From Variation of Influenza Viral Proteins to Vaccine Development
Abstract
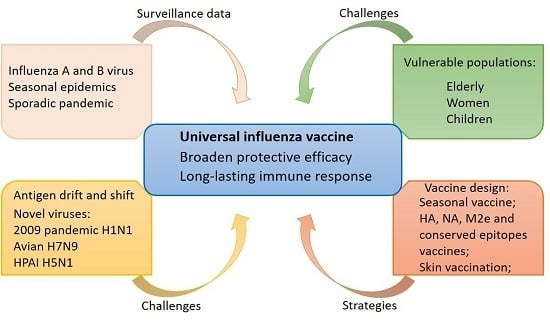
1. Introduction
2. Relationship between Influenza Virus Proteins and Vaccine Development
2.1. Hemagglutinin
2.2. Neuraminidase
2.3. Mutations in Other Influenza Virus Proteins
3. Strategy for Influenza Vaccine
3.1. Current Influenza Vaccine
3.2. New Strategy of Influenza Vaccine
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hause, B.M.; Collin, E.A.; Liu, R.X.; Huang, B.; Sheng, Z.Z.; Lu, W.X.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the orthomyxoviridae family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [PubMed]
- Andrew, M.Q.; King, E.L.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2012; pp. 749–761. [Google Scholar]
- Taubenberger, J.K.; Kash, J.C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010, 7, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Olivier, A.K.; Genova, S.; Epperson, W.B.; Smith, D.R.; Schneider, L.; Barton, K.; McCuan, K.; Webby, R.J.; Wan, X.F. Pathogenesis of influenza D virus in cattle. J. Virol. 2016, 90, 5636–5642. [Google Scholar] [CrossRef] [PubMed]
- Lambert, T.K.K.; Leedom Larson, K.R. Influenza C and influenza D viruses. Swine Health Information Center and Center for Food Security and Public Health, 2016. Available online: http://www.cfsph.iastate.edu/pdf/shic-factsheet-influenza-cd (accessed on 15 June 2017).
- WHO. Influenza (Seasonal). Fact Sheet. 2016. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 15 June 2017).
- Luckhaupt, S.E.; Sweeney, M.H.; Funk, R.; Calvert, G.M.; Nowell, M.; D’Mello, T.; Reingold, A.; Meek, J.; Yousey-Hindes, K.; Arnold, K.E.; et al. Influenza-associated hospitalizations by industry, 2009–10 influenza season, United States. Emerg. Infect. Dis. 2012, 18, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.W.; Shay, D.K.; Weintraub, E.; Brammer, L.; Cox, N.; Anderson, L.J.; Fukuda, K. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 2003, 289, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Anar, C.; Bicmen, C.; Yapicioglu, S.; Unsal, I.; Halilcolar, H.; Yilmaz, U. Evaluation of clinical data and antibody response following influenza vaccination in patients with chronic obstructive pulmonary disease. New Microbiol. 2010, 33, 117–127. [Google Scholar] [PubMed]
- Baldo, V.; Baldovin, T.; Pellegrini, M.; Angiolelli, G.; Majori, S.; Floreani, A.; Busana, M.C.; Bertoncello, C.; Trivello, R. Immunogenicity of three different influenza vaccines against homologous and heterologous strains in nursing home elderly residents. Clin. Dev. Immunol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; McVernon, J.; Skeljo, M.; Richmond, P.; Wadia, U.; Lambert, S.; Nissen, M.; Marshall, H.; Booy, R.; Heron, L.; et al. Immunogenicity of a monovalent 2009 influenza A(H1N1) vaccine in infants and children: A randomized trial. JAMA 2010, 303, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Santibanez, T.A.; Jamieson, D.J.; Weinbaum, C.M.; Euler, G.L.; Grohskopf, L.A.; Lu, P.J.; Singleton, J.A. Influenza vaccination coverage among pregnant women—National 2009 H1N1 Flu Survey (NHFS). Am. J. Obstet. Gynecol. 2011, 204, S96–S106. [Google Scholar] [CrossRef] [PubMed]
- Advisory Committee on Immunization Practices; Smith, N.M.; Bresee, J.S.; Shay, D.K.; Uyeki, T.M.; Cox, N.J.; Strikas, R.A. Prevention and control of influenza: Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR. Recomm. Rep. 2006, 55, 1–42. [Google Scholar] [PubMed]
- Taubenberger, J.K.; Morens, D.M. Pandemic influenza—Including a risk assessment of H5N1. Rev. Sci. Tech. 2009, 28, 187–202. [Google Scholar] [CrossRef] [PubMed]
- WHO/GIP. Cumulative Number of Confirmed Human Cases for Avian influenza A(H5N1) Reported to WHO. 2003–2017. Available online: http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/ (accessed on 15 June 2017).
- Alexander, D.J. An overview of the epidemiology of avian influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Fouchier, R.A.; Schneeberger, P.M.; Rozendaal, F.W.; Broekman, J.M.; Kemink, S.A.; Munster, V.; Kuiken, T.; Rimmelzwaan, G.F.; Schutten, M.; van Doornum, G.J.; et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 1356–1361. [Google Scholar] [CrossRef] [PubMed]
- Tweed, S.A.; Skowronski, D.M.; David, S.T.; Larder, A.; Petric, M.; Lees, W.; Li, Y.; Katz, J.; Krajden, M.; Tellier, R.; et al. Human illness from avian influenza H7N3, British Columbia. Emerg. Infect. Dis. 2004, 10, 2196–2199. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, H. Global concern regarding the fifth epidemic of human infection with avian influenza A (H7N9) virus in China. Biosci. Trends 2017, 11, 120–121. [Google Scholar] [CrossRef] [PubMed]
- Marshall, N.; Priyamvada, L.; Ende, Z.; Steel, J.; Lowen, A.C. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Govorkova, E.A. Continuing challenges in influenza. Ann. N. Y. Acad. Sci. 2014, 1323, 115–139. [Google Scholar] [CrossRef] [PubMed]
- Ndifon, W.; Wingreen, N.S.; Levin, S.A. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc. Natl. Acad. Sci. USA 2009, 106, 8701–8706. [Google Scholar] [CrossRef] [PubMed]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Basler, C.F.; Crowe, J.E. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 2011, 85, 10905–10908. [Google Scholar] [CrossRef] [PubMed]
- Whittle, J.R.R.; Zhang, R.J.; Khurana, S.; King, L.R.; Manischewitz, J.; Golding, H.; Dormitzer, P.R.; Haynes, B.F.; Walter, E.B.; Moody, M.A.; et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 2011, 108, 14216–14221. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Palese, P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013, 3, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.A.; Friesen, R.H.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody recognition of a highly conserved influenza virus epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Isegawa, Y.; Sasao, F.; Ueda, S. A common neutralizing epitope conserved between the hemagglutinins of influenza—A virus H1 and H2 strains. J. Virol. 1993, 67, 2552–2558. [Google Scholar] [PubMed]
- Friesen, R.H.; Lee, P.S.; Stoop, E.J.; Hoffman, R.M.; Ekiert, D.C.; Bhabha, G.; Yu, W.; Juraszek, J.; Koudstaal, W.; Jongeneelen, M.; et al. A common solution to group 2 influenza virus neutralization. Proc. Natl. Acad. Sci. USA 2014, 111, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef] [PubMed]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.S.; Lee, P.S.; Hoffman, R.M.B.; Mazel-Sanchez, B.; Krammer, F.; Leon, P.E.; Ward, A.B.; Wilson, I.A.; Palese, P. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza A virus hemagglutinin. J. Virol. 2014, 88, 13580–13592. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, B.; Koudstaal, W.; Goudsmit, J.; Klaren, V.; Tang, C.; Bujny, M.V.; Korse, H.J.; Kwaks, T.; Otterstrom, J.J.; Juraszek, J.; et al. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- DiLillo, D.J.; Tan, G.S.; Palese, P.; Ravetch, J.V. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014, 20, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Jegaskanda, S.; Reading, P.C.; Kent, S.J. Influenza-specific antibody-dependent cellular cytotoxicity: Toward a universal influenza vaccine. J. Immunol. 2014, 193, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; Garcia-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [PubMed]
- Valkenburg, S.A.; Mallajosyula, V.V.; Li, O.T.; Chin, A.W.; Carnell, G.; Temperton, N.; Varadarajan, R.; Poon, L.L. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci. Rep. 2016, 6, 22666. [Google Scholar] [CrossRef] [PubMed]
- Mallajosyula, V.V.; Citron, M.; Ferrara, F.; Temperton, N.J.; Liang, X.; Flynn, J.A.; Varadarajan, R. Hemagglutinin sequence conservation guided stem immunogen design from influenza A H3 subtype. Front. Immunol. 2015, 6, 329. [Google Scholar] [CrossRef] [PubMed]
- Klausberger, M.; Tscheliessnig, R.; Neff, S.; Nachbagauer, R.; Wohlbold, T.J.; Wilde, M.; Palmberger, D.; Krammer, F.; Jungbauer, A.; Grabherr, R. Globular head-displayed conserved influenza H1 hemagglutinin stalk epitopes confer protection against heterologous H1N1 virus. PLoS ONE 2016, 11, e0153579. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.; Rao, S.S.; Kong, W.P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Job, E.R.; Deng, Y.M.; Gunalan, V.; Maurer-Stroh, S.; Reading, P.C. Playing hide and seek: How glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014, 6, 1294–1316. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.J.; Chen, S.J.; Jiang, Y.; Huang, K.; Huang, J.; Yang, D.; Zhu, J.J.; Zhu, Y.B.; Shi, S.H.; Peng, D.X.; et al. Hemagglutinin glycosylation modulates the pathogenicity and antigenicity of the H5N1 avian influenza virus. Vet. Microbiol. 2015, 175, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.A.; Stertz, S.; Manicassamy, B.; Zimmermann, P.; Sun, X.; Albrecht, R.A.; Uusi-Kerttula, H.; Zagordi, O.; Belshe, R.B.; Frey, S.E.; et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013, 5, 187ra170. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jayaraman, A.; Maniprasad, P.; Raman, R.; Houser, K.V.; Pappas, C.; Zeng, H.; Sasisekharan, R.; Katz, J.M.; Tumpey, T.M. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J. Virol. 2013, 87, 8756–8766. [Google Scholar] [CrossRef] [PubMed]
- Tate, M.D.; Job, E.R.; Brooks, A.G.; Reading, P.C. Gl ycosylation of the hemagglutinin modulates the sensitivity of H3N2 influenza viruses to innate proteins in airway secretions and virulence in mice. Virology 2011, 413, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Alymova, I.V.; York, I.A.; Air, G.M.; Cipollo, J.F.; Gulati, S.; Baranovich, T.; Kumar, A.; Zeng, H.; Gansebom, S.; McCullers, J.A. Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate receptor binding without affecting virus virulence. Sci. Rep. 2016, 6, 36216. [Google Scholar] [CrossRef] [PubMed]
- Eggink, D.; Goff, P.H.; Palese, P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 2014, 88, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.C.; Garten, W.; Klenk, H.D. Role of conserved glycosylation sites in maturation and transport of influenza A virus hemagglutinin. J. Virol. 1993, 67, 3048–3060. [Google Scholar] [PubMed]
- Daniels, R.; Kurowski, B.; Johnson, A.E.; Hebert, D.N. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol. Cell 2003, 11, 79–90. [Google Scholar] [CrossRef]
- Ohuchi, R.; Ohuchi, M.; Garten, W.; Klenk, H.D. Oligosaccharides in the stem region maintain the influenza virus hemagglutinin in the metastable form required for fusion activity. J. Virol. 1997, 71, 3719–3725. [Google Scholar] [PubMed]
- Nao, N.; Yamagishi, J.; Miyamoto, H.; Igarashi, M.; Manzoor, R.; Ohnuma, A.; Tsuda, Y.; Furuyama, W.; Shigeno, A.; Kajihara, M.; et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Y.P.; Sun, H.L.; Pu, J.; Bi, Y.H.; Shi, Y.; Lu, X.S.; Li, J.; Zhu, Q.Y.; Gao, G.F.; et al. A single amino acid at the hemagglutinin cleavage site contributes to the pathogenicity and neurovirulence of H5N1 influenza virus in mice. J. Virol. 2012, 86, 6924–6931. [Google Scholar] [CrossRef] [PubMed]
- Stech, J.; Garn, H.; Wegmann, M.; Wagner, R.; Klenk, H.D. A new approach to an influenza live vaccine: Modification of the cleavage site of hemagglutinin. Nat. Med. 2005, 11, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, E.; Liang, X.; Ingallinella, P.; Finotto, M.; Chastain, M.A.; Fan, J.; Fu, T.M.; Song, H.C.; Horton, M.S.; Freed, D.C.; et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 2005, 79, 7380–7388. [Google Scholar] [CrossRef] [PubMed]
- Chambers, B.S.; Li, Y.; Hodinka, R.L.; Hensley, S.E. Recent H3N2 influenza virus clinical isolates rapidly acquire hemagglutinin or neuraminidase mutations when propagated for antigenic analyses. J. Virol. 2014, 88, 10986–10989. [Google Scholar] [CrossRef] [PubMed]
- Bradley, K.C.; Galloway, S.E.; Lasanajak, Y.; Song, X.; Heimburg-Molinaro, J.; Yu, H.; Chen, X.; Talekar, G.R.; Smith, D.F.; Cummings, R.D.; et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J. Virol. 2011, 85, 12387–12398. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.L.; Wetzel, K.S.; Linderman, S.L.; Li, Y.; Sullivan, C.B.; Hensley, S.E. Compensatory hemagglutinin mutations alter antigenic properties of influenza viruses. J. Virol. 2013, 87, 11168–11172. [Google Scholar] [CrossRef] [PubMed]
- Air, G.M. Influenza neuraminidase. Influ. Other Respir. Viruses 2012, 6, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Y.; Steukers, L.; Forier, K.; Xiong, R.H.; Braeckmans, K.; van Reeth, K.; Nauwynck, H. A beneficiary role for neuraminidase in influenza virus penetration through the respiratory mucus. PLoS ONE 2014, 9, e110026. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Zhang, X.Q.; Senaati, H.P.; Chen, H.W.; Varki, N.M.; Schooley, R.T.; Gagneux, P. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol. J. 2013, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Zanin, M.; Baviskar, P.; Webster, R.; Webby, R. The Interaction between respiratory pathogens and mucus. Cell Host Microbe 2016, 19, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Rockman, S.; Brown, L.E.; Barr, I.G.; Gilbertson, B.; Lowther, S.; Kachurin, A.; Kachurina, O.; Klippel, J.; Bodle, J.; Pearse, M.; et al. Neuraminidase-inhibiting antibody is a correlate of cross-protection against lethal H5N1 influenza virus in ferrets immunized with seasonal influenza vaccine. J. Virol. 2013, 87, 3053–3061. [Google Scholar] [CrossRef] [PubMed]
- Wohlbold, T.J.; Nachbagauer, R.; Xu, H.M.; Tan, G.S.; Hirsh, A.; Brokstad, K.A.; Cox, R.J.; Palese, P.; Krammer, F. Vaccination with Adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 2015, 6, e02556-14. [Google Scholar] [CrossRef] [PubMed]
- Easterbrook, J.D.; Schwartzman, L.M.; Gao, J.; Kash, J.C.; Morens, D.M.; Couzens, L.; Wan, H.; Eichelberger, M.C.; Taubenberger, J.K. Protection against a lethal H5N1 influenza challenge by intranasal immunization with virus-like particles containing 2009 pandemic H1N1 neuraminidase in mice. Virology 2012, 432, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Akram, A.; Inman, R.D. Immunodominance: A pivotal principle in host response to viral infections. Clin. Immunol. 2012, 143, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.E.; Bucher, D.J.; Kilbourne, E.D. Purified influenza virus hemagglutinin and neuraminidase are equivalent in stimulation of antibody response but induce contrasting types of immunity to infection. J. Virol. 1989, 63, 1239–1246. [Google Scholar] [PubMed]
- Shoji, Y.; Chichester, J.A.; Palmer, G.A.; Farrance, C.E.; Stevens, R.; Stewart, M.; Goldschmidt, L.; Deyde, V.; Gubareva, L.; Klimov, A.; et al. An influenza N1 neuraminidase-specific monoclonal antibody with broad neuraminidase inhibition activity against H5N1 HPAI viruses. Hum. Vaccines 2011, 7, 199–204. [Google Scholar] [CrossRef]
- Wu, C.Y.; Yeh, Y.C.; Chan, J.T.; Yang, Y.C.; Yang, J.R.; Liu, M.T.; Wu, H.S.; Hsiao, P.W. A VLP vaccine induces broad-spectrum cross-protective antibody immunity against H5N1 and H1N1 subtypes of influenza A virus. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Li, X.; Gao, J.; Lin, Z.S.; Jing, X.H.; Plant, E.; Zoueva, O.; Eichelberger, M.C.; Ye, Z.P. Revisiting the 1976 “Swine Flu” vaccine clinical trials: Cross-reactive hemagglutinin and neuraminidase antibodies and their role in protection against the 2009 H1N1 pandemic virus in mice. Clin. Infect. Dis. 2011, 53, 1179–1187. [Google Scholar] [CrossRef] [PubMed]
- Blumenkrantz, D.; Roberts, K.L.; Shelton, H.; Lycett, S.; Barclay, W.S. The short stalk length of highly pathogenic avian influenza H5N1 virus neuraminidase limits transmission of pandemic H1N1 virus in ferrets. J. Virol. 2013, 87, 10539–10551. [Google Scholar] [CrossRef] [PubMed]
- Stech, O.; Veits, J.; Abdelwhab, E.S.M.; Wessels, U.; Mettenleiter, T.C.; Stech, J. The neuraminidase stalk deletion serves as major virulence determinant of H5N1 highly pathogenic avian influenza viruses in chicken. Sci. Rep. 2015, 5, 13493. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Chen, S.J.; Zhang, X.J.; Fu, Q.; Zhang, Z.Y.; Shi, S.H.; Zhu, Y.B.; Gu, M.; Peng, D.X.; Liu, X.F. A 20-amino-acid deletion in the neuraminidase stalk and a five-amino-acid deletion in the NS1 protein both contribute to the pathogenicity of H5N1 avian influenza viruses in mallard ducks. PLoS ONE 2014, 9, e95539. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Lin, C.W.; Tsai, T.I.; Lee, C.D.; Chuang, H.Y.; Chen, J.B.; Tsai, M.H.; Chen, B.R.; Lo, P.W.; Liu, C.P.; et al. Influenza A surface glycosylation and vaccine design. Proc. Natl. Acad. Sci. USA 2017, 114, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Zanin, M.; Duan, S.; Wong, S.S.; Kumar, G.; Baviskar, P.; Collin, E.; Russell, C.; Barman, S.; Hause, B.; Webby, R. An amino acid in the stalk domain of N1 neuraminidase is critical for enzymatic activity. J. Virol. 2017, 91, e00868. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Hashem, A.M.; Li, C.G.; Van Domselaar, G.; Larocque, L.; Wang, J.Z.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.T.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antivir. Res. 2013, 100, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Li, C.; Bucher, D.J.; Hashem, A.M.; van Domselaar, G.; Wang, J.; Farnsworth, A.; She, Y.M.; Cyr, T.; He, R.; et al. A monoclonal antibody targeting a highly conserved epitope in influenza B neuraminidase provides protection against drug resistant strains. Biochem. Biophys. Res. Commun. 2013, 441, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Zebedee, S.L.; Richardson, C.D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 1985, 40, 627–633. [Google Scholar] [CrossRef]
- Roberts, K.L.; Leser, G.P.; Ma, C.L.; Lamb, R.A. The amphipathic helix of influenza A virus M2 protein is required for filamentous bud formation and scission of filamentous and spherical particles. J. Virol. 2013, 87, 9973–9982. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Kim, K.H.; Ko, E.J.; Lee, Y.N.; Kim, M.C.; Kwon, Y.M.; Tang, Y.; Cho, M.K.; Lee, Y.J.; Kang, S.M. New vaccines against influenza virus. Clin. Exp. Vaccine Res. 2014, 3, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Cady, S.D.; Schmidt-Rohr, K.; Wang, J.; Soto, C.S.; Degrado, W.F.; Hong, M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 2010, 463, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Furuse, Y.; Suzuki, A.; Oshitani, H. Large-scale sequence analysis of M gene of influenza A viruses from different species: Mechanisms for emergence and spread of amantadine resistance. Antimicrob. Agents Chemother. 2009, 53, 4457–4463. [Google Scholar] [CrossRef] [PubMed]
- Krumbholz, A.; Schmidtke, M.; Bergmann, S.; Motzke, S.; Bauer, K.; Stech, J.; Durrwald, R.; Wutzler, P.; Zell, R. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J. Gen. Virol. 2009, 90, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, Y.; Ma, C.; Fiorin, G.; Wang, J.; Pinto, L.H.; Lamb, R.A.; Klein, M.L.; Degrado, W.F. Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus. Proc. Natl. Acad. Sci. USA 2013, 110, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Balgi, A.D.; Wang, J.; Cheng, D.Y.H.; Ma, C.L.; Pfeifer, T.A.; Shimizu, Y.; Anderson, H.J.; Pinto, L.H.; Lamb, R.A.; DeGrado, W.F.; et al. Inhibitors of the influenza A virus M2 proton channel discovered using a high-throughput yeast growth restoration assay. PLoS ONE 2013, 8, e5527. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, C.; DeGrado, W.F.; Wang, J. Discovery of highly potent inhibitors targeting the predominant drug-resistant S31N mutant of the influenza A virus M2 proton channel. J. Med. Chem. 2016, 59, 1207–1216. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhang, J.; Wang, J. Pharmacological characterization of the spectrum of antiviral activity and genetic barrier to drug resistance of M2-S31N channel blockers. Mol. Pharmacol. 2016, 90, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Kawaoka, Y.; Krauss, S.; Webster, R.G. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 1989, 63, 4603–4608. [Google Scholar] [PubMed]
- Danzy, S.; Studdard, L.R.; Manicassamy, B.; Solorzano, A.; Marshall, N.; Garcia-Sastre, A.; Steel, J.; Lowen, A.C. Mutations to PB2 and NP proteins of an avian influenza virus combine to confer efficient growth in primary human respiratory cells. J. Virol. 2014, 88, 13436–13446. [Google Scholar] [CrossRef] [PubMed]
- Subbarao, E.K.; London, W.; Murphy, B.R. A single amino-acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 1993, 67, 1761–1764. [Google Scholar] [PubMed]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef] [PubMed]
- Arai, Y.; Kawashita, N.; Daidoji, T.; Ibrahim, M.S.; El-Gendy, E.M.; Takagi, T.; Takahashi, K.; Suzuki, Y.; Ikuta, K.; Nakaya, T.; et al. Novel polymerase gene mutations for human adaptation in clinical isolates of avian H5N1 influenza viruses. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Czudai-Matwich, V.; Otte, A.; Matrosovich, M.; Gabriel, G.; Klenka, H.D. PB2 mutations D701N and S714R promote adaptation of an influenza H5N1 virus to a mammalian host. J. Virol. 2014, 88, 8735–8742. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Li, L.; Yan, Z.; Gan, T.; Li, L.; Chen, R.; Chen, R.; Zheng, Z.; Hong, W.; Wang, J.; et al. Dual E627K and D701N mutations in the PB2 protein of A(H7N9) influenza virus increased its virulence in mammalian models. Sci. Rep. 2015, 5, 14170. [Google Scholar] [CrossRef] [PubMed]
- Taft, A.S.; Ozawa, M.; Fitch, A.; Depasse, J.V.; Halfmann, P.J.; Hill-Batorski, L.; Hatta, M.; Friedrich, T.C.; Lopes, T.J.; Maher, E.A.; et al. Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat. Commun. 2015, 6, 7491. [Google Scholar] [CrossRef] [PubMed]
- Mehle, A.; Dugan, V.G.; Taubenberger, J.K.; Doudna, J.A. Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 2012, 86, 1750–1757. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Feng, H.; Xu, J.; Zhao, D.; Shi, J.; Li, Y.; Deng, G.; Jiang, Y.; Li, X.; Zhu, P.; et al. The PA protein directly contributes to the virulence of H5N1 avian influenza viruses in domestic ducks. J. Virol. 2011, 85, 2180–2188. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.F.; Hatta, M.; Kim, J.H.; Le, M.Q.; Neumann, G.; Kawaoka, Y. Amino acid changes in the influenza A virus PA protein that attenuate avian H5N1 viruses in mammals. J. Virol. 2014, 88, 13737–13746. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.; Sausset, A.; Sedano, L.; Da Costa, B.; Le Goffic, R.; Delmas, B. Codon deletions in the influenza A virus PA Gene generate temperature-sensitive viruses. J. Virol. 2016, 90, 3684–3693. [Google Scholar] [CrossRef] [PubMed]
- Selman, M.; Dankar, S.K.; Forbes, N.E.; Jia, J.J.; Brown, E.G. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 2012, 1, e42. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nogales, A.; Lambert-Emo, K.; Martinez-Sobrido, L.; Topham, D.J. NS1 protein mutation I64T affects interferon responses and virulence of circulating H3N2 human influenza A viruses. J. Virol. 2016, 90, 9693–9711. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Jiao, P.; Wang, A.; Zhao, F.; Tian, G.; Wang, X.; Yu, K.; Bu, Z.; Chen, H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 2006, 80, 11115–11123. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Hossain, M.J.; Hickman, D.; Perez, D.R.; Lamb, R.A. A new influenza virus virulence determinant: The NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 4381–4386. [Google Scholar] [CrossRef] [PubMed]
- Dankar, S.K.; Wang, S.; Ping, J.; Forbes, N.E.; Keleta, L.; Li, Y.; Brown, E.G. Influenza A virus NS1 gene mutations F103L and M106I increase replication and virulence. Virol. J. 2011, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. NS1 protein amino acid changes D189N and V194I affect interferon responses, thermosensitivity, and virulence of circulating H3N2 human influenza A viruses. J. Virol. 2017, 91, e01930. [Google Scholar] [CrossRef] [PubMed]
- Pica, N.; Langlois, R.A.; Krammer, F.; Margine, I.; Palese, P. NS1-truncated live attenuated virus vaccine provides robust protection to aged mice from viral challenge. J. Virol. 2012, 86, 10293–10301. [Google Scholar] [CrossRef] [PubMed]
- Hai, R.; Martinez-Sobrido, L.; Fraser, K.A.; Ayllon, J.; Garcia-Sastre, A.; Palese, P. Influenza B virus NS1-truncated mutants: Live-attenuated vaccine approach. J. Virol. 2008, 82, 10580–10590. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Chen, S.; Han, W.; Wu, B.; Zhang, X.; Tang, Y.; Wang, X.; Zhu, Y.; Peng, D.; Liu, X. Cross-clade protective immune responses of NS1-truncated live attenuated H5N1 avian influenza vaccines. Vaccine 2016, 34, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Tosh, P.K.; Jacobson, R.M.; Poland, G.A. Influenza vaccines: From surveillance through production to protection. Mayo Clin. Proc. 2010, 85, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Ninomiya, A.; Ogasawara, K.; Kajino, K.; Takada, A.; Kida, H. Intranasal administration of a synthetic peptide vaccine encapsulated in liposome together with an anti-CD40 antibody induces protective immunity against influenza A virus in mice. Vaccine 2002, 20, 3123–3129. [Google Scholar] [CrossRef]
- Taneichi, M.; Tanaka, Y.; Kakiuchi, T.; Uchida, T. Liposome-coupled peptides induce long-lived memory CD8 T cells without CD4 T cells. PLoS ONE 2010, 5, e15091. [Google Scholar] [CrossRef] [PubMed]
- Day, E.B.; Zeng, W.G.; Doherty, P.C.; Jackson, D.C.; Kedzierska, K.; Turner, S.J. The context of epitope presentation can influence functional quality of recalled influenza A virus-specific memory CD8+ T cells. J. Immunol. 2007, 179, 2187–2194. [Google Scholar] [CrossRef] [PubMed]
- Ichihashi, T.; Satoh, T.; Sugimoto, C.; Kajino, K. Emulsified phosphatidylserine, simple and effective peptide carrier for induction of potent epitope-specific T cell responses. PLoS ONE 2013, 8, e60068. [Google Scholar] [CrossRef] [PubMed]
- Soema, P.C.; Huber, S.K.R.; Willems, G.J.; Jiskoot, W.; Kersten, G.F.A.; Amorij, J.P. Influenza T-cell epitope-loaded virosomes adjuvanted with CpG as a potential influenza vaccine. Pharm. Res. 2015, 32, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Atsmon, J.; Caraco, Y.; Ziv-Sefer, S.; Shaikevich, D.; Abramov, E.; Volokhov, I.; Bruzil, S.; Haima, K.Y.; Gottlieb, T.; Ben-Yedidia, T. Priming by a novel universal influenza vaccine (Multimeric-001)-A gateway for improving immune response in the elderly population. Vaccine 2014, 32, 5816–5823. [Google Scholar] [CrossRef] [PubMed]
- Francis, J.N.; Bunce, C.J.; Horlock, C.; Watson, J.M.; Warrington, S.J.; Georges, B.; Brown, C.B. A novel peptide-based pan-influenza A vaccine: A double blind, randomised clinical trial of immunogenicity and safety. Vaccine 2015, 33, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.H.; Yang, F.R.; Yu, H.; Zhou, Y.J.; Li, G.X.; Huang, M.; Wen, F.; Tong, G. An M2e-based synthetic peptide vaccine for influenza A virus confers heterosubtypic protection from lethal virus challenge. Virol. J. 2013, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Clausen, B.E.; Stoitzner, P. Functional specialization of skin dendritic cell subsets in regulating T cell responses. Front. Immunol. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed]
- Heath, W.R.; Carbone, F.R. The skin-resident and migratory immune system in steady state and memory: Innate lymphocytes, dendritic cells and T cells. Nat. Immunol. 2013, 14, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.P.; Koutsonanos, D.G.; Del Pilar Martin, M.; Lee, J.W.; Zarnitsyn, V.; Choi, S.O.; Murthy, N.; Compans, R.W.; Skountzou, I.; Prausnitz, M.R. Dissolving polymer microneedle patches for influenza vaccination. Nat. Med. 2010, 16, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Z.; Gill, H.S.; He, C.; Ou, C.; Wang, L.; Wang, Y.C.; Feng, H.; Zhang, H.; Prausnitz, M.R.; Compans, R.W. Microneedle delivery of an M2e–TLR5 ligand fusion protein to skin confers broadly cross-protective influenza immunity. J. Controll. Release 2014, 178, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Esser, E.S.; Romanyuk, A.; Vassilieva, E.V.; Jacob, J.; Prausnitz, M.R.; Compans, R.W.; Skountzou, I. Tetanus vaccination with a dissolving microneedle patch confers protective immune responses in pregnancy. J. Controll. Release 2016, 236, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Pewin, W.; Wang, C.; Luo, Y.; Gonzalez, G.X.; Mohan, T.; Prausnitz, M.R.; Wang, B.Z. A boosting skin vaccination with dissolving microneedle patch encapsulating M2e vaccine broadens the protective efficacy of conventional influenza vaccines. J. Controll. Release 2017. [Google Scholar] [CrossRef] [PubMed]

| Influenza Viral Protein | Functions | Mutation Sites | Conservation Sites | Applications in Vaccines |
|---|---|---|---|---|
| Hemagglutinin (HA) | Receptor binding, Membrane fusion | Receptor binding site, Glycosylation site, Proteolytic cleavage site | Stalk domain | HA stalk based vaccines, HA head COBRAs * Live attenuated vaccine strains |
| Neuraminidase (NA) | Virus releasing, Prevention aggregation, Penetration through mucus layer | Deletion in stalk domain, Glycosylation in stalk domain, Surface loops surrounding the enzyme active site | Enzymatic active site | Induction of NA immunity, Conserved epitopes in enzymatic site |
| Matrix 2 (M2) | Ion channel protein, Viral uncoating, Maintaining HA configuration, Virion budding and scission | Amantadine-resistant mutations V27A, S31N, and L26F | Ectodomain, An amphipathic helix in cytoplasmic tail | M2e ectodomain based vaccines |
| Viral ribonucleoprotein complex (vRNP) | NP-single strand RNA binding protein | NP-309K, 50G Temperature sensitive mutations | Live attenuated vaccine strains Conserved peptides in NP, PA and PB | |
| PB1-RNA dependent RNA polymerase | PB1-105S | |||
| PB2-binding host mRNA caps | PB2-627K, 701N, 591K | |||
| PA-essential for polymerase function | PA-552S, 224P, 383D | |||
| Nonstructural 1 protein (NS1) | RNA binding, Type I interferon antagonism, Enhancing viral RNA translation, Inhibition of host mRNA processing | NS1-S42P, D92E, V149A NS1-103L, 106I | Live attenuated vaccine strains |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.; Wang, C.; Wang, B.-Z. From Variation of Influenza Viral Proteins to Vaccine Development. Int. J. Mol. Sci. 2017, 18, 1554. https://doi.org/10.3390/ijms18071554
Zhu W, Wang C, Wang B-Z. From Variation of Influenza Viral Proteins to Vaccine Development. International Journal of Molecular Sciences. 2017; 18(7):1554. https://doi.org/10.3390/ijms18071554
Chicago/Turabian StyleZhu, Wandi, Chao Wang, and Bao-Zhong Wang. 2017. "From Variation of Influenza Viral Proteins to Vaccine Development" International Journal of Molecular Sciences 18, no. 7: 1554. https://doi.org/10.3390/ijms18071554
APA StyleZhu, W., Wang, C., & Wang, B.-Z. (2017). From Variation of Influenza Viral Proteins to Vaccine Development. International Journal of Molecular Sciences, 18(7), 1554. https://doi.org/10.3390/ijms18071554