Mapping QTLs for polyphenol oxidase activity in a DH population from common wheat. Acta Agron. Sin. 2005, 31, 7–10. [Google Scholar]
- Watanabe, N.; Takeuchi, A.; Nakayama, A. Inheritance and chromosomal location of the homoeologous genes affecting phenol colour reaction of kernels in durum wheat. Euphytica 2004, 139, 87–93. [Google Scholar] [CrossRef]
- Mares, D.J.; Campbell, A.W. Mapping components of flour and noodle colour in Australian wheat. Aust. J. Agric. Res. 2001, 52, 1297–1309. [Google Scholar] [CrossRef]
- Jukanti, A.K.; Bruckner, P.L.; Fischer, A.M. Evaluation of wheat polyphenol oxidase genes. Cereal Chem. 2004, 81, 481–485. [Google Scholar] [CrossRef]
- Beecher, B.S.; Carter, H.A.; See, D.E. Genetic mapping of new seed-expressed polyphenol oxidase genes in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2012, 124, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Beecher, B.S.; Skinner, D.Z. Molecular cloning and expression analysis of multiple polyphenol oxidase genes in developing wheat (Triticum aestivum L.) kernels. J. Cereal Sci. 2011, 53, 371–378. [Google Scholar] [CrossRef]
- Martin, J.M.; Berg, J.E.; Hofer, P.; Kephart, K.D.; Nash, D.; Bruckner, P.L. Allelic variation of polyphenol oxidase genes impacts on Chinese raw noodle color. J. Cereal Sci. 2011, 54, 387–394. [Google Scholar] [CrossRef]
- Taranto, F.; Mangini, G.; Pasqualone, A.; Gadaleta, A.; Blanco, A. Mapping and allelic variations of Ppo-B1 and Ppo-B2 gene-related polyphenol oxidase activity in durum wheat. Mol. Breed. 2015, 35, 80. [Google Scholar] [CrossRef]
- Sun, Y.; He, Z.; Ma, W.; Xia, X. Alternative splicing in the coding of Ppo-A1 directly influences the polyphenol oxidase activity in common wheat (Triticum aestivum L.). Funct. Integr. Genom. 2011, 11, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.J.; He, Z.H.; Xia, X.C.; Zhang, L.P.; Morris, C.F.; Appels, R.; Ma, W.J.; Wang, H. A novel STS marker for polyphenol oxidase activity in bread wheat. Mol. Breed. 2005, 16, 209–218. [Google Scholar] [CrossRef]
- Mangini, G.; Taranto, F.; Delvecchio, L.N.; Pasqualone, A.; Blanco, A. Development and validation of a new Ppo-A1 marker useful for marker-assisted selection in tetraploid wheats. Mol. Breed. 2014, 34, 385–392. [Google Scholar] [CrossRef]
- He, X.Y.; He, Z.H.; Zhang, L.P.; Sun, D.J.; Morris, C.F.; Fuerst, E.P.; Xia, X.C. Allelic variation of polyphenol oxidase (PPO) genes located on chromosomes 2A and 2D and development of functional markers for the PPO genes in common wheat. Theor. Appl. Genet. 2007, 115, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Taketa, S.; Matsuki, K.; Amano, S.; Saisho, D.; Himi, E.; Shitsukawa, N.; You, T.; Noda, K.; Takeda, K. Duplicate polyphenol oxidase genes on barley chromosome 2H and their functional differentiation in the phenol reaction of spikes and grains. J. Exp. Bot. 2010, 61, 3983–3993. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Suárez, C.; Atienza, S. Polyphenol oxidase genes in Hordeum chilense and implications in tritordeum breeding. Mol. Breed. 2014, 34, 1867. [Google Scholar] [CrossRef]
- Yu, Y.; Tang, T.; Qian, Q.; Wang, Y.; Yan, M.; Zeng, D.; Han, B.; Wu, C.I.; Shi, S.; Li, J. Independent loss of function in a polyphenol oxidase in rice: Differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 2008, 20, 2946–2959. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, M.; Tadiello, A.; Farneti, B.; Lorenz, G.; Masuero, D.; Vrhovsek, U.; Costa, G.; Velasco, R.; Costa, F. A multidisciplinary approach providing new insight into fruit flesh browning physiology in apple (Malus × domestica Borkh). PLoS ONE 2013, 8, e78004. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, U.K.S.; Ganapathi, T.R. Transgenic banana plants overexpressing MusabZIP53 display severe growth retardation with enhanced sucrose and polyphenol oxidase activity. Plant Cell Tissue Organ Cult. 2014, 116, 387–402. [Google Scholar] [CrossRef]
- Chevalier, T.; de Rigal, D.; Mbéguié-A-Mbéguié, D.; Gauillard, F.; Richard-Forget, F.; Fils-Lycaon, B.R. Molecular cloning and characterization of apricot fruit polyphenol oxidase. Plant Physiol. 1999, 119, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Gardner, R.C.; Janssen, B.J.; Ross, G.S. An apple polyphenol oxidase cDNA is up-regulated in wounded tissues. Plant Mol. Biol. 1995, 27, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Seo, Y.S.; Kim, J.E.; Sung, S.K.; Song, K.J.; An, G.H.; Kim, W.T. Two polyphenol oxidases are differentially expressed during vegetative and reproductive development and in response to wounding in the Fuji apple. Plant Sci. 2001, 161, 1145–1152. [Google Scholar] [CrossRef]
- Quarta, A.; Mita, G.; Durante, M.; Arlorio, M.; de Paolis, A. Isolation of a polyphenol oxidase (PPO) cDNA from artichoke and expression analysis in wounded artichoke heads. Plant Physiol. Biochem. 2013, 68, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of nonstructural polyphenols. Plant J. 2016, 5, 507–532. [Google Scholar] [CrossRef] [PubMed]
- Muir, R.M.; Ibáñez, A.M.; Uratsu, S.L.; Ingham, E.S.; Leslie, C.A.; McGranahan, G.H.; Batra, N.; Goyal, S.; Joseph, J.; Jemmis, E.D.; et al. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol. Biol. 2011, 75, 555–565. [Google Scholar] [CrossRef]
- Cook, M.T.; Bassett, H.P.; Thompson, F.; Taubenhaus, J.J. Protective enzymes. Science 1911, 33, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Khodadadi, F.; Tohidfar, M.; Mohayeji, M.; Dandekar, A.M.; Leslie, C.A.; Kluepfel, D.A.; Butterfield, T.; Vahdati, K. Induction of polyphenol oxidase in walnut and its relationship to the pathogenic response to bacterial blight. J. Am. Soc. Hortic. Sci. 2016, 141, 119–124. [Google Scholar]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taranto, F.; Pasqualone, A.; Mangini, G.; Tripodi, P.; Miazzi, M.M.; Pavan, S.; Montemurro, C. Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects. Int. J. Mol. Sci. 2017, 18, 377. https://doi.org/10.3390/ijms18020377
Taranto F, Pasqualone A, Mangini G, Tripodi P, Miazzi MM, Pavan S, Montemurro C. Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects. International Journal of Molecular Sciences. 2017; 18(2):377. https://doi.org/10.3390/ijms18020377
Chicago/Turabian StyleTaranto, Francesca, Antonella Pasqualone, Giacomo Mangini, Pasquale Tripodi, Monica Marilena Miazzi, Stefano Pavan, and Cinzia Montemurro. 2017. "Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects" International Journal of Molecular Sciences 18, no. 2: 377. https://doi.org/10.3390/ijms18020377
APA StyleTaranto, F., Pasqualone, A., Mangini, G., Tripodi, P., Miazzi, M. M., Pavan, S., & Montemurro, C. (2017). Polyphenol Oxidases in Crops: Biochemical, Physiological and Genetic Aspects. International Journal of Molecular Sciences, 18(2), 377. https://doi.org/10.3390/ijms18020377