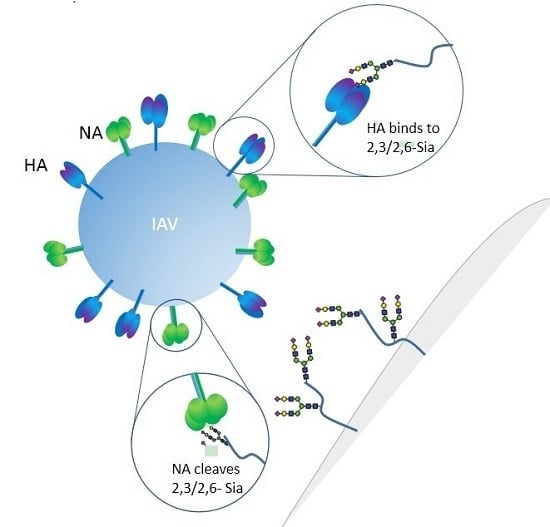
The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase
Abstract
:1. Introduction
2. Influenza Virus and Host Receptor Interactions
3. Carbohydrates Recognized by Influenza A Virus
4. Presentation of Receptors in Avian, Swine, and Human Hosts
4.1. Birds
4.2. Pigs
4.3. Humans
5. Interspecies Transmission
6. Functional Balance of HA and NA
Acknowledgments
Conflicts of Interest
References
- Shope, R.E. Swine influenza: I. Experimental transmission and pathology. J. Exp. Med. 1931, 54, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Geraci, J.R.; St Aubin, D.J.; Barker, I.K.; Webster, R.G.; Hinshaw, V.S.; Bean, W.J.; Ruhnke, H.L.; Prescott, J.H.; Early, G.; Baker, A.S.; et al. Mass mortality of harbor seals: Pneumonia associated with influenza a virus. Science 1982, 215, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Sovinova, O.; Tumova, B.; Pouska, F.; Nemec, J. Isolation of a virus causing respiratory disease in horses. Acta Virol. 1958, 2, 52–61. [Google Scholar] [PubMed]
- Organization, W.H. Fact Sheet 211—Influenza (Seasonal). Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 10 November 2016).
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007, 3, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Deyle, E.R.; Maher, M.C.; Hernandez, R.D.; Basu, S.; Sugihara, G. Global environmental drivers of influenza. Proc. Natl. Acad. Sci. USA 2016, 113, 13081–13086. [Google Scholar] [CrossRef] [PubMed]
- Kash, J.C.; Tumpey, T.M.; Proll, S.C.; Carter, V.; Perwitasari, O.; Thomas, M.J.; Basler, C.F.; Palese, P.; Taubenberger, J.K.; Garcia-Sastre, A.; et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006, 443, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Kim, J.H.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Morens, D.M.; Taubenberger, J.K.; Fauci, A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J. Infect. Dis. 2008, 198, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lee, K.H.; Steinhauer, D.A.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 1998, 95, 409–417. [Google Scholar] [CrossRef]
- Bosch, F.X.; Garten, W.; Klenk, H.D.; Rott, R. Proteolytic cleavage of influenza virus hemagglutinins: Primary structure of the connecting peptide between ha1 and ha2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology 1981, 113, 725–735. [Google Scholar] [CrossRef]
- Garten, W.; Bosch, F.X.; Linder, D.; Rott, R.; Klenk, H.D. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology 1981, 115, 361–374. [Google Scholar] [CrossRef]
- Claas, E.C.; Osterhaus, A.D.; van Beek, R.; De Jong, J.C.; Rimmelzwaan, G.F.; Senne, D.A.; Krauss, S.; Shortridge, K.F.; Webster, R.G. Human influenza a h5n1 virus related to a highly pathogenic avian influenza virus. Lancet 1998, 351, 472–477. [Google Scholar] [CrossRef]
- Subbarao, K.; Klimov, A.; Katz, J.; Regnery, H.; Lim, W.; Hall, H.; Perdue, M.; Swayne, D.; Bender, C.; Huang, J.; et al. Characterization of an avian influenza a (h5n1) virus isolated from a child with a fatal respiratory illness. Science 1998, 279, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Normile, D. Avian influenza. New h5n1 strain emerges in southern china. Science 2006, 314, 742. [Google Scholar] [CrossRef] [PubMed]
- Ungchusak, K.; Auewarakul, P.; Dowell, S.F.; Kitphati, R.; Auwanit, W.; Puthavathana, P.; Uiprasertkul, M.; Boonnak, K.; Pittayawonganon, C.; Cox, N.J.; et al. Probable person-to-person transmission of avian influenza a (h5n1). N. Engl. J. Med. 2005, 352, 333–340. [Google Scholar] [CrossRef] [PubMed]
- A revision of the system of nomenclature for influenza viruses: A who memorandum. Bull. World Health Organ. 1980, 58, 585–591.
- Tong, S.; Li, Y.; Rivailler, P.; Conrardy, C.; Castillo, D.A.; Chen, L.M.; Recuenco, S.; Ellison, J.A.; Davis, C.T.; York, I.A.; et al. A distinct lineage of influenza a virus from bats. Proc. Natl. Acad. Sci. USA 2012, 109, 4269–4274. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Garcia-Sastre, A.; Schwemmle, M. Expected and unexpected features of the newly discovered bat influenza a-like viruses. PLoS Pathog. 2015, 11, e1004819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New world bats harbor diverse influenza a viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gamblin, S.J.; Haire, L.F.; Stevens, D.J.; Xiao, B.; Ha, Y.; Skehel, J.J. H1 and h7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 2004, 325, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Cao, B.; Hu, Y.; Feng, Z.; Wang, D.; Hu, W.; Chen, J.; Jie, Z.; Qiu, H.; Xu, K.; et al. Human infection with a novel avian-origin influenza a (h7n9) virus. N. Engl. J. Med. 2013, 368, 1888–1897. [Google Scholar] [CrossRef] [PubMed]
- Peiris, M.; Yuen, K.Y.; Leung, C.W.; Chan, K.H.; Ip, P.L.; Lai, R.W.; Orr, W.K.; Shortridge, K.F. Human infection with influenza h9n2. Lancet 1999, 354, 916–917. [Google Scholar] [CrossRef]
- Bulai, T.; Bratosin, D.; Pons, A.; Montreuil, J.; Zanetta, J.P. Diversity of the human erythrocyte membrane sialic acids in relation with blood groups. FEBS Lett. 2003, 534, 185–189. [Google Scholar] [CrossRef]
- Bradley, K.C.; Jones, C.A.; Tompkins, S.M.; Tripp, R.A.; Russell, R.J.; Gramer, M.R.; Heimburg-Molinaro, J.; Smith, D.F.; Cummings, R.D.; Steinhauer, D.A. Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic h1n1 isolates (novel 2009 h1n1). Virology 2011, 413, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.M.; Rivailler, P.; Hossain, J.; Carney, P.; Balish, A.; Perry, I.; Davis, C.T.; Garten, R.; Shu, B.; Xu, X.; et al. Receptor specificity of subtype h1 influenza a viruses isolated from swine and humans in the united states. Virology 2011, 412, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Connor, R.J.; Kawaoka, Y.; Webster, R.G.; Paulson, J.C. Receptor specificity in human, avian, and equine h2 and h3 influenza virus isolates. Virology 1994, 205, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Gamblin, S.J.; Haire, L.F.; Russell, R.J.; Stevens, D.J.; Xiao, B.; Ha, Y.; Vasisht, N.; Steinhauer, D.A.; Daniels, R.S.; Elliot, A.; et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 2004, 303, 1838–1842. [Google Scholar] [CrossRef] [PubMed]
- Ibricevic, A.; Pekosz, A.; Walter, M.J.; Newby, C.; Battaile, J.T.; Brown, E.G.; Holtzman, M.J.; Brody, S.L. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006, 80, 7469–7480. [Google Scholar] [CrossRef] [PubMed]
- Kumari, K.; Gulati, S.; Smith, D.F.; Gulati, U.; Cummings, R.D.; Air, G.M. Receptor binding specificity of recent human h3n2 influenza viruses. Virol. J. 2007, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stevens, D.J.; Haire, L.F.; Walker, P.A.; Coombs, P.J.; Russell, R.J.; Gamblin, S.J.; Skehel, J.J. Structures of receptor complexes formed by hemagglutinins from the asian influenza pandemic of 1957. Proc. Natl. Acad. Sci. USA 2009, 106, 17175–17180. [Google Scholar] [CrossRef] [PubMed]
- Maines, T.R.; Chen, L.M.; Van Hoeven, N.; Tumpey, T.M.; Blixt, O.; Belser, J.A.; Gustin, K.M.; Pearce, M.B.; Pappas, C.; Stevens, J.; et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza h5n1 viruses. Virology 2011, 413, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.N.; D’Souza, B.L. Receptor binding properties of human and animal h1 influenza virus isolates. Virology 1989, 173, 317–322. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the h3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Russell, R.J.; Stevens, D.J.; Haire, L.F.; Gamblin, S.J.; Skehel, J.J. Avian and human receptor binding by hemagglutinins of influenza a viruses. Glycoconj. J. 2006, 23, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Shelton, H.; Ayora-Talavera, G.; Ren, J.; Loureiro, S.; Pickles, R.J.; Barclay, W.S.; Jones, I.M. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J. Virol. 2011, 85, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Blixt, O.; Glaser, L.; Taubenberger, J.K.; Palese, P.; Paulson, J.C.; Wilson, I.A. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 2006, 355, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and receptor specificity of the hemagglutinin from an h5n1 influenza virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; McBride, R.; Paulson, J.C.; Basler, C.F.; Wilson, I.A. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 h2n2 pandemic. J. Virol. 2010, 84, 1715–1721. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Suzuki, Y.; Suzuki, T.; Le, M.Q.; Nidom, C.A.; Sakai-Tagawa, Y.; Muramoto, Y.; Ito, M.; Kiso, M.; Horimoto, T.; et al. Haemagglutinin mutations responsible for the binding of h5n1 influenza a viruses to human-type receptors. Nature 2006, 444, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Wei, C.J.; Kong, W.P.; Wu, L.; Xu, L.; Smith, D.F.; Nabel, G.J. Immunization by avian h5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 2007, 317, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Shinya, K.; Ebina, M.; Yamada, S.; Ono, M.; Kasai, N.; Kawaoka, Y. Avian flu: Influenza virus receptors in the human airway. Nature 2006, 440, 435–436. [Google Scholar] [CrossRef] [PubMed]
- Kimble, B.; Nieto, G.R.; Perez, D.R. Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J. 2010, 7, 365. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Couceiro, J.N.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza a viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar] [PubMed]
- Wilson, I.A.; Skehel, J.J.; Wiley, D.C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 a resolution. Nature 1981, 289, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.; Brown, J.H.; Cusack, S.; Paulson, J.C.; Skehel, J.J.; Wiley, D.C. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.N.; Paulson, J.C.; Daniels, R.S.; Skehel, J.J.; Wilson, I.A.; Wiley, D.C. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 1983, 304, 76–78. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, K.; Koh, X.; Chandrasekaran, A.; Pappas, C.; Raman, R.; Srinivasan, A.; Shriver, Z.; Tumpey, T.M.; Sasisekharan, R. Determinants of glycan receptor specificity of h2n2 influenza a virus hemagglutinin. PLoS ONE 2010, 5, e13768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, Y.; Stevens, D.J.; Skehel, J.J.; Wiley, D.C. X-ray structure of the hemagglutinin of a potential h3 avian progenitor of the 1968 hong kong pandemic influenza virus. Virology 2003, 309, 209–218. [Google Scholar] [CrossRef]
- Glaser, L.; Stevens, J.; Zamarin, D.; Wilson, I.A.; Garcia-Sastre, A.; Tumpey, T.M.; Basler, C.F.; Taubenberger, J.K.; Palese, P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 2005, 79, 11533–11536. [Google Scholar] [CrossRef] [PubMed]
- Tumpey, T.M.; Maines, T.R.; Van Hoeven, N.; Glaser, L.; Solorzano, A.; Pappas, C.; Cox, N.J.; Swayne, D.E.; Palese, P.; Katz, J.M.; et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 2007, 315, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Chen, L.M.; Carney, P.J.; Garten, R.; Foust, A.; Le, J.; Pokorny, B.A.; Manojkumar, R.; Silverman, J.; Devis, R.; et al. Receptor specificity of influenza a h3n2 viruses isolated in mammalian cells and embryonated chicken eggs. J. Virol. 2010, 84, 8287–8299. [Google Scholar] [CrossRef] [PubMed]
- Maines, T.R.; Jayaraman, A.; Belser, J.A.; Wadford, D.A.; Pappas, C.; Zeng, H.; Gustin, K.M.; Pearce, M.B.; Viswanathan, K.; Shriver, Z.H.; et al. Transmission and pathogenesis of swine-origin 2009 a (h1n1) influenza viruses in ferrets and mice. Science 2009, 325, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Bradley, K.C.; Galloway, S.E.; Lasanajak, Y.; Song, X.; Heimburg-Molinaro, J.; Yu, H.; Chen, X.; Talekar, G.R.; Smith, D.F.; Cummings, R.D.; et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J. Virol. 2011, 85, 12387–12398. [Google Scholar] [CrossRef] [PubMed]
- Palese, P.; Tobita, K.; Ueda, M.; Compans, R.W. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 1974, 61, 397–410. [Google Scholar] [CrossRef]
- Colman, P.M.; Tulip, W.R.; Varghese, J.N.; Tulloch, P.A.; Baker, A.T.; Laver, W.G.; Air, G.M.; Webster, R.G. Three-dimensional structures of influenza virus neuraminidase-antibody complexes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1989, 323, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Haire, L.F.; Stevens, D.J.; Collins, P.J.; Lin, Y.P.; Blackburn, G.M.; Hay, A.J.; Gamblin, S.J.; Skehel, J.J. The structure of h5n1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006, 443, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Tulip, W.R.; Varghese, J.N.; Baker, A.T.; van Donkelaar, A.; Laver, W.G.; Webster, R.G.; Colman, P.M. Refined atomic structures of n9 subtype influenza virus neuraminidase and escape mutants. J. Mol. Biol. 1991, 221, 487–497. [Google Scholar] [CrossRef]
- Bossart-Whitaker, P.; Carson, M.; Babu, Y.S.; Smith, C.D.; Laver, W.G.; Air, G.M. Three-dimensional structure of influenza a n9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-n-acetyl neuraminic acid. J. Mol. Biol. 1993, 232, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.N.; Colman, P.M.; van Donkelaar, A.; Blick, T.J.; Sahasrabudhe, A.; McKimm-Breschkin, J.L. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA 1997, 94, 11808–11812. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wharton, S.A.; Martin, S.R.; McCauley, J.W. Role of neuraminidase in influenza a (h7n9) virus receptor binding. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Castrucci, M.R.; Kawaoka, Y. Biologic importance of neuraminidase stalk length in influenza a virus. J. Virol. 1993, 67, 759–764. [Google Scholar] [PubMed]
- Els, M.C.; Air, G.M.; Murti, K.G.; Webster, R.G.; Laver, W.G. An 18-amino acid deletion in an influenza neuraminidase. Virology 1985, 142, 241–247. [Google Scholar] [CrossRef]
- Li, J.; zu Dohna, H.; Anchell, N.L.; Adams, S.C.; Dao, N.T.; Xing, Z.; Cardona, C.J. Adaptation and transmission of a duck-origin avian influenza virus in poultry species. Virus Res. 2010, 147, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Okazaki, K.; Ozaki, H.; Sakoda, Y.; Wu, Q.; Chen, F.; Kida, H. H9n2 influenza viruses prevalent in poultry in china are phylogenetically distinct from a/quail/hong kong/g1/97 presumed to be the donor of the internal protein genes of the h5n1 hong kong/97 virus. Avian Pathol. 2003, 32, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zu Dohna, H.; Cardona, C.J.; Miller, J.; Carpenter, T.E. Emergence and genetic variation of neuraminidase stalk deletions in avian influenza viruses. PLoS ONE 2011, 6, e14722. [Google Scholar] [CrossRef] [PubMed]
- Baigent, S.J.; McCauley, J.W. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001, 79, 177–185. [Google Scholar] [CrossRef]
- Wagner, R.; Wolff, T.; Herwig, A.; Pleschka, S.; Klenk, H.D. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: A study by reverse genetics. J. Virol. 2000, 74, 6316–6323. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J. Virol. 2004, 78, 12665–12667. [Google Scholar] [CrossRef] [PubMed]
- Gagneux, P.; Cheriyan, M.; Hurtado-Ziola, N.; van der Linden, E.C.; Anderson, D.; McClure, H.; Varki, A.; Varki, N.M. Human-specific regulation of α 2–6-linked sialic acids. J. Biol. Chem. 2003, 278, 48245–48250. [Google Scholar] [CrossRef] [PubMed]
- Button, B.; Cai, L.H.; Ehre, C.; Kesimer, M.; Hill, D.B.; Sheehan, J.K.; Boucher, R.C.; Rubinstein, M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science 2012, 337, 937–941. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I.; Schachter, H.; Stanley, P. O-galnac glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009. [Google Scholar]
- Roy, M.G.; Livraghi-Butrico, A.; Fletcher, A.A.; McElwee, M.M.; Evans, S.E.; Boerner, R.M.; Alexander, S.N.; Bellinghausen, L.K.; Song, A.S.; Petrova, Y.M.; et al. Muc5b is required for airway defence. Nature 2014, 505, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Ehre, C.; Worthington, E.N.; Liesman, R.M.; Grubb, B.R.; Barbier, D.; O’Neal, W.K.; Sallenave, J.M.; Pickles, R.J.; Boucher, R.C. Overexpressing mouse model demonstrates the protective role of muc5ac in the lungs. Proc. Natl. Acad. Sci. USA 2012, 109, 16528–16533. [Google Scholar] [CrossRef] [PubMed]
- Lillehoj, E.P.; Kato, K.; Lu, W.; Kim, K.C. Cellular and molecular biology of airway mucins. Int. Rev. Cell Mol. Biol. 2013, 303, 139–202. [Google Scholar] [PubMed]
- Campbell, P.J.; Danzy, S.; Kyriakis, C.S.; Deymier, M.J.; Lowen, A.C.; Steel, J. The m segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to a/puerto rico/8/1934-based reassortant viruses. J. Virol. 2014, 88, 3802–3814. [Google Scholar] [CrossRef] [PubMed]
- Lakdawala, S.S.; Lamirande, E.W.; Suguitan, A.L., Jr.; Wang, W.; Santos, C.P.; Vogel, L.; Matsuoka, Y.; Lindsley, W.G.; Jin, H.; Subbarao, K. Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic h1n1 influenza virus. PLoS Pathog. 2011, 7, e1002443. [Google Scholar] [CrossRef] [PubMed]
- Booy, F.P.; Ruigrok, R.W.; van Bruggen, E.F. Electron microscopy of influenza virus. A comparison of negatively stained and ice-embedded particles. J. Mol. Biol. 1985, 184, 667–676. [Google Scholar] [CrossRef]
- Nermut, M.V. Further investigation on the fine structure of influenza virus. J. Gen. Virol. 1972, 17, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Elleman, C.J.; Barclay, W.S. The m1 matrix protein controls the filamentous phenotype of influenza a virus. Virology 2004, 321, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.C.; Lamb, R.A.; Compans, R.W. The m1 and m2 proteins of influenza a virus are important determinants in filamentous particle formation. Virology 1998, 240, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Burleigh, L.M.; Calder, L.J.; Skehel, J.J.; Steinhauer, D.A. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 2005, 79, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Choppin, P.W.; Tamm, I. Studies of two kinds of virus particles which comprise influenza a2 virus strains. Ii. Reactivity with virus inhibitors in normal sera. J. Exp. Med. 1960, 112, 921–944. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Dawson, I.M.; Elford, W.J. Filamentous forms associated with newly isolated influenza virus. Lancet 1949, 1, 602. [Google Scholar] [CrossRef]
- Campbell, P.J.; Kyriakis, C.S.; Marshall, N.; Suppiah, S.; Seladi-Schulman, J.; Danzy, S.; Lowen, A.C.; Steel, J. Residue 41 of the eurasian avian-like swine influenza a virus matrix protein modulates virion filament length and efficiency of contact transmission. J. Virol. 2014, 88, 7569–7577. [Google Scholar] [CrossRef] [PubMed]
- Bourmakina, S.V.; Garcia-Sastre, A. Reverse genetics studies on the filamentous morphology of influenza a virus. J. Gen. Virol. 2003, 84, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Seladi-Schulman, J.; Campbell, P.J.; Suppiah, S.; Steel, J.; Lowen, A.C. Filament-producing mutants of influenza a/puerto rico/8/1934 (h1n1) virus have higher neuraminidase activities than the spherical wild-type. PLoS ONE 2014, 9, e112462. [Google Scholar] [CrossRef] [PubMed]
- Mosley, V.M.; Wyckoff, R.W. Election micrography of the virus of influenza. Nature 1946, 157, 263. [Google Scholar] [CrossRef] [PubMed]
- Calder, L.J.; Wasilewski, S.; Berriman, J.A.; Rosenthal, P.B. Structural organization of a filamentous influenza a virus. Proc. Natl. Acad. Sci. USA 2010, 107, 10685–10690. [Google Scholar] [CrossRef] [PubMed]
- Seladi-Schulman, J.; Steel, J.; Lowen, A.C. Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo. J. Virol. 2013, 87, 13343–13353. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.; Cardone, G.; Winkler, D.C.; Heymann, J.B.; Brecher, M.; White, J.M.; Steven, A.C. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. USA 2006, 103, 19123–19127. [Google Scholar] [CrossRef] [PubMed]
- Burnet, F.M. Enzymic action of influenza viruses on glandular mucin and on purified blood group substances. Aust. J. Sci. 1947, 10, 21. [Google Scholar] [PubMed]
- Gottschalk, A.; Lind, P.E. Product of interaction between influenza virus enzyme and ovomucin. Nature 1949, 164, 232. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, M.; Asaoka, N.; Sakai, T.; Ohuchi, R. Roles of neuraminidase in the initial stage of influenza virus infection. Microbes Infect. 2006, 8, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Zhang, X.Q.; Senaati, H.P.; Chen, H.W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza a penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Zanin, M.; Marathe, B.; Wong, S.S.; Yoon, S.W.; Collin, E.; Oshansky, C.; Jones, J.; Hause, B.; Webby, R. Pandemic swine h1n1 influenza viruses with almost undetectable neuraminidase activity are not transmitted via aerosols in ferrets and are inhibited by human mucus but not swine mucus. J. Virol. 2015, 89, 5935–5948. [Google Scholar] [CrossRef] [PubMed]
- Hirst, G.K. The agglutination of red cells by allantoic fluid of chick embryos infected with influenza virus. Science 1941, 94, 22–23. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic acid receptors of viruses. Top. Curr. Chem. 2015, 367, 1–28. [Google Scholar] [PubMed]
- Klenk, E.; Faillard, H.; Lempfrid, H. Enzymatic effect of the influenza virus. Hoppe Seylers Z Physiol. Chem. 1955, 301, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Schauer, R. Sialic acids. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009. [Google Scholar]
- Kelm, S.; Schauer, R. Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 1997, 175, 137–240. [Google Scholar] [PubMed]
- Byrd-Leotis, L.; Liu, R.; Bradley, K.C.; Lasanajak, Y.; Cummings, S.F.; Song, X.; Heimburg-Molinaro, J.; Galloway, S.E.; Culhane, M.R.; Smith, D.F.; et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc. Natl. Acad. Sci. USA 2014, 111, E2241–E2250. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P.; Schachter, H.; Taniguchi, N. N-glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009. [Google Scholar]
- Helenius, A.; Aebi, M. Roles of n-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004, 73, 1019–1049. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Schachter, H. Genetic disorders of glycosylation. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: New York, NY, USA, 2009. [Google Scholar]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza a viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [PubMed]
- Shibuya, N.; Goldstein, I.J.; Broekaert, W.F.; Nsimba-Lubaki, M.; Peeters, B.; Peumans, W.J. The elderberry (Sambucus nigra L.) bark lectin recognizes the neu5ac (α2–6) gal/galnac sequence. J. Biol. Chem. 1987, 262, 1596–1601. [Google Scholar] [PubMed]
- Wang, W.C.; Cummings, R.D. The immobilized leukoagglutinin from the seeds of maackia amurensis binds with high affinity to complex-type asn-linked oligosaccharides containing terminal sialic acid-linked α-2,3 to penultimate galactose residues. J. Biol. Chem. 1988, 263, 4576–4585. [Google Scholar] [PubMed]
- Konami, Y.; Yamamoto, K.; Osawa, T.; Irimura, T. Strong affinity of maackia amurensis hemagglutinin (mah) for sialic acid-containing ser/thr-linked carbohydrate chains of n-terminal octapeptides from human glycophorin A. FEBS Lett. 1994, 342, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Geisler, C.; Jarvis, D.L. Effective glycoanalysis with maackia amurensis lectins requires a clear understanding of their binding specificities. Glycobiology 2011, 21, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Franca, M.; Stallknecht, D.E.; Howerth, E.W. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian Pathol. 2013, 42, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Nelli, R.K.; Kuchipudi, S.V.; White, G.A.; Perez, B.B.; Dunham, S.P.; Chang, K.C. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 2010, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.; Chaves, A.J.; Valle, R.; Darji, A.; van Riel, D.; Kuiken, T.; Majo, N.; Ramis, A. Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species. Vet. Res. 2012, 43, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambaryan, A.; Webster, R.; Matrosovich, M. Differences between influenza virus receptors on target cells of duck and chicken. Arch. Virol. 2002, 147, 1197–1208. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, J.M.; Bourne, A.J.; Chen, H.; Guan, Y.; Peiris, J.S. Sialic acid receptor detection in the human respiratory tract: Evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir. Res. 2007, 8, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donatelli, I.; Castrucci, M.R.; De Marco, M.A.; Delogu, M.; Webster, R.G. Human-animal interface: The case for influenza interspecies transmission. Adv. Exp. Med. Biol. 2016, 972, 17–33. [Google Scholar]
- Nelson, M.I.; Wentworth, D.E.; Culhane, M.R.; Vincent, A.L.; Viboud, C.; LaPointe, M.P.; Lin, X.; Holmes, E.C.; Detmer, S.E. Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J. Virol. 2014, 88, 10110–10119. [Google Scholar] [CrossRef] [PubMed]
- Trebbien, R.; Larsen, L.E.; Viuff, B.M. Distribution of sialic acid receptors and influenza a virus of avian and swine origin in experimentally infected pigs. Virol. J. 2011, 8, 434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisen, M.B.; Sabesan, S.; Skehel, J.J.; Wiley, D.C. Binding of the influenza a virus to cell-surface receptors: Structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology 1997, 232, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Gambaryan, A.S.; Karasin, A.I.; Tuzikov, A.B.; Chinarev, A.A.; Pazynina, G.V.; Bovin, N.V.; Matrosovich, M.N.; Olsen, C.W.; Klimov, A.I. Receptor-binding properties of swine influenza viruses isolated and propagated in mdck cells. Virus Res. 2005, 114, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.C.; Karamanska, R.; Busch, M.G.; Dell, A.; Olsen, C.W.; Haslam, S.M. Glycan analysis and influenza a virus infection of primary swine respiratory epithelial cells: The importance of neuacα2–6 glycans. J. Biol. Chem. 2010, 285, 34016–34026. [Google Scholar] [CrossRef] [PubMed]
- Couceiro, J.N.; Paulson, J.C.; Baum, L.G. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993, 29, 155–165. [Google Scholar] [CrossRef]
- Van Riel, D.; Munster, V.J.; de Wit, E.; Rimmelzwaan, G.F.; Fouchier, R.A.; Osterhaus, A.D.; Kuiken, T. H5n1 virus attachment to lower respiratory tract. Science 2006, 312, 399. [Google Scholar] [CrossRef] [PubMed]
- Scholtissek, C. Molecular evolution of influenza viruses. Virus Genes 1995, 11, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.W.; Karamanska, R.; Van Poucke, S.; Van Reeth, K.; Chan, I.W.; Chan, M.C.; Dell, A.; Peiris, J.S.; Haslam, S.M.; Guan, Y.; et al. Infection of swine ex vivo tissues with avian viruses including h7n9 and correlation with glycomic analysis. Influenza Respir. Viruses 2013, 7, 1269–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Punyadarsaniya, D.; Liang, C.H.; Winter, C.; Petersen, H.; Rautenschlein, S.; Hennig-Pauka, I.; Schwegmann-Wessels, C.; Wu, C.Y.; Wong, C.H.; Herrler, G. Infection of differentiated porcine airway epithelial cells by influenza virus: Differential susceptibility to infection by porcine and avian viruses. PLoS ONE 2011, 6, e28429. [Google Scholar] [CrossRef] [PubMed]
- Van Poucke, S.G.; Nicholls, J.M.; Nauwynck, H.J.; Van Reeth, K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol. J. 2010, 7, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemae, N.; Ruttanapumma, R.; Parchariyanon, S.; Yoneyama, S.; Hayashi, T.; Hiramatsu, H.; Sriwilaijaroen, N.; Uchida, Y.; Kondo, S.; Yagi, H.; et al. Alterations in receptor-binding properties of swine influenza viruses of the h1 subtype after isolation in embryonated chicken eggs. J. Gen. Virol. 2010, 91, 938–948. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.H.; Takematsu, H.; Diaz, S.; Iber, J.; Nickerson, E.; Wright, K.L.; Muchmore, E.A.; Nelson, D.L.; Warren, S.T.; Varki, A. A mutation in human cmp-sialic acid hydroxylase occurred after the homo-pan divergence. Proc. Natl. Acad. Sci. USA 1998, 95, 11751–11756. [Google Scholar] [CrossRef] [PubMed]
- Irie, A.; Koyama, S.; Kozutsumi, Y.; Kawasaki, T.; Suzuki, A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J. Biol. Chem. 1998, 273, 15866–15871. [Google Scholar] [CrossRef] [PubMed]
- Sillanaukee, P.; Ponnio, M.; Jaaskelainen, I.P. Occurrence of sialic acids in healthy humans and different disorders. Eur. J. Clin. Investig. 1999, 29, 413–425. [Google Scholar] [CrossRef]
- Sriwilaijaroen, N.; Kondo, S.; Yagi, H.; Takemae, N.; Saito, T.; Hiramatsu, H.; Kato, K.; Suzuki, Y. N-glycans from porcine trachea and lung: Predominant neuacα2–6gal could be a selective pressure for influenza variants in favor of human-type receptor. PLoS ONE 2011, 6, e16302. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Horiike, G.; Yamazaki, Y.; Kawabe, K.; Masuda, H.; Miyamoto, D.; Matsuda, M.; Nishimura, S.I.; Yamagata, T.; Ito, T.; et al. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997, 404, 192–196. [Google Scholar] [CrossRef]
- Karasin, A.I.; Schutten, M.M.; Cooper, L.A.; Smith, C.B.; Subbarao, K.; Anderson, G.A.; Carman, S.; Olsen, C.W. Genetic characterization of h3n2 influenza viruses isolated from pigs in north america, 1977–1999: Evidence for wholly human and reassortant virus genotypes. Virus Res. 2000, 68, 71–85. [Google Scholar] [CrossRef]
- Matrosovich, M.N.; Matrosovich, T.Y.; Gray, T.; Roberts, N.A.; Klenk, H.D. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 4620–4624. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.; Karamanska, R.; Chan, R.W.; Chan, M.C.; Jia, N.; Air, G.; Hopton, C.; Wong, M.P.; Dell, A.; Malik Peiris, J.S.; et al. Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLoS Pathog. 2013, 9, e1003223. [Google Scholar] [CrossRef] [PubMed]
- van Riel, D.; Munster, V.J.; de Wit, E.; Rimmelzwaan, G.F.; Fouchier, R.A.; Osterhaus, A.D.; Kuiken, T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007, 171, 1215–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hare, R.; Curl, M.; Mc, C.L. The efficiency of the red cell adsorption and elution method for the preparation of influenza vaccine. Can. J. Public Health 1946, 37, 284–291. [Google Scholar] [PubMed]
- Paulson, J.C.; Rogers, G.N. Resialylated erythrocytes for assessment of the specificity of sialyloligosaccharide binding proteins. Methods Enzymol. 1987, 138, 162–168. [Google Scholar] [PubMed]
- Burnet, F.M.; Bull, D.R. Changes in influenza virus associated with adaptation to passage in chick embryos. Aust. J. Exp. Biol. Med. Sci. 1943, 21, 55–69. [Google Scholar] [CrossRef]
- Hoyle, L. Virology Monographs; Springer-Verlag: New York, NY, USA; Berlin, Germany, 1968; Volume 4. [Google Scholar]
- Ito, T.; Suzuki, Y.; Mitnaul, L.; Vines, A.; Kida, H.; Kawaoka, Y. Receptor specificity of influenza a viruses correlates with the agglutination of erythrocytes from different animal species. Virology 1997, 227, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.M.; Higa, H.H.; Paulson, J.C. Different cell-surface receptor determinants of antigenically similar influenza virus hemagglutinins. J. Biol. Chem. 1981, 256, 8357–8363. [Google Scholar] [PubMed]
- Chu, V.C.; Whittaker, G.R. Influenza virus entry and infection require host cell n-linked glycoprotein. Proc. Natl. Acad. Sci. USA 2004, 101, 18153–18158. [Google Scholar] [CrossRef] [PubMed]
- Gambaryan, A.S.; Matrosovich, M.N. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J. Virol. Methods 1992, 39, 111–123. [Google Scholar] [CrossRef]
- Gambaryan, A.S.; Tuzikov, A.B.; Piskarev, V.E.; Yamnikova, S.S.; Lvov, D.K.; Robertson, J.S.; Bovin, N.V.; Matrosovich, M.N. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: Non-egg-adapted human h1 and h3 influenza a and influenza b viruses share a common high binding affinity for 6′-sialyl (N-acetyllactosamine). Virology 1997, 232, 345–350. [Google Scholar] [PubMed]
- Blixt, O.; Head, S.; Mondala, T.; Scanlan, C.; Huflejt, M.E.; Alvarez, R.; Bryan, M.C.; Fazio, F.; Calarese, D.; Stevens, J.; et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17033–17038. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Xiong, X.; Wharton, S.A.; Martin, S.R.; Coombs, P.J.; Vachieri, S.G.; Christodoulou, E.; Walker, P.A.; Liu, J.; Skehel, J.J.; et al. Evolution of the receptor binding properties of the influenza a(h3n2) hemagglutinin. Proc. Natl. Acad. Sci. USA 2012, 109, 21474–21479. [Google Scholar] [CrossRef] [PubMed]
- Childs, R.A.; Palma, A.S.; Wharton, S.; Matrosovich, T.; Liu, Y.; Chai, W.; Campanero-Rhodes, M.A.; Zhang, Y.; Eickmann, M.; Kiso, M.; et al. Receptor-binding specificity of pandemic influenza a (h1n1) 2009 virus determined by carbohydrate microarray. Nat. Biotechnol. 2009, 27, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Childs, R.A.; Matrosovich, T.; Wharton, S.; Palma, A.S.; Chai, W.; Daniels, R.; Gregory, V.; Uhlendorff, J.; Kiso, M.; et al. Altered receptor specificity and cell tropism of d222g hemagglutinin mutants isolated from fatal cases of pandemic a (h1n1) 2009 influenza virus. J. Virol. 2010, 84, 12069–12074. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yu, H.; Chen, X.; Lasanajak, Y.; Tappert, M.M.; Air, G.M.; Tiwari, V.K.; Cao, H.; Chokhawala, H.A.; Zheng, H.; et al. A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J. Biol. Chem. 2011, 286, 31610–31622. [Google Scholar] [CrossRef] [PubMed]
- Yen, H.L.; Liang, C.H.; Wu, C.Y.; Forrest, H.L.; Ferguson, A.; Choy, K.T.; Jones, J.; Wong, D.D.; Cheung, P.P.; Hsu, C.H.; et al. Hemagglutinin-neuraminidase balance confers respiratory-droplet transmissibility of the pandemic h1n1 influenza virus in ferrets. Proc. Natl. Acad. Sci. USA 2011, 108, 14264–14269. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Lasanajak, Y.; Xia, B.; Heimburg-Molinaro, J.; Rhea, J.M.; Ju, H.; Zhao, C.; Molinaro, R.J.; Cummings, R.D.; Smith, D.F. Shotgun glycomics: A microarray strategy for functional glycomics. Nat. Methods 2011, 8, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; de Vries, R.P.; Grant, O.C.; Thompson, A.J.; McBride, R.; Tsogtbaatar, B.; Lee, P.S.; Razi, N.; Wilson, I.A.; Woods, R.J.; et al. Recent h3n2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 2016, 21, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Perez, D.R. Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 2006, 346, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Nelli, R.; White, G.A.; Bain, M.; Chang, K.C.; Dunham, S. Differences in influenza virus receptors in chickens and ducks: Implications for interspecies transmission. J. Mol. Genet. Med. 2009, 3, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Shortridge, K.F. Poultry and the influenza h5n1 outbreak in hong kong, 1997: Abridged chronology and virus isolation. Vaccine 1999, 17, S26–S29. [Google Scholar] [CrossRef]
- Xiong, X.; Coombs, P.J.; Martin, S.R.; Liu, J.; Xiao, H.; McCauley, J.W.; Locher, K.; Walker, P.A.; Collins, P.J.; Kawaoka, Y.; et al. Receptor binding by a ferret-transmissible h5 avian influenza virus. Nature 2013, 497, 392–396. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.P.; Zhu, X.; McBride, R.; Rigter, A.; Hanson, A.; Zhong, G.; Hatta, M.; Xu, R.; Yu, W.; Kawaoka, Y.; et al. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible h5n1 viruses. J. Virol. 2014, 88, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shi, Y.; Zhang, W.; Zhang, Y.; Qi, J.; Gao, G.F. Structure and receptor-binding properties of an airborne transmissible avian influenza a virus hemagglutinin h5 (vn1203mut). Protein Cell 2013, 4, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Martin, S.R.; Haire, L.F.; Wharton, S.A.; Daniels, R.S.; Bennett, M.S.; McCauley, J.W.; Collins, P.J.; Walker, P.A.; Skehel, J.J.; et al. Receptor binding by an h7n9 influenza virus from humans. Nature 2013, 499, 496–499. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; McCauley, J.W.; Steinhauer, D.A. Receptor binding properties of the influenza virus hemagglutinin as a determinant of host range. Curr. Top. Microbiol. Immunol. 2014, 385, 63–91. [Google Scholar] [PubMed]
- Chutinimitkul, S.; Herfst, S.; Steel, J.; Lowen, A.C.; Ye, J.; van Riel, D.; Schrauwen, E.J.; Bestebroer, T.M.; Koel, B.; Burke, D.F.; et al. Virulence-associated substitution d222g in the hemagglutinin of 2009 pandemic influenza a (h1n1) virus affects receptor binding. J. Virol. 2010, 84, 11802–11813. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Tuzikov, A.; Bovin, N.; Gambaryan, A.; Klimov, A.; Castrucci, M.R.; Donatelli, I.; Kawaoka, Y. Early alterations of the receptor-binding properties of h1, h2, and h3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 2000, 74, 8502–8512. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ibrahim, M.S.; Ellakany, H.F.; Kawashita, N.; Mizuike, R.; Hiramatsu, H.; Sriwilaijaroen, N.; Takagi, T.; Suzuki, Y.; Ikuta, K. Acquisition of human-type receptor binding specificity by new h5n1 influenza virus sublineages during their emergence in birds in egypt. PLoS Pathog. 2011, 7, e1002068. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Xiao, H.; Martin, S.R.; Coombs, P.J.; Liu, J.; Collins, P.J.; Vachieri, S.G.; Walker, P.A.; Lin, Y.P.; McCauley, J.W.; et al. Enhanced human receptor binding by h5 haemagglutinins. Virology 2014, 456–457, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, W.; Wang, F.; Qi, J.; Wu, Y.; Song, H.; Gao, F.; Bi, Y.; Zhang, Y.; Fan, Z.; et al. Structures and receptor binding of hemagglutinins from human-infecting h7n9 influenza viruses. Science 2013, 342, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Belser, J.A.; Gustin, K.M.; Pearce, M.B.; Maines, T.R.; Zeng, H.; Pappas, C.; Sun, X.; Carney, P.J.; Villanueva, J.M.; Stevens, J.; et al. Pathogenesis and transmission of avian influenza a (h7n9) virus in ferrets and mice. Nature 2013, 501, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, D.; Kelvin, D.J.; Li, L.; Zheng, Z.; Yoon, S.W.; Wong, S.S.; Farooqui, A.; Wang, J.; Banner, D.; et al. Infectivity, transmission, and pathology of human-isolated h7n9 influenza virus in ferrets and pigs. Science 2013, 341, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, D.; Gao, R.; Zhao, B.; Song, J.; Qi, X.; Zhang, Y.; Shi, Y.; Yang, L.; Zhu, W.; et al. Biological features of novel avian influenza a (h7n9) virus. Nature 2013, 499, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kiso, M.; Fukuyama, S.; Nakajima, N.; Imai, M.; Yamada, S.; Murakami, S.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Sakoda, Y.; et al. Characterization of h7n9 influenza a viruses isolated from humans. Nature 2013, 501, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Tharakaraman, K.; Jayaraman, A.; Raman, R.; Viswanathan, K.; Stebbins, N.W.; Johnson, D.; Shriver, Z.; Sasisekharan, V.; Sasisekharan, R. Glycan receptor binding of the influenza a virus h7n9 hemagglutinin. Cell 2013, 153, 1486–1493. [Google Scholar] [CrossRef] [PubMed]
- Steinhauer, D.A. Influenza: Pathways to human adaptation. Nature 2013, 499, 412–413. [Google Scholar] [CrossRef] [PubMed]
- Herfst, S.; Schrauwen, E.J.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne transmission of influenza a/h5n1 virus between ferrets. Science 2012, 336, 1534–1541. [Google Scholar] [CrossRef] [PubMed]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza h5 ha confers respiratory droplet transmission to a reassortant h5 ha/h1n1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef] [PubMed]
- McFadden, E.R., Jr.; Pichurko, B.M.; Bowman, H.F.; Ingenito, E.; Burns, S.; Dowling, N.; Solway, J. Thermal mapping of the airways in humans. J. Appl. Physiol. 1985, 58, 564–570. [Google Scholar] [PubMed]
- Massin, P.; van der Werf, S.; Naffakh, N. Residue 627 of pb2 is a determinant of cold sensitivity in rna replication of avian influenza viruses. J. Virol. 2001, 75, 5398–5404. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, J.; Leiacker, R.; Rettinger, G.; Keck, T. Nasal mucosal temperature during respiration. Clin. Otolaryngol. Allied Sci. 2002, 27, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by pb2 amino acids 627k or 627e/701n. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, E.K.; Kawaoka, Y.; Murphy, B.R. Rescue of an influenza a virus wild-type pb2 gene and a mutant derivative bearing a site-specific temperature-sensitive and attenuating mutation. J. Virol. 1993, 67, 7223–7228. [Google Scholar] [PubMed]
- Zaraket, H.; Bridges, O.A.; Duan, S.; Baranovich, T.; Yoon, S.W.; Reed, M.L.; Salomon, R.; Webby, R.J.; Webster, R.G.; Russell, C.J. Increased acid stability of the hemagglutinin protein enhances h5n1 influenza virus growth in the upper respiratory tract but is insufficient for transmission in ferrets. J. Virol. 2013, 87, 9911–9922. [Google Scholar] [CrossRef] [PubMed]
- Galloway, S.E.; Reed, M.L.; Russell, C.J.; Steinhauer, D.A. Influenza ha subtypes demonstrate divergent phenotypes for cleavage activation and ph of fusion: Implications for host range and adaptation. PLoS Pathog. 2013, 9, e1003151. [Google Scholar] [CrossRef] [PubMed]
- Byrd-Leotis, L.; Galloway, S.E.; Agbogu, E.; Steinhauer, D.A. Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple ha subtypes. J. Virol. 2015, 89, 4504–4516. [Google Scholar] [CrossRef] [PubMed]
- Baum, L.G.; Paulson, J.C. The n2 neuraminidase of human influenza virus has acquired a substrate specificity complementary to the hemagglutinin receptor specificity. Virology 1991, 180, 10–15. [Google Scholar] [CrossRef]
- Kobasa, D.; Kodihalli, S.; Luo, M.; Castrucci, M.R.; Donatelli, I.; Suzuki, Y.; Suzuki, T.; Kawaoka, Y. Amino acid residues contributing to the substrate specificity of the influenza a virus neuraminidase. J. Virol. 1999, 73, 6743–6751. [Google Scholar] [PubMed]
- Xu, R.; Zhu, X.; McBride, R.; Nycholat, C.M.; Yu, W.; Paulson, J.C.; Wilson, I.A. Functional balance of the hemagglutinin and neuraminidase activities accompanies the emergence of the 2009 h1n1 influenza pandemic. J. Virol. 2012, 86, 9221–9232. [Google Scholar] [CrossRef] [PubMed]
- Ginting, T.E.; Shinya, K.; Kyan, Y.; Makino, A.; Matsumoto, N.; Kaneda, S.; Kawaoka, Y. Amino acid changes in hemagglutinin contribute to the replication of oseltamivir-resistant h1n1 influenza viruses. J. Virol. 2012, 86, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Blick, T.J.; Sahasrabudhe, A.; McDonald, M.; Owens, I.J.; Morley, P.J.; Fenton, R.J.; McKimm-Breschkin, J.L. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-neu5ac2en. Virology 1998, 246, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Kaverin, N.V.; Gambaryan, A.S.; Bovin, N.V.; Rudneva, I.A.; Shilov, A.A.; Khodova, O.M.; Varich, N.L.; Sinitsin, B.V.; Makarova, N.V.; Kropotkina, E.A. Postreassortment changes in influenza a virus hemagglutinin restoring ha-na functional match. Virology 1998, 244, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Hughes, M.T.; Matrosovich, M.; Rodgers, M.E.; McGregor, M.; Kawaoka, Y. Influenza a viruses lacking sialidase activity can undergo multiple cycles of replication in cell culture, eggs, or mice. J. Virol. 2000, 74, 5206–5212. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.; Erny, A.; Care, B.; Traversier, A.; Barthelemy, M.; Hay, A.; Lin, Y.P.; Ferraris, O.; Lina, B. Rescue of a h3n2 influenza virus containing a deficient neuraminidase protein by a hemagglutinin with a low receptor-binding affinity. PLoS ONE 2012, 7, e33880. [Google Scholar] [CrossRef] [PubMed]
- Mitnaul, L.J.; Matrosovich, M.N.; Castrucci, M.R.; Tuzikov, A.B.; Bovin, N.V.; Kobasa, D.; Kawaoka, Y. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza a virus. J. Virol. 2000, 74, 6015–6020. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Bansal, A.; Liu, C.; Air, G.M. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology 1997, 229, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Diederich, S.; Berhane, Y.; Embury-Hyatt, C.; Hisanaga, T.; Handel, K.; Cottam-Birt, C.; Ranadheera, C.; Kobasa, D.; Pasick, J. Hemagglutinin-neuraminidase balance influences the virulence phenotype of a recombinant h5n3 influenza a virus possessing a polybasic ha0 cleavage site. J. Virol. 2015, 89, 10724–10734. [Google Scholar] [CrossRef] [PubMed]
- De Vries, E.; de Vries, R.P.; Wienholts, M.J.; Floris, C.E.; Jacobs, M.S.; van den Heuvel, A.; Rottier, P.J.; de Haan, C.A. Influenza a virus entry into cells lacking sialylated N-glycans. Proc. Natl. Acad. Sci. USA 2012, 109, 7457–7462. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Martin, S.R.; Wharton, S.A.; McCauley, J.W. Biophysical measurement of the balance of influenza a hemagglutinin and neuraminidase activities. J. Biol. Chem. 2015, 290, 6516–6521. [Google Scholar] [CrossRef] [PubMed]




| Species | Tissue | 2,3Sia | 2,6Sia | Reference |
|---|---|---|---|---|
| Ducks | Trachea | + | + | Franca et al. [111], Costa et al. [114] |
| + | Kuchipudi et al. [112] | |||
| Bronchus/lung | + | + | Franca et al. [111], Kuchipudi et al. [112], Costa et al. [114] | |
| Intestine | + | + | Franca et al. [111] | |
| + | Kuchipudi et al. [112], Gambaryan et al. [116], Costa et al. [114] | |||
| Gulls | Trachea | + | + | Franca et al. [111] |
| Bronchus/lung | + | + | Franca et al. [111] | |
| Intestine | + | + | Franca et al. [111] | |
| Quail | Trachea | + | + | Wan et al. [113] |
| Intestine | + | + | Wan et al. [113] | |
| Chickens | Trachea | + | Wan et al. [113] | |
| + | Kuchipudi et al. [112], Trebbien et al. [117] | |||
| + | + | Gambaryan et al. [116], Costa et al. [114] | ||
| Bronchus/lung | + | + | Kuchipudi et al. [112], Gambaryan et al. [116], Costa et al. [114] | |
| Intestine | + | Kuchipudi et al. [112], Costa et al. [114], Wan et al. [113] | ||
| + | + | Gambaryan et al. [116], Trebbien et al. [117] | ||
| Pigs | Trachea | + | Nelli et al. [118], Chan et al. [119], VanPoucke et al. [120] | |
| Bronchus/lung | + | + | VanPoucke et al. [120], Chan et al. [119], Punyadarsaniya et al. [121], VanPoucke et al. [120] | |
| + | Nelli et al. [118], Trebbien et al. [117] | |||
| Humans | Trachea | inmucindroplets | + | Couceiro et al. [122] |
| Bronchus/lung | + | + | Barkhordari et al. [123] | |
| +lower | +upper | Shinya et al. [42] | ||
| variable | + | Nicholls et al. [124] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byrd-Leotis, L.; Cummings, R.D.; Steinhauer, D.A. The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. Int. J. Mol. Sci. 2017, 18, 1541. https://doi.org/10.3390/ijms18071541
Byrd-Leotis L, Cummings RD, Steinhauer DA. The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. International Journal of Molecular Sciences. 2017; 18(7):1541. https://doi.org/10.3390/ijms18071541
Chicago/Turabian StyleByrd-Leotis, Lauren, Richard D. Cummings, and David A. Steinhauer. 2017. "The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase" International Journal of Molecular Sciences 18, no. 7: 1541. https://doi.org/10.3390/ijms18071541
APA StyleByrd-Leotis, L., Cummings, R. D., & Steinhauer, D. A. (2017). The Interplay between the Host Receptor and Influenza Virus Hemagglutinin and Neuraminidase. International Journal of Molecular Sciences, 18(7), 1541. https://doi.org/10.3390/ijms18071541