MDG-1, a Potential Regulator of PPARα and PPARγ, Ameliorates Dyslipidemia in Mice
Abstract
:1. Introduction
2. Results
2.1. MDG-1 Blocks Obesity in DIO Mice
2.2. MDG-1 Attenuates Dyslipidaemia in DIO Mice
2.3. MDG-1 Improves Glucose Tolerance and Insulin Resistance in Obese Mice
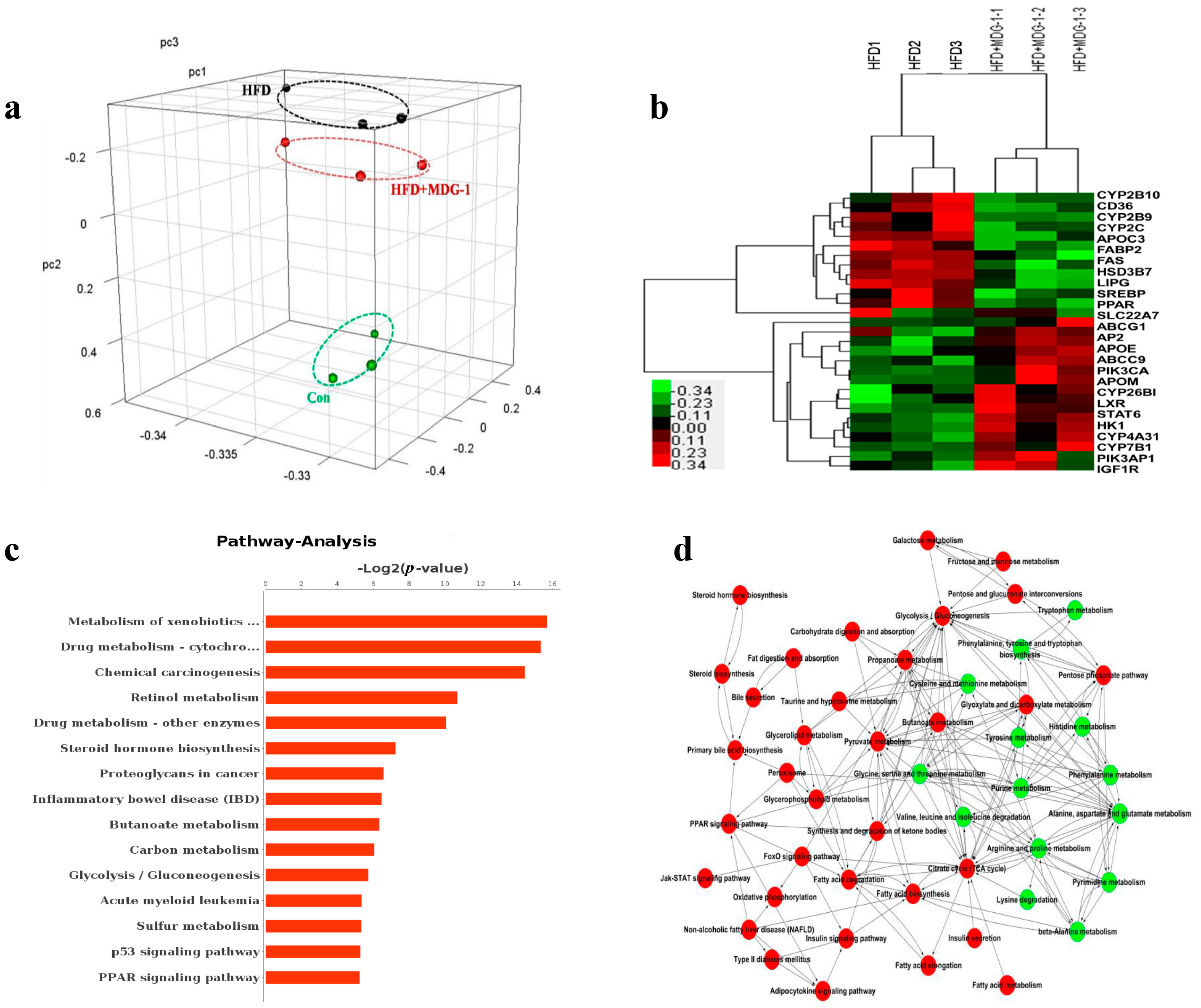
2.4. MDG-1 Effects the Expression of Lipid Genes in HFD-Fed Mice by Gene Chip Technology
2.5. MDG-1 Regulates Gene Expression of PPARs, LXR, and the Target Genes
3. Discussion
4. Materials and Methods
4.1. Chemical and Diet
4.2. Animals and Treatment
4.3. Serum Chemistry Analysis
4.4. Liver and Fecal Lipid Contents Analysis
4.5. Hematoxylin and Eosin (H&E) Staining and Scanning Electron Microscopy
4.6. Glucose Tolerance and Insulin Tolerance Tests
4.7. Gene Array Experiment
4.7.1. RNA Preparation and cDNA Generation
4.7.2. Gene Array Experiment and Gene Array Data Analysis
4.8. Quantitative Real-Time PCR Analysis of Animal Tissues
4.9. Stastical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| HFD | High-fat diet |
| DIO | Diet-induced obese |
| TC | Total cholesterol |
| TG | Total triglyceride |
| HDL-c | High-density lipoprotein cholesterol |
| LDL-c | Low-density lipoprotein cholesterol |
| PPARs | Peroxisome proliferator-activated receptors |
| SCFAs | Short-chain fatty acids |
| qPCR | Quantitative real-time PCR |
| H&E staining | Hematoxylin and eosin staining |
References
- Bourbon, M.; Alves, A.C.; Alonso, R.; Mata, N.; Aguiar, P.; Padro, T.; Mata, P. Mutational analysis and genotype-phenotype relation in familial hypercholesterolemia: The SAFEHEART registry. Atherosclerosis 2017, 262, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Iacocca, M.A.; Hegele, R.A. Recent advances in genetic testing for familial hypercholesterolemia. Expert. Rev. Mol. Diagn. 2017, 17, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Jian, D.Y.; Lin, M.W.; Zhao, J.Z.; Ho, L.T.; Juan, C.C. Evidence in obese children: Contribution of hyperlipidemia, obesity-inflammation, and insulin sensitivity. PLoS ONE 2015, 10, e0125935. [Google Scholar] [CrossRef] [PubMed]
- Navar-Boggan, A.M.; Peterson, E.D.; D’agostino, R.B.; Neely, B.; Sniderman, A.D.; Pencina, M.J. Hyperlipidemia in early adulthood increases long-term risk of coronary heart disease. Circulation 2015, 131, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.; Bhatnagar, S. Hyperlipidemia, disease associations, and top 10 potential drug targets: A network View. OMICS 2016, 20, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, W.; Liu, X.; Zhang, W.; Li, Y. Interrelationship between diabetes and periodontitis: Role of hyperlipidemia. Arch. Oral. Biol. 2015, 60, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Zhao, Y.; Dong, F.; Yan, Z.; Zheng, W.; Fan, J.; Sun, G. Meta-analysis of the effect and safety of berberine in the treatment of type 2 diabetes mellitus, hyperlipemia and hypertension. J. Ethnopharmacol. 2015, 161, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Parikh, N.H.; Parikh, P.K.; Kothari, C. Indigenous plant medicines for health care: Treatment of diabetes mellitus and hyperlipidemia. Chin. J. Nat. Med. 2014, 12, 335–344. [Google Scholar] [CrossRef]
- Wu, J.B.; Kuo, Y.H.; Lin, C.H.; Ho, H.Y.; Shih, C.C. Tormentic acid, a major component of suspension cells of Eriobotrya japonica, suppresses high-fat diet-induced diabetes and hyperlipidemia by glucose transporter 4 and AMP-activated protein kinase phosphorylation. J. Agric. Food Chem. 2014, 62, 10717–10726. [Google Scholar] [CrossRef] [PubMed]
- Kidani, Y.; Bensinger, S.J. Liver X receptor and peroxisome proliferator-activated receptor as integrators of lipid homeostasis and immunity. Immunol. Rev. 2012, 249, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Grygiel-Gorniak, B. Peroxisome proliferator-activated receptors and their ligands: Nutritional and clinical implications—A review. Nutr. J. 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Mahajan, V.K.; Mehta, K.S.; Chauhan, P.S.; Rawat, R. Peroxisome proliferator-activated receptors (PPARs) and PPAR agonists: The “future” in dermatology therapeutics? Arch. Dermatol. Res. 2015, 307, 767–780. [Google Scholar] [CrossRef] [PubMed]
- Laganà, A.S.; Vitale, S.G.; Nigro, A.; Sofo, V.; Salmeri, F.M.; Rossetti, P.; Rapisarda, A.M.; la Vignera, S.; Condorelli, R.A.; Rizzo, G.; et al. Pleiotropic actions of peroxisome proliferator-activated receptors (PPARs) in dysregulated metabolic homeostasis, inflammation and cancer: Current evidence and future perspectives. Int. J. Mol. Sci. 2016, 17, 999. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M. The roles of peroxisome proliferator-activated receptors in the metabolic syndrome. Prog. Mol. Biol. Transl. Sci. 2014, 121, 217–266. [Google Scholar] [PubMed]
- Fan, S.; Guo, L.; Zhang, Y.; Sun, Q.; Yang, B.; Huang, C. Okra polysaccharide improves metabolic disorders in high-fat diet-induced obese C57BL/6 mice. Mol. Nutr. Food Res. 2013, 57, 2075–2078. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Xia, Y.P.; Chen, W.J.; Yu, M.H.; Li, Y.M.; Ye, H.Y. Improvement of myocardial glycolipid metabolic disorder in diabetic hamster with Astragalus polysaccharides treatment. Mol. Biol. Rep. 2012, 39, 7609–7615. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Dai, X.Y.; Chen, Q.; Zang, J.N.; Deng, L.L.; Liu, Y.H.; Ying, H.Z. Hypolipidemic and antioxidant activities of polysaccharides from Rosae Laevigatae Fructus in rats. Carbohydr. Polym. 2013, 94, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.S.; Feng, Y.; Lin, X.; Deng, H.L.; Fang, J.N.; Dong, Q. Isolation: Purification and structural analysis of a polysaccharide MDG-1 from Ophiopogon japonicas. Acta Pharm. Sin. B 2005, 40, 636–639. [Google Scholar]
- Shi, L.; Wang, J.; Wang, Y.; Feng, Y. MDG-1, an Ophiopogon polysaccharide, alleviates hyperlipidemia in mice based on metabolic profile of bile acids. Carbohydr. Polym. 2016, 150, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, Y.; Ruan, K.; Wei, H.; Feng, Y. MDG-1, a polysaccharide from Ophiopogon japonicus, prevents high fat diet-induced obesity and increases energy expenditure in mice. Carbohydr. Polym. 2014, 114, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, Y.; Xu, D.S.; Ruan, K.F.; Feng, Y.; Wang, S. Hypoglycemic effects of MDG-1, a polysaccharide derived from Ophiopogon japonicas, in the ob/ob mouse model of type 2 diabetes mellitus. Int. J. Biol. Macromol. 2011, 49, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Graham, I.M.; Catapano, A.L.; Wong, N.D. Current guidelines on prevention with a focus on dyslipidemias. Cardiovasc. Diagn. Ther. 2017, 7 (Suppl. S1), S4–S10. [Google Scholar] [CrossRef] [PubMed]
- Han, B.H.; Sutin, D.; Williamson, J.D.; Davis, B.R.; Piller, L.B.; Pervin, H.; Pressel, S.L.; Blaum, C.S. Effect of statin treatment vs usual care on primary cardiovascular prevention among older adults: The ALLHAT-LLT randomized clinical trial. JAMA Intern. Med. 2017, 177, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Cong, W.; Shen, L.; Wei, H.; Wang, Y.; Wang, L.; Ruan, K.; Wu, F.; Feng, Y. Fecal metabonomic study of a polysaccharide, MDG-1 from Ophiopogon japonicus on diabetic mice based on gas chromatography/time-of-flight mass spectrometry (GC TOF/MS). Mol. Biosyst. 2014, 10, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lee, Y.J.; Chung, Y.H.; Nam, Y.; Kim, S.T.; Park, E.S.; Hong, S.M.; Yang, Y.K.; Kim, H.C.; Jeong, J.H. Beneficial effects of red yeast rice on high-fat diet-induced obesity, hyperlipidemia, and fatty liver in mice. J. Med. Food 2015, 18, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.M.; Lopez-Maside, L.; Donapetry-Garcia, C.; Fernandez-Fernandez, C.; Sixto-Leal, C. Enzymes involved in branched-chain amino acid metabolism in humans. Amino Acids 2017, 49, 1005–1028. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Jung, J.; Park, H.S.; Lee, S.M.; Jeong, N.J.; Kim, S.H.; Lee, K.W.; Lee, J.A.; Kim, M.S. Shaofu Zhuyu decoction ameliorates obesity-mediated hepatic steatosis and systemic inflammation by regulating metabolic pathways. PLoS ONE 2017, 12, e0178514. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Zhang, Y.; Fan, S.; Ding, X.; Ji, G.; Huang, C. Extracts of Rhizoma polygonati odorati prevent high-fat diet-induced metabolic disorders in C57BL/6 mice. PLoS ONE 2013, 8, e81724. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Guo, L.; Zhang, Y.; Fan, S.; Gu, M.; Lu, Y.; Jiang, D.; Li, Y.; Huang, C.; Zhou, Z. Extracts of pomelo peels prevent high-fat diet-induced metabolic disorders in C57BL/6 mice through activating the PPARα and GLUT4 pathway. PLoS ONE 2013, 8, e77915. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chu, E.S.; Hui, A.Y.; Cheung, K.F.; Chan, H.L.; Leung, W.K.; Farrell, G.C.; Sung, J.J. Lipoprotein lipase activator ameliorates the severity of dietary steatohepatitis. Biochem. Biophys. Res. Commun. 2007, 356, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Fan, S.; Liu, G.; Guo, L.; Ding, X.; Lu, Y.; Zhang, Y.; Ji, G.; Huang, C. Extract of wax gourd peel prevents high-fat diet-induced hyperlipidemia in C57BL/6 mice via the inhibition of the PPARγ pathway. Evid. Based Complement Alternat. Med. 2013, 2013, 342561. [Google Scholar] [CrossRef] [PubMed]
- Morán-Salvador, E.; López-Parra, M.; García-Alonso, V.; Titos, E.; Martínez-Clemente, M.; González-Périz, A.; López-Vicario, C.; Barak, Y.; Arroyo, V.; Clària, J. Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 2011, 25, 2538–2550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kim, J.H.; Jang, Y.S.; Kim, C.H.; Lee, J.Y.; Lim, S.S. Anti-obesity effect of Solidago virgaurea var. gigantea extract through regulation of adipogenesis and lipogenesis pathways in high-fat diet-induced obese mice (C57BL/6N). Food Nutr. Res. 2017, 61, 1273479. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.T.; Cheng, W.Z.; Xu, Q.; Shao, L.X. Dietary restriction reduces blood lipids and ameliorates liver function of mice with hyperlipidemia. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2017, 37, 79–86. [Google Scholar] [PubMed]
- Chiang, J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [PubMed]
- Pannu, P.S.; Allahverdian, S.; Francis, G.A. Oxysterol generation and liver X receptor-dependent reverse cholesterol transport: Not all roads lead to Rome. Mol. Cell Endocrinol. 2013, 368, 99–107. [Google Scholar] [CrossRef] [PubMed]
- He, X.W.; Yu, D.; Li, W.L.; Zheng, Z.; Lv, C.L.; Li, C.; Liu, P.; Xu, C.Q.; Hu, X.F.; Jin, X.P. Anti-atherosclerotic potential of baicalin mediated by promoting cholesterol efflux from macrophages via the PPARγ–LXRα–ABCA1/ABCG1 pathway. Biomed. Pharmacother. 2016, 83, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Sang, G.; Zhu, Y.; Yang, G.; Zhang, H. Preparation and characterization of high porosity cement-based foam material. Construct. Build. Mater. 2015, 91, 133–137. [Google Scholar] [CrossRef]
- Qi, Y.; Jiang, C.; Cheng, J.; Krausz, K.W.; Li, T.; Ferrell, J.M.; Gonzalez, F.J.; Chiang, J.Y. Bile acid signaling in lipid metabolism: Metabolomic and lipidomic analysis of lipid and bile acid markers linked to anti-obesity and anti-diabetes in mice. Biochim. Biophys. Acta 2015, 1851, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Ducheix, S.; Montagner, A.; Theodorou, V.; Ferrier, L.; Guillou, H. The liver X receptor: A master regulator of the gut-liver axis and a target for non-alcoholic fatty liver disease. Biochem. Pharmacol. 2013, 86, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Iichiro Shimomura, Y.B.; Jay, D. Horton, Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biolog. Chem. 1999, 274, 30028–30032. [Google Scholar] [CrossRef]
- Foufelle, P.F.F. Hepatic steatosis: A role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes. Metab. 2010, 12 (Suppl. S2), 83–92. [Google Scholar]
- Li, G.; Zhou, F.; Chen, Y.; Zhang, W.; Wang, N. Kukoamine A attenuates insulin resistance and fatty liver through downregulation of Srebp-1c. Biomed. Pharmacother. 2017, 89, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.A. The SCAP/SREBP pathway: A mediator of hepatic steatosis. Endocrinol. Metab. 2017, 32, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, Y.; Hu, N.; Sun, Q.; Ding, X.; Li, G.; Zheng, B.; Gu, M.; Huang, F.; Sun, Y.Q.; et al. Extract of Kuding tea prevents high-fat diet-induced metabolic disorders in C57BL/6 mice via liver X receptor (LXR) β antagonism. PLoS ONE 2012, 7, e51007. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Cai, W.; Zhang, Y.; Feng, L.; Huang, C. Red bayberry extract prevents high-fat diet-induced metabolic disorders in C57BL/6 mice. J. Funct. Foods 2015, 14, 278–288. [Google Scholar] [CrossRef]
- Chun, T.H.; Inoue, M.; Morisaki, H.; Yamanaka, I.; Miyamoto, Y.; Okamura, T.; Sato-Kusubata, K.; Weiss, S.J. Genetic link between obesity and MMP14-dependent adipogenic collagen turnover. Diabetes 2010, 59, 2484–2494. [Google Scholar] [CrossRef] [PubMed]
- Le, R.; Kou, Z.; Jiang, Y.; Li, M.; Huang, B.; Liu, W.; Li, H.; Kou, X.; He, W.; Rudolph, K.L.; et al. Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell 2014, 14, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, J.; Li, K.; Wu, T.; Huang, B.; Liu, W.; Kou, X.; Zhang, Y.; Huang, H.; Jiang, Y.; et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 2013, 12, 453–469. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| β-Actin | TGTCCACCTTCCAGCAGATGT | AGCTCAGTAACAGTCCGCCTAGA |
| PPARα | AGGCTGTAAGGGCTTCTTTCG | GGCATTTGTTCCGGTTCTTC |
| PPARβ | AGTGACCTGGCGCTCTTCAT | CGCAGAATGGTGTCCTGGAT |
| PPARγ | CGCTGATGCACTGCCTATGA | AGAGGTCCACAGAGCTGATTCC |
| LXRα | GAGTGTCGACTTCGCAAATGC | CCTCTTCTTGCCGCTTCAGT |
| LXRβ | CAGGCTTGCAGGTGGAATTC | ATGGCGATAAGCAAGGCATACT |
| Cyp7a1 | GTGGTAGTGAGCTGTTGCATATGG | CACAGCCCAGGTATGGAATCA |
| CYP8B1 | GGACAGCCTATCCTTGGTGA | GACGGAACTTCCTGAACAGC |
| SREBP-1c | GGCTATTCCGTGAACATCTCCTA | ATCCAAGGGCATCTGAGAACTC |
| FAS | CTGAGATCCCAGCACTTCTTGA | GCCTCCGAAGCCAAATGAG |
| ACC-1 | GAATCTCCTGGTGACAATGCTTATT | GGTCTTGCTGAGTTGGGTTAGCT |
| aP2 | CATGGCCAAGCCCAACAT | CGCCCAGTTTGAAGGAAATC |
| ApoE | GAACCGCTTCTGGGATTACCT | TCAGTGCCGTCAGTTCTTGTG |
| CD36 | GCTTGCAACTGTCAGCACAT | GCCTTGCTGTAGCCAAGAAC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Shi, L.; Joyce, S.; Wang, Y.; Feng, Y. MDG-1, a Potential Regulator of PPARα and PPARγ, Ameliorates Dyslipidemia in Mice. Int. J. Mol. Sci. 2017, 18, 1930. https://doi.org/10.3390/ijms18091930
Wang X, Shi L, Joyce S, Wang Y, Feng Y. MDG-1, a Potential Regulator of PPARα and PPARγ, Ameliorates Dyslipidemia in Mice. International Journal of Molecular Sciences. 2017; 18(9):1930. https://doi.org/10.3390/ijms18091930
Chicago/Turabian StyleWang, Xu, Linlin Shi, Sun Joyce, Yuan Wang, and Yi Feng. 2017. "MDG-1, a Potential Regulator of PPARα and PPARγ, Ameliorates Dyslipidemia in Mice" International Journal of Molecular Sciences 18, no. 9: 1930. https://doi.org/10.3390/ijms18091930
APA StyleWang, X., Shi, L., Joyce, S., Wang, Y., & Feng, Y. (2017). MDG-1, a Potential Regulator of PPARα and PPARγ, Ameliorates Dyslipidemia in Mice. International Journal of Molecular Sciences, 18(9), 1930. https://doi.org/10.3390/ijms18091930




