Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis
Abstract
:1. Introduction
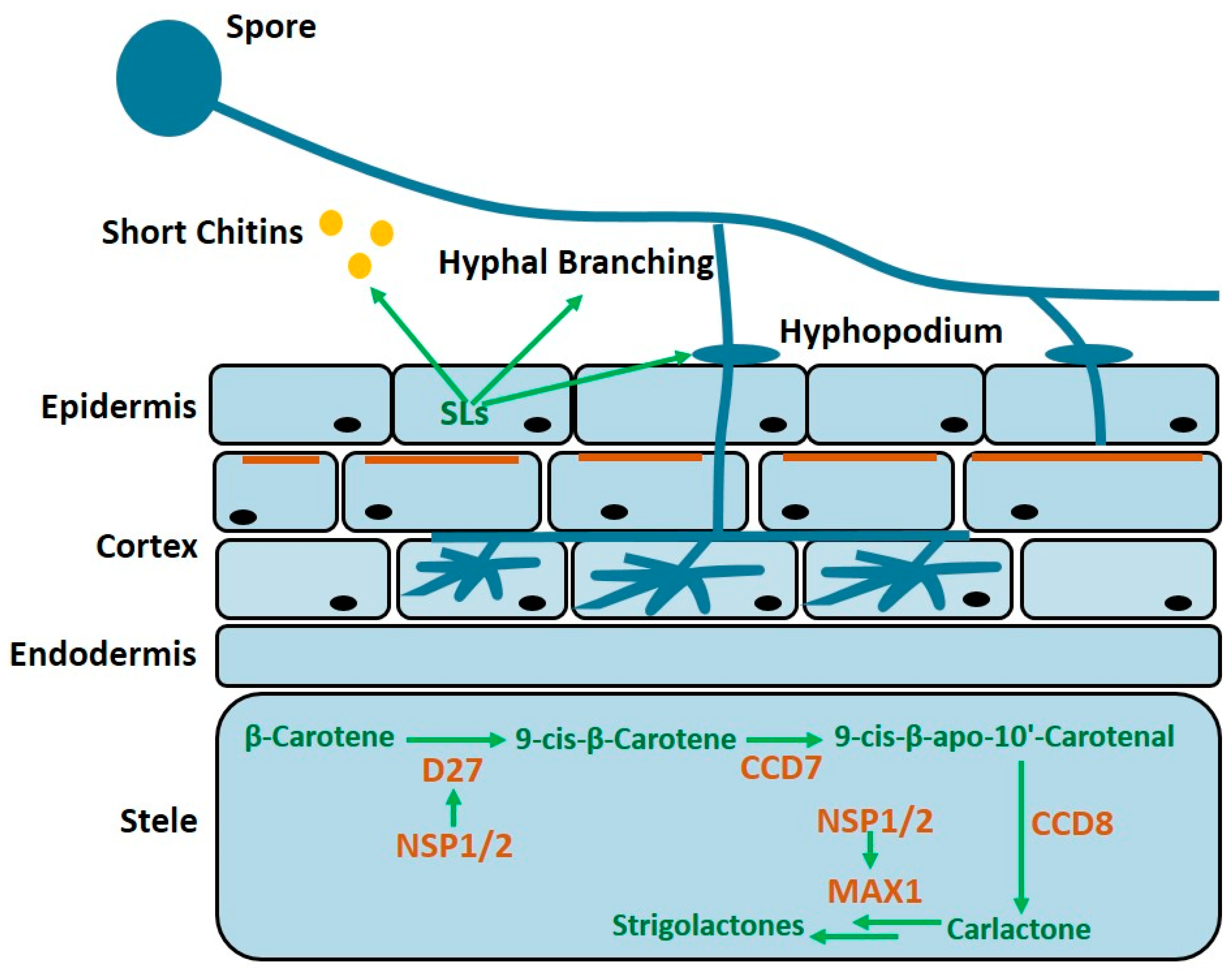
2. Strigolactones
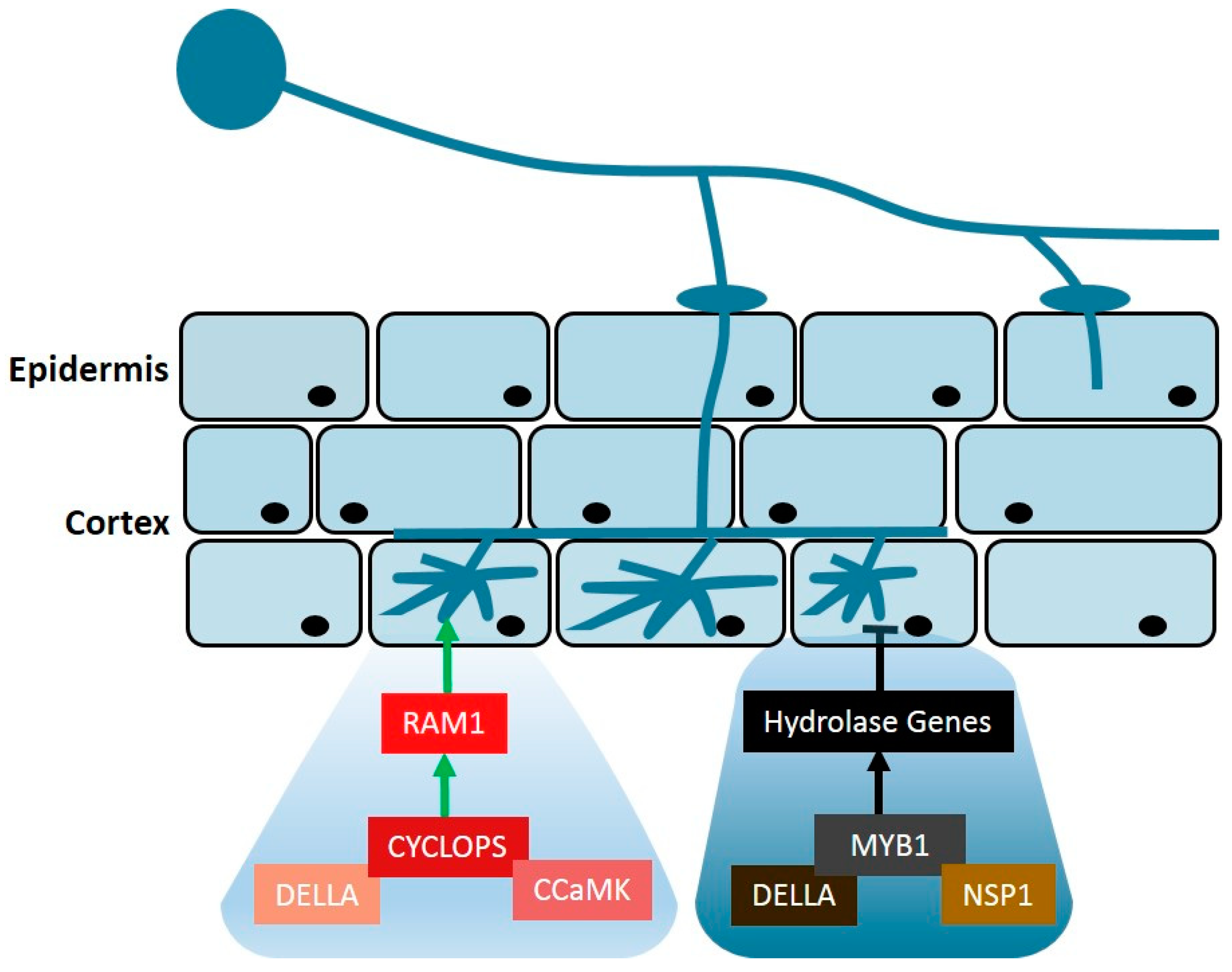
3. Gibberellin
4. Auxin
5. Abscisic Acid and Ethylene
6. Jasmonic Acid, Salicylic Acid, Cytokinins, and Brassinosteroids
7. Conclusions and Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gutjahr, C.; Parniske, M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef] [PubMed]
- Cosme, M.; Fernandez, I.; van der Heijden, M.G.A.; Pieterse, C.M.J. Non-Mycorrhizal Plants: The Exceptions that Prove the Rule. Trend. Plant Sci. 2018, 23, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Roth, R.; Paszkowski, U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017, 39, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Remy, W.; Taylor, T.N.; Hass, H.; Kerp, H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 1994, 91, 11841–11843. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.C. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002, 154, 275–304. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Qiu, Y.L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef] [PubMed]
- Werner, G.D.A.; Cornelissen, J.H.C.; Cornwell, W.K.; Soudzilovskaia, N.A.; Kattge, J.; West, S.A.; Kiers, E.T. Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc. Natl. Acad. Sci. USA 2018, 15, 5229–5234. [Google Scholar] [CrossRef] [PubMed]
- Dickson, S. The Arum-Paris continuum of mycorrhizal symbioses. New Phytol. 2004, 163, 187–200. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.E.; Read, D.J. Mycorrhizal symbiosis; Academic Press: New York, NY, USA, 2008. [Google Scholar]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant Signaling and Metabolic Pathways Enabling Arbuscular Mycorrhizal Symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Al-Babili, S.; van der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trend. Plant Sci. 2013, 18, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, K.; Ruelens, P.; Geuten, K.; Rohde, A.; van der Straeten, D. Exploiting DELLA Signaling in Cereals. Trend. Plant Sci. 2017, 22, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Ross, J.J.; Jones, W.T.; Reid, J.B. Plant hormones in arbuscular mycorrhizal symbioses: An emerging role for gibberellins. Ann. Bot. 2013, 111, 769–779. [Google Scholar] [CrossRef] [PubMed]
- Gutjahr, C. Phytohormone signaling in arbuscular mycorhiza development. Curr. Opin. Plant Biol. 2014, 20, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; López-Ráez, J.A.; Azcón-Aguilar, C.; García-Garrido, J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a Novel Carotenoid-Derived Plant Hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, L.; Huguet, S.; Wipf, D.; Pauly, N.; Truong, H.N. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. 2013, 199, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.W.; Tao, J.Y.; Liu, S.J.; Huang, S.J.; Chen, S.; Xie, X.N.; Yoneyama, K.; Zhang, Y.L.; Xu, G.H. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Puech-Pages, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Becard, G.; Sejalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, e226. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Becard, G.; Jauneau, A.; Roux, C.; Sejalon-Delmas, N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008, 148, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Nishiuma, K.; Sugiyama, T.; Hayashi, H.; Akiyama, K. Carlactone-type strigolactones and their synthetic analogues as inducers of hyphal branching in arbuscular mycorrhizal fungi. Phytochemistry 2016, 130, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pages, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Becard, G.; Bonfante, P.; et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013, 198, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kohlen, W.; Lillo, A.; Op den Camp, R.; Ivanov, S.; Hartog, M.; Limpens, E.; Jamil, M.; Smaczniak, C.; Kaufmann, K.; et al. Strigolactone Biosynthesis in Medicago truncatula and Rice Requires the Symbiotic GRAS-Type Transcription Factors NSP1 and NSP2. Plant Cell 2011, 23, 3853–3865. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Charnikhova, T.; Lammers, M.; Pollina, T.; Toth, P.; Haider, I.; Pozo, M.J.; de Maagd, R.A.; Ruyter-Spira, C.; Bouwmeester, H.J.; et al. The tomato CAROTENOID CLEAVAGE DIOXYGENASE8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 2012, 196, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; Yoneyama, K.; Hugill, C.J.; Quittenden, L.J.; Reid, J.B. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant 2013, 6, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, T.; Kohlen, W.; Sasse, J.; Borghi, L.; Schlegel, M.; Bachelier, J.B.; Reinhardt, D.; Bours, R.; Bouwmeester, H.J.; Martinoia, E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 2012, 483, 341–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasse, J.; Simon, S.; Gübeli, C.; Liu, G.-W.; Cheng, X.; Friml, J.; Bouwmeester, H.; Martinoia, E.; Borghi, L. Asymmetric Localizations of the ABC Transporter PaPDR1 Trace Paths of Directional Strigolactone Transport. Curr. Biol. 2015, 25, 647–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobae, Y.; Kameoka, H.; Sugimura, Y.; Saito, K.; Ohtomo, R.; Fujiwara, T.; Kyozuka, J. Strigolactone Biosynthesis Genes of Rice are Required for the Punctual Entry of Arbuscular Mycorrhizal Fungi into the Roots. Plant Cell Physiol. 2018, 59, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Tsuzuki, S.; Suzaki, T.; Parniske, M.; Kawaguchi, M. CERBERUS and NSP1 of Lotus japonicus are Common Symbiosis Genes That Modulate Arbuscular Mycorrhiza Development. Plant Cell Physiol. 2013, 54, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 7, 12433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonouni-Farde, C.; Tan, S.; Baudin, M.; Brault, M.; Wen, J.Q.; Mysore, K.S.; Niebel, A.; Frugier, F.; Diet, A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016, 7, 12636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, S.; Kameoka, H.; Tempo, M.; Akiyama, K.; Umehara, M.; Yamaguchi, S.; Hayashi, H.; Kyozuka, J.; Shirasu, K. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. New Phytol. 2012, 196, 1208–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutjahr, C.; Gobbato, E.; Choi, J.; Riemann, M.; Johnston, M.G.; Summers, W.; Carbonnel, S.; Mansfield, C.; Yang, S.Y.; Nadal, M.; et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 2015, 350, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Nadal, M.; Sawers, R.; Naseem, S.; Bassin, B.; Kulicke, C.; Sharman, A.; An, G.; An, K.; Ahern, K.R.; Romag, A.; et al. An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 2017, 3, 17073. [Google Scholar] [CrossRef] [PubMed]
- Shaul-Keinan, O.; Gadkar, V.; Ginzberg, I.; Grunzweig, J.M.; Chet, I.; Elad, Y.; Wininger, S.; Belausov, E.; Eshed, Y.; Arzmon, N.; et al. Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomus intraradices. New Phytol. 2002, 154, 501–507. [Google Scholar] [CrossRef]
- Floss, D.S.; Levy, J.G.; Levesque-Tremblay, V.; Pumplin, N.; Harrison, M.J. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, E5025–E5034. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Luo, D.; Zhang, X.; Liu, J.; Wang, W.; Jin, Y.; Dong, W.; Liu, J.; Liu, H.; Yang, W.; et al. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res. 2013, 24, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Ghachtouli, N.; Martin-Tanguy, J.; Paynot, M.; Gianinazzi, S. First-report of the inhibition of arbuscular mycorrhizal infection of Pisum sativum by specific and irreversible inhibition of polyamine biosynthesis or by gibberellic acid treatment. FEBS lett. 1996, 385, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Takeda, N.; Handa, Y.; Tsuzuki, S.; Kojima, M.; Sakakibara, H.; Kawaguchi, M. Gibberellins Interfere with Symbiosis Signaling and Gene Expression and Alter Colonization by Arbuscular Mycorrhizal Fungi in Lotus japonicus. Plant Physiol. 2015, 167, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Lévesque-Tremblay, V.; Park, H.-J.; Harrison, M.J. DELLA proteins regulate expression of a subset of AM symbiosis-induced genes in Medicago truncatula. Plant Signal. Behav. 2016, 11, e1162369. [Google Scholar] [CrossRef] [PubMed]
- Pimprikar, P.; Carbonnel, S.; Paries, M.; Katzer, K.; Klingl, V.; Bohmer, M.J.; Karl, L.; Floss, D.S.; Harrison, M.J.; Parniske, M.; et al. A CCaMK-CYCLOPS-DELLA Complex Activates Transcription of RAM1 to Regulate Arbuscule Branching. Curr. Biol. 2016, 26, 987–998. [Google Scholar] [CrossRef] [PubMed]
- Heck, C.; Kuhn, H.; Heidt, S.; Walter, S.; Rieger, N.; Requena, N. Symbiotic Fungi Control Plant Root Cortex Development through the Novel GRAS Transcription Factor MIG1. Curr. Biol. 2016, 26, 2770–2778. [Google Scholar] [CrossRef] [PubMed]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef] [PubMed]
- Floss, D.S.; Gomez, S.K.; Park, H.-J.; MacLean, A.M.; Müller, L.M.; Bhattarai, K.K.; Lévesque-Tremblay, V.; Maldonado-Mendoza, I.E.; Harrison, M.J. A Transcriptional Program for Arbuscule Degeneration during AM Symbiosis Is Regulated by MYB1. Curr. Biol. 2017, 27, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Perez-Torres, C.A.; Lopez-Bucio, J.; Cruz-Ramirez, A.; Ibarra-Laclette, E.; Dharmasiri, S.; Estelle, M.; Herrera-Estrella, L. Phosphate Availability Alters Lateral Root Development in Arabidopsis by Modulating Auxin Sensitivity via a Mechanism Involving the TIR1 Auxin Receptor. Plant Cell 2008, 20, 3258–3272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.L.; Ma, G.J.; Zhang, M.L.; Xiong, H.; Wu, H.; Zhao, C.Z.; Liu, C.S.; Jia, H.X.; Chen, L.; Kjorven, J.O.; et al. The ARF7 and ARF19 Transcription Factors Positively Regulate PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis Roots. Plant Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Stoeckle, D.; Thellmann, M.; Vermeer, J.E.M. Breakout—Lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2018, 41, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Fitze, D.; Wiepning, A.; Kaldorf, M.; Ludwig-Muller, J. Auxins in the development of an arbuscular mycorrhizal symbiosis in maize. J. Plant Physiol. 2005, 162, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Jentschel, K.; Thiel, D.; Rehn, F.; Ludwig-Muller, J. Arbuscular mycorrhiza enhances auxin levels and alters auxin biosynthesis in Tropaeolum majus during early stages of colonization. Physiol. Plant. 2007, 129, 320–333. [Google Scholar] [CrossRef]
- Campanella, J.J.; Smith, S.M.; Leibu, D.; Wexler, S.; Ludwig-Muller, J. The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 2008, 27, 26–38. [Google Scholar] [CrossRef]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.-M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.-P. Auxin Perception Is Required for Arbuscule Development in Arbuscular Mycorrhizal Symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanlon, M.T.; Coenen, C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 2011, 189, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Foo, E. Auxin influences strigolactones in pea mycorrhizal symbiosis. J. Plant Physiol. 2013, 170, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Stirnberg, P.; Beveridge, C.; Leyser, O. Interactions between Auxin and Strigolactone in Shoot Branching Control. Plant Physiol. 2009, 151, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agusti, J.; Herold, S.; Schwarz, M.; Sanchez, P.; Ljung, K.; Dun, E.A.; Brewer, P.B.; Beveridge, C.A.; Sieberer, T.; Sehr, E.M.; et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 20242–20247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koltai, H. Cellular events of strigolactone signalling and their crosstalk with auxin in roots. J. Exp. Bot. 2015, 66, 4855–4861. [Google Scholar] [CrossRef] [PubMed]
- Liao, D.; Chen, X.; Chen, A.; Wang, H.; Liu, J.; Liu, J.; Gu, M.; Sun, S.; Xu, G. The characterization of six auxin-induced tomato GH3 genes uncovers a member, SlGH3.4, strongly responsive to arbuscular mycorrhizal symbiosis. Plant Cell Physiol. 2015, 56, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liao, D.; Yang, X.; Ji, M.; Wang, S.; Gu, M.; Chen, A.; Xu, G. Three cis-Regulatory Motifs, AuxRE, MYCRS1 and MYCRS2, are Required for Modulating the Auxin- and Mycorrhiza-Responsive Expression of a Tomato GH3 Gene. Plant Cell Physiol. 2017, 58, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Melcher, K.; Xu, Y.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Chinnusamy, V.; Suino-Powell, K.M.; Kovach, A.; Tham, F.S.; Cutler, S.R.; et al. Identification and mechanism of ABA receptor antagonism. Nat. Struct. Mol. Biol. 2010, 17, 1102. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lv, Z.; Liu, Y.; Li, L.; Zhang, L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 2018, 41, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Medina, M.J.; Steinkellner, S.; Vierheilig, H.; Bote, J.A.O.; Garrido, J.M.G. Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 2007, 175, 554–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herde, O.; Pena Cortes, H.; Wasternack, C.; Willmitzer, L.; Fisahn, J. Electric signaling and pin2 gene expression on different abiotic stimuli depend on a distinct threshold level of endogenous abscisic acid in several abscisic acid-deficient tomato mutants. Plant Physiol. 1999, 119, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rodriguez, J.A.; Leon-Morcillo, R.; Vierheilig, H.; Ocampo, J.A.; Ludwig-Muller, J.; Garcia-Garrido, J.M. Ethylene-dependent/ethylene-independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol 2011, 190, 193–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Akune, M.; Kogiso, M.; Imagama, Y.; Osuki, K.; Uchiumi, T.; Higashi, S.; Han, S.Y.; Yoshida, S.; Asami, T.; et al. Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol. 2004, 45, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.L.; Kalo, P.; Yendrek, C.; Sun, J.H.; Liang, Y.; Marsh, J.F.; Harris, J.M.; Oldroyd, G.E.D. Abscisic Acid Coordinates Nod Factor and Cytokinin Signaling during the Regulation of Nodulation in Medicago truncatula. Plant Cell 2008, 20, 2681–2695. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, M.; Sun, J.H.; Wen, J.Q.; Mysore, K.S.; Oldroyd, G.E.D. Abscisic Acid Promotion of Arbuscular Mycorrhizal Colonization Requires a Component of the PROTEIN PHOSPHATASE 2A Complex. Plant Physiol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; de Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; van der Straeten, D.; Peng, J.R.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef] [PubMed]
- De Los Santos, R.T.; Vierheilig, H.; Ocampo, J.A.; Garrido, J.M. Altered pattern of arbuscular mycorrhizal formation in tomato ethylene mutants. Plant Signal. Behav. 2011, 6, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foo, E.; McAdam, E.L.; Weller, J.L.; Reid, J.B. Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J. Exp. Bot. 2016, 67, 2413–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De los Santos, R.T.; Rosales, N.M.; Ocampo, J.A.; Garcia-Garrido, J.M. Ethylene Alleviates the Suppressive Effect of Phosphate on Arbuscular Mycorrhiza Formation. J. Plant Growth Regul. 2016, 35, 611–617. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef] [PubMed]
- Segarra, G.; van der Ent, S.; Trillas, I.; Pieterse, C.M.J. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 2009, 11, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutjahr, C.; Paszkowski, U. Weights in the Balance: Jasmonic Acid and Salicylic Acid Signaling in Root-Biotroph Interactions. Mol. Plant-Microbe Interact. 2009, 22, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Tejeda-Sartorius, M.; de la Vega, O.M.; Delano-Frier, J.P. Jasmonic acid influences mycorrhizal colonization in tomato plants by modifying the expression of genes involved in carbohydrate partitioning. Physiol. Plant. 2008, 133, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Medina, M.J.; Tamayo, M.I.; Vierheilig, H.; Ocampo, J.A.; Garcia-Garrido, J.M. The jasmonic acid signalling pathway restricts the development of the arbuscular mycorrhizal association in tomato. J. Plant Growth Regul. 2008, 27, 221–230. [Google Scholar] [CrossRef]
- Gutjahr, C.; Siegler, H.; Haga, K.; Iino, M.; Paszkowski, U. Full Establishment of Arbuscular Mycorrhizal Symbiosis in Rice Occurs Independently of Enzymatic Jasmonate Biosynthesis. PLoS ONE 2015, 10, e0123422. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Ding, L.; Yu, H. Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 2013, 32, 1067–1074. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.; Mrosk, C.; Isayenkov, S.; Strack, D. Jasmonates in arbuscular mycorrhizal interactions. Phytochemistry 2007, 68, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W.; Somerville, S.C. Perception of conserved pathogen elicitors at the plasma membrane leads to relocalization of the Arabidopsis PEN3 transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 12492–12497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, M.J.H.; Gagnon, H.; Piche, Y.; Ocampo, J.A.; Garrido, J.M.G.; Vierheilig, H. Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci. 2003, 164, 993–998. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed]
- Gamas, P.; Brault, M.; Jardinaud, M.F.; Frugier, F. Cytokinins in Symbiotic Nodulation: When, Where, What For? Trend. Plant Sci. 2017, 22, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Cosme, M.; Wurst, S. Interactions between arbuscular mycorrhizal fungi, rhizobacteria, soil phosphorus and plant cytokinin deficiency change the root morphology, yield and quality of tobacco. Soil Biol. Biochem. 2013, 57, 436–443. [Google Scholar] [CrossRef]
- Cosme, M.; Ramireddy, E.; Franken, P.; Schmulling, T.; Wurst, S. Shoot- and root-borne cytokinin influences arbuscular mycorrhizal symbiosis. Mycorrhiza 2016, 26, 709–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.M.; Clairmont, L.; Macdonald, E.S.; Weiner, C.A.; Emery, R.J.; Guinel, F.C. E151 (sym15), a pleiotropic mutant of pea (Pisum sativum L.), displays low nodule number, enhanced mycorrhizae, delayed lateral root emergence, and high root cytokinin levels. J. Exp. Bot. 2015, 66, 4047–4059. [Google Scholar] [CrossRef] [PubMed]
- Plet, J.; Wasson, A.; Ariel, F.; Le Signor, C.; Baker, D.; Mathesius, U.; Crespi, M.; Frugier, F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011, 65, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmulling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Moore, S.; Chen, C.; Lindsey, K. Crosstalk Complexities between Auxin, Cytokinin, and Ethylene in Arabidopsis Root Development: From Experiments to Systems Modeling, and Back Again. Mol. Plant 2017, 10, 1480–1496. [Google Scholar] [CrossRef] [PubMed]
- Fridman, Y.; Savaldi-Goldstein, S. Brassinosteroids in growth control: How, when and where. Plant Sci. 2013, 209, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Bitterlich, M.; Krugel, U.; Boldt-Burisch, K.; Franken, P.; Kuhn, C. The sucrose transporter SlSUT2 from tomato interacts with brassinosteroid functioning and affects arbuscular mycorrhiza formation. Plant J. 2014, 78, 877–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, H.; Xiao, Y.; Liu, D.; Gao, S.; Liu, L.; Yin, Y.; Jin, Y.; Qian, Q.; Chu, C. Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 2014, 26, 4376–4393. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Rozhon, W.; Papacek, M.; Ciomas, J.; Lange, T.; Kugler, K.G.; Mayer, K.F.; Sieberer, T.; Poppenberger, B. Brassinosteroids Are Master Regulators of Gibberellin Biosynthesis in Arabidopsis. Plant Cell 2015, 27, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.F.; Moore, T.S., Jr.; Christensen, M. Phytohormone changes in Bouteloua gracilis infected by vesicular-arbuscular mycorrhizae: I. Cytokinin increases in the host plant. Can. J. Bot. 1980, 58, 371–374. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, D.; Wang, S.; Cui, M.; Liu, J.; Chen, A.; Xu, G. Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis. Int. J. Mol. Sci. 2018, 19, 3146. https://doi.org/10.3390/ijms19103146
Liao D, Wang S, Cui M, Liu J, Chen A, Xu G. Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis. International Journal of Molecular Sciences. 2018; 19(10):3146. https://doi.org/10.3390/ijms19103146
Chicago/Turabian StyleLiao, Dehua, Shuangshuang Wang, Miaomiao Cui, Jinhui Liu, Aiqun Chen, and Guohua Xu. 2018. "Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis" International Journal of Molecular Sciences 19, no. 10: 3146. https://doi.org/10.3390/ijms19103146
APA StyleLiao, D., Wang, S., Cui, M., Liu, J., Chen, A., & Xu, G. (2018). Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis. International Journal of Molecular Sciences, 19(10), 3146. https://doi.org/10.3390/ijms19103146




