The Role of Focal Adhesion Kinase in Keratinocyte Fibrogenic Gene Expression
Abstract
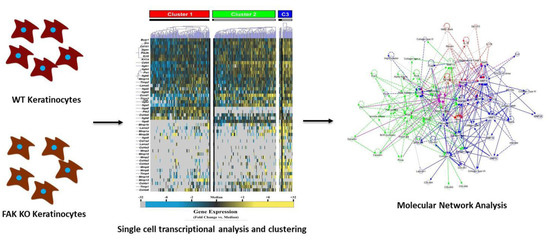
:1. Introduction
2. Results
2.1. Knockout of Keratinocyte FAK (Focal Adhesion Kinase) Alters the Expression of Numerous Genes Integral to Tissue Repair
2.2. FAK-Deleted Keratinocytes Demonstrate Alterations in Key Mechanotransduction and Collagen Signaling Pathways
2.3. FAK Deletion Affects Keratinocyte Gene Expression Asymmetrically and Induces a Transcriptionally Activated Subpopulation
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Keratinocyte Harvest and Culture
4.3. Single-Cell Transcriptional Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gurtner, G.C.; Werner, S.; Barrandon, Y.; Longaker, M.T. Wound repair and regeneration. Nature 2008, 453, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Horch, R.E.; Popescu, L.M.; Polykandriotis, E. History of Regenerative Medicine. In Regenerative Medicine, 1st ed.; Steinhoff, G., Ed.; Springer: Dordrecht, The Netherlands, 2011; Volume 1, p. 1. [Google Scholar]
- Walmsley, G.G.; Maan, Z.N.; Wong, V.W.; Duscher, D.; Hu, M.S.; Zielins, E.R.; Wearda, T.; Muhonen, E.; McArdle, A.; Tevlin, R.; et al. Scarless wound healing: Chasing the holy grail. Plast. Reconstr. Surg. 2015, 135, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Zielins, E.R.; Atashroo, D.A.; Maan, Z.N.; Duscher, D.; Walmsley, G.G.; Hu, M.; Senarath-Yapa, K.; McArdle, A.; Tevlin, R.; Wearda, T.; et al. Wound healing: An update. Regen. Med. 2014, 9, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Demidova-Rice, T.N.; Hamblin, M.R.; Herman, I.M. Acute and impaired wound healing: Pathophysiology and current methods for drug delivery, part 1: Normal and chronic wounds: Biology, causes, and approaches to care. Adv. Skin Wound Care 2012, 25, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen. 2009, 17, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Van De Water, L.; Varney, S.; Tomasek, J.J. Mechanoregulation of the myofibroblast in wound contraction, scarring, and fibrosis: Opportunities for new therapeutic intervention. Adv. Wound Care 2013, 2, 122–141. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Cellular mechanotransduction: Putting all the pieces together again. FASEB J. 2006, 20, 811–827. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Duscher, D.; Maan, Z.N.; Wong, V.W.; Rennert, R.C.; Januszyk, M.; Rodrigues, M.; Hu, M.; Whitmore, A.J.; Whittam, A.J.; Longaker, M.T.; et al. Mechanotransduction and fibrosis. J. Biomech. 2014, 47, 1997–2005. [Google Scholar] [CrossRef] [PubMed]
- Edlich, R.F.; Carl, B.A. Predicting scar formation: From ritual practice (langer’s lines) to scientific discipline (static and dynamic skin tensions). J. Emerg. Med. 1998, 16, 759–760. [Google Scholar] [PubMed]
- Evans, N.D.; Oreffo, R.O.; Healy, E.; Thurner, P.J.; Man, Y.H. Epithelial mechanobiology, skin wound healing, and the stem cell niche. J. Mech. Behav. Biomed. Mater. 2013, 28, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.Q.; Zhang, X.L.; Liu, L.; Yang, C.C. Relationship between focal adhesion kinase and hepatic stellate cell proliferation during rat hepatic fibrogenesis. World J. Gastroenterol. 2004, 10, 3001–3005. [Google Scholar] [CrossRef] [PubMed]
- Clemente, C.F.; Tornatore, T.F.; Theizen, T.H.; Deckmann, A.C.; Pereira, T.C.; Lopes-Cendes, I.; Souza, J.R.; Franchini, K.G. Targeting focal adhesion kinase with small interfering RNA prevents and reverses load-induced cardiac hypertrophy in mice. Circ. Res. 2007, 101, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Morla, A.O.; Mogford, J.E. Control of smooth muscle cell proliferation and phenotype by integrin signaling through focal adhesion kinase. Biochem. Biophys. Res. Commun. 2000, 272, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Thannickal, V.J. Protein kinase inhibitors in the treatment of pulmonary fibrosis. Curr. Med. Chem. 2008, 15, 2632–2640. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Ouyang, G.; Bao, S. The activation of akt/pkb signaling pathway and cell survival. J. Cell. Mol. Med. 2005, 9, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.R.; Toker, A. Function of akt/pkb signaling to cell motility, invasion and the tumor stroma in cancer. Cell. Signal. 2009, 21, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Li, W.; Hoshino, Y.; Kadohama, T.; Asada, H.; Ohgi, S.; Sumpio, B.E. Role of akt in cyclic strain-induced endothelial cell proliferation and survival. Am. J. Physiol. Cell Physiol. 2006, 290, C812–C821. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.J.; Pedraza, L.G.; Xu, T. Akt regulates growth by directly phosphorylating tsc2. Nat. Cell Biol. 2002, 4, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Lessan, K.; Kahm, J.; Kleidon, J.; Henke, C. Beta 1 integrin regulates fibroblast viability during collagen matrix contraction through a phosphatidylinositol 3-kinase/AKT/protein kinase b signaling pathway. J. Biol. Chem. 2002, 277, 24667–24675. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Gladson, C.L.; Wu, H.; Hayasaka, H.; Olman, M.A. Focal adhesion kinase (FAK)-related non-kinase inhibits myofibroblast differentiation through differential mapk activation in a FAK-dependent manner. J. Biol. Chem. 2008, 283, 26839–26849. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, T.; Wu, M.H.; Pierce, A.; Poncelet, A.C.; Varga, J.; Schnaper, H.W. Map-kinase activity necessary for tgfbeta1-stimulated mesangial cell type I collagen expression requires adhesion-dependent phosphorylation of FAK tyrosine 397. J. Cell Sci. 2007, 120, 4230–4240. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Rustad, K.C.; Akaishi, S.; Sorkin, M.; Glotzbach, J.P.; Januszyk, M.; Nelson, E.R.; Levi, K.; Paterno, J.; Vial, I.N.; et al. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat. Med. 2012, 18, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Garg, R.K.; Sorkin, M.; Rustad, K.C.; Akaishi, S.; Levi, K.; Nelson, E.R.; Tran, M.; Rennert, R.; Liu, W.; et al. Loss of keratinocyte focal adhesion kinase stimulates dermal proteolysis through upregulation of mmp9 in wound healing. Ann. Surg. 2014, 260, 1138–1146. [Google Scholar] [CrossRef] [PubMed]
- Bernard, F.X.; Pedretti, N.; Rosdy, M.; Deguercy, A. Comparison of gene expression profiles in human keratinocyte mono-layer cultures, reconstituted epidermis and normal human skin; transcriptional effects of retinoid treatments in reconstituted human epidermis. Exp. Dermatol. 2002, 11, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.S.; Januszyk, M.; Hong, W.X.; Walmsley, G.G.; Zielins, E.R.; Atashroo, D.A.; Maan, Z.N.; McArdle, A.; Takanishi, D.M., Jr.; Gurtner, G.C.; et al. Gene expression in fetal murine keratinocytes and fibroblasts. J. Surg. Res. 2014, 190, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Januszyk, M.; Gurtner, G.C. High-throughput single-cell analysis for wound healing applications. Adv. Wound Care 2013, 2, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Glotzbach, J.P.; Januszyk, M.; Vial, I.N.; Wong, V.W.; Gelbard, A.; Kalisky, T.; Thangarajah, H.; Longaker, M.T.; Quake, S.R.; Chu, G.; et al. An information theoretic, microfluidic-based single cell analysis permits identification of subpopulations among putatively homogeneous stem cells. PLoS ONE 2011, 6, e21211. [Google Scholar] [CrossRef] [PubMed]
- Varkey, M.; Ding, J.; Tredget, E.E. Fibrotic remodeling of tissue-engineered skin with deep dermal fibroblasts is reduced by keratinocytes. Tissue Eng. Part A 2014, 20, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Kippenberger, S.; Loitsch, S.; Guschel, M.; Muller, J.; Knies, Y.; Kaufmann, R.; Bernd, A. Mechanical stretch stimulates protein kinase b/akt phosphorylation in epidermal cells via angiotensin ii type 1 receptor and epidermal growth factor receptor. J. Biol. Chem. 2005, 280, 3060–3067. [Google Scholar] [CrossRef] [PubMed]
- Kippenberger, S.; Loitsch, S.; Guschel, M.; Muller, J.; Kaufmann, R.; Bernd, A. Hypotonic stress induces e-cadherin expression in cultured human keratinocytes. FEBS Lett. 2005, 579, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Knies, Y.; Bernd, A.; Kaufmann, R.; Bereiter-Hahn, J.; Kippenberger, S. Mechanical stretch induces clustering of beta1-integrins and facilitates adhesion. Exp. Dermatol. 2006, 15, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Lavker, R.M.; Sun, T.T. Heterogeneity in epidermal basal keratinocytes: Morphological and functional correlations. Science 1982, 215, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Haustead, D.J.; Stevenson, A.; Saxena, V.; Marriage, F.; Firth, M.; Silla, R.; Martin, L.; Adcroft, K.F.; Rea, S.; Day, P.J.; et al. Transcriptome analysis of human ageing in male skin shows mid-life period of variability and central role of nf-kappab. Sci. Rep. 2016, 6, 26846. [Google Scholar] [CrossRef] [PubMed]
- Shirakihara, T.; Kawasaki, T.; Fukagawa, A.; Semba, K.; Sakai, R.; Miyazono, K.; Miyazawa, K.; Saitoh, M. Identification of integrin α3 as a molecular marker of cells undergoing epithelial-mesenchymal transition and of cancer cells with aggressive phenotypes. Cancer Sci. 2013, 104, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Longaker, M.T.; Gurtner, G.C. Soft tissue mechanotransduction in wound healing and fibrosis. Semin. Cell Dev. Biol. 2012, 23, 981–986. [Google Scholar] [CrossRef] [PubMed]
- Young, S.R.; Gerard-O’Riley, R.; Kim, J.B.; Pavalko, F.M. Focal adhesion kinase is important for fluid shear stress-induced mechanotransduction in osteoblasts. J. Bone Miner. Res. 2009, 24, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, L.S.; Marsh, H.M.; Basson, M.D. Src and focal adhesion kinase mediate mechanical strain-induced proliferation and erk1/2 phosphorylation in human h441 pulmonary epithelial cells. Am. J. Physiol. Cell Physiol. 2007, 292, C1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, A.P.; Clemente, C.F.; Carvalho, H.F.; Carvalheira, J.B.; Nadruz, W., Jr.; Franchini, K.G. Fak mediates the activation of cardiac fibroblasts induced by mechanical stress through regulation of the mtor complex. Cardiovasc. Res. 2010, 86, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Zebda, N.; Dubrovskyi, O.; Birukov, K.G. Focal adhesion kinase regulation of mechanotransduction and its impact on endothelial cell functions. Microvasc. Res. 2012, 83, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.; Akaishi, S.; Longaker, M.T.; Gurtner, G.C. Pushing back: Wound mechanotransduction in repair and regeneration. J. Investig. Dermatol. 2011, 131, 2186–2196. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Leavitt, T.; Bayer, L.R.; Orgill, D.P. Effect of negative pressure wound therapy on wound healing. Curr. Probl. Surg. 2014, 51, 301–331. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.E.; Fu, W.; Schaller, M.D. Focal adhesion kinase: Exploring fak structure to gain insight into function. Int. Rev. Cell Mol. Biol. 2011, 288, 185–225. [Google Scholar] [PubMed]
- Jahed, Z.; Shams, H.; Mehrbod, M.; Mofrad, M.R. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int. Rev. Cell Mol. Biol. 2014, 310, 171–220. [Google Scholar] [PubMed]
- Margadant, C.; Sonnenberg, A. Integrin-tgf-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010, 11, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Golbert, D.C.; Correa-de-Santana, E.; Ribeiro-Alves, M.; de Vasconcelos, A.T.; Savino, W. Itga6 gene silencing by rna interference modulates the expression of a large number of cell migration-related genes in human thymic epithelial cells. BMC Genom. 2013, 14 (Suppl. S6), S3. [Google Scholar]
- Muller, U.; Wang, D.; Denda, S.; Meneses, J.J.; Pedersen, R.A.; Reichardt, L.F. Integrin α8β1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell 1997, 88, 603–613. [Google Scholar] [CrossRef]
- Bieritz, B.; Spessotto, P.; Colombatti, A.; Jahn, A.; Prols, F.; Hartner, A. Role of α8 integrin in mesangial cell adhesion, migration, and proliferation. Kidney Int. 2003, 64, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Bedal, K.B.; Grassel, S.; Oefner, P.J.; Reinders, J.; Reichert, T.E.; Bauer, R. Collagen xvi induces expression of mmp9 via modulation of ap-1 transcription factors and facilitates invasion of oral squamous cell carcinoma. PLoS ONE 2014, 9, e86777. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Tang, Y.A.; Huang, S.M.; Juan, H.F.; Wu, L.W.; Sun, Y.C.; Wang, S.C.; Wu, K.W.; Balraj, G.; Chang, T.T.; et al. A novel sialyltransferase inhibitor suppresses fak/paxillin signaling and cancer angiogenesis and metastasis pathways. Cancer Res. 2011, 71, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Lichti, U.; Anders, J.; Yuspa, S.H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 2008, 3, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Rennert, R.C.; Januszyk, M.; Sorkin, M.; Rodrigues, M.; Maan, Z.N.; Duscher, D.; Whittam, A.J.; Kosaraju, R.; Chung, M.T.; Paik, K.; et al. Microfluidic single-cell transcriptional analysis rationally identifies novel surface marker profiles to enhance cell-based therapies. Nat. Commun. 2016, 7, 11945. [Google Scholar] [CrossRef] [PubMed]
- Januszyk, M.; Sorkin, M.; Glotzbach, J.P.; Vial, I.N.; Maan, Z.N.; Rennert, R.C.; Duscher, D.; Thangarajah, H.; Longaker, M.T.; Butte, A.J.; et al. Diabetes irreversibly depletes bone marrow-derived mesenchymal progenitor cell subpopulations. Diabetes 2014, 63, 3047–3056. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Januszyk, M.; Kwon, S.H.; Wong, V.W.; Padmanabhan, J.; Maan, Z.N.; Whittam, A.J.; Major, M.R.; Gurtner, G.C. The Role of Focal Adhesion Kinase in Keratinocyte Fibrogenic Gene Expression. Int. J. Mol. Sci. 2017, 18, 1915. https://doi.org/10.3390/ijms18091915
Januszyk M, Kwon SH, Wong VW, Padmanabhan J, Maan ZN, Whittam AJ, Major MR, Gurtner GC. The Role of Focal Adhesion Kinase in Keratinocyte Fibrogenic Gene Expression. International Journal of Molecular Sciences. 2017; 18(9):1915. https://doi.org/10.3390/ijms18091915
Chicago/Turabian StyleJanuszyk, Michael, Sun Hyung Kwon, Victor W. Wong, Jagannath Padmanabhan, Zeshaan N. Maan, Alexander J. Whittam, Melanie R. Major, and Geoffrey C. Gurtner. 2017. "The Role of Focal Adhesion Kinase in Keratinocyte Fibrogenic Gene Expression" International Journal of Molecular Sciences 18, no. 9: 1915. https://doi.org/10.3390/ijms18091915
APA StyleJanuszyk, M., Kwon, S. H., Wong, V. W., Padmanabhan, J., Maan, Z. N., Whittam, A. J., Major, M. R., & Gurtner, G. C. (2017). The Role of Focal Adhesion Kinase in Keratinocyte Fibrogenic Gene Expression. International Journal of Molecular Sciences, 18(9), 1915. https://doi.org/10.3390/ijms18091915