Abstract
Glutathione peroxidases (GPXs) are important enzymes in the glutathione-ascorbate cycle for catalyzing the reduction of H2O2 or organic hydroperoxides to water. GPXs play an essential role in plant growth and development by participating in photosynthesis, respiration, and stress tolerance. Rhodiola crenulata is a popular traditional Chinese medicinal plant which displays an extreme energy of tolerance to harsh alpine climate. The GPXs gene family might provide R. crenulata for extensively tolerance to environment stimulus. In this study, five GPX genes were isolated from R. crenulata. The protein amino acid sequences were analyzed by bioinformation softwares with the results that RcGPXs gene sequences contained three conserve cysteine residues, and the subcellular location predication were in the chloroplast, endoplasmic reticulum, or cytoplasm. Five RcGPXs members presented spatial and temporal specific expression with higher levels in young and green organs. And the expression patterns of RcGPXs in response to stresses or plant hormones were investigated by quantitative real-time PCR. In addition, the putative interaction proteins of RcGPXs were obtained by yeast two-hybrid with the results that RcGPXs could physically interact with specific proteins of multiple pathways like transcription factor, calmodulin, thioredoxin, and abscisic acid signal pathway. These results showed the regulation mechanism of RcGPXs were complicated and they were necessary for R. crenulata to adapt to the treacherous weather in highland.
1. Introduction
Rhodiola crenulata belongs to the Rhodiola (Crassulaceae) species which is a popular medicinal plant and has been used in traditional Chinese medicine for thousands of years [1]. The roots and rhizomes of Rhodiola crenulata contained several active ingredients like salidrosid, tyrosol, and their derivates which have been examined for adaptogenic and stress protection [2,3,4]. Rhodiola crenulata is widespread in Qinghai-Tibet Plateau (QTP) and its adjacent area with high elevations of 3500–5000 m [5]. R. crenualta often grows on less weathered regions with covered by gravel slopes or rock outcrops of limestones and granites [6]. The R. crenulata growing environment is alpine climate with characteristics of cold, drought, strong ultraviolet rays, and hypoxia [7]. The harsh circumstance made it develop slowly and promoted many changes in physiology, morphology, and genetics. In appearance, R. crenulata is a perennial herb which could adapt to multiple stresses [8]. In addition, R. crenulata employed a different carbon dioxide fixation pathway by crassulacean acid metabolism which could maintain the photosynthesis in extreme environments. However, little is known about the adaptive mechanism of genetics until now.
Plants, as sessile organisms, were susceptible to injury because they persistently accepted multiple external stimulus [9]. For the ever-changing environmental factors, abiotic stresses are the major cause to stunt plant growth by generating osmotic and oxidative stresses [10,11]. In order to survive from these stresses, plants should evolve an enormous capacity to maintain a steady state cellular circumstance by adjusting metabolism and growth [12]. Osmotic stress is induced by intracellular water deficiency, plant cells increase ctyoplasmic concentration and water retention to avoid disruption of homeostasis and ion distribution [13]. There are many molecules involved in osmotic regulation contained inorganic electrolytes like K+, Na+, Ca2+, and organic solutes like proline, glycine betaine, polyols, and non-reducing sugars [11]. Oxidative stress is induced by reactive oxygen species (ROS) which include oxygen ions, free radicals, and peroxides [14]. ROS are generated and accumulated at the early event of stresses and worked as a secondary messenger to trigger subsequent gene expression and defense system, however, exceeding ROS levels are damaging to cells by membrane phospholipid layers disturbation and biological molecules denaturation [15,16].
Considering the function and danger of ROS, a deliberate mechanism would be used to control ROS level [17]. The lifetime of ROS is mainly determined by various antioxidants like glutathione, ascorbate, and associated enzymes including superoxide dismutases (SODs), catalases (CATs), and other enzymes in glutathione-ascorbate cycle contained ascorbate peroxidases (APXs), monodehydroascorbates (MDHARs), glutathione peroxidases (GPXs), and glutathione reductases (GRs) [18]. Among them, GPXs are key enzymes for the antioxidant network. At first, unlike CATs or APX, GPXs have low substrate specificity and they are able to reduce a wide spectrum of peroxides besides H2O2 [19,20,21]. Second, some GPx isoforms exhibit the unique capacity to reduce lipid hydroperoxides thus protecting membranes oxidative stress [22,23]. At last, it has been reported that GPXs have precedence over APX to scavenge ROS under severe and persistently stressful condition [24]. With the progress of biochemical technology, the functions of GPXs have been elaborated on for crystal structures and validated through transgenic plants [19,25,26,27,28,29]. As the reports showed, GPXs were important for plants in response to abiotic stresses by multiple metabolic pathways. However, there were not any reports about Rhodiola crenulata GPXs.
In recent years, RNA sequencing (RNA-seq) technology has been widely used to reveal the difference in gene transcriptional levels for many plants with the development of next generation sequencing [30]. Especially for no-model plants, these organisms were the absence of detailed genetic information or closest reference genome [31]. In order to avoid the problem of homogenetic amplification caused by non-whole genome sequencing, RNA sequencing (RNA-seq) is a quick way to obtain coding sequences, discover new genes by constructing the genomic library (cDNA) or the transcription database [32]. Based on de novo assembly technology, many Chinese medicinal plants have been carried out in transcriptome studies successfully such as Pelargonium graveolens [33], Salvia miltiorrhiza [34], Forsythia koreana [35], and Panax japonicas [36]. Conducted transcirptome analysis for different organs or developmental stages, many new genes were identified related to secondary metabolites biosynthetic pathway or cultivation and breeding process [37,38,39,40]. In addition, RNA-seq was a convenient method for intensive studies in gene families like AP2/ERF transcription factor of Petroselinum crispum and cytochrome P450 of Salvia miltiorrhiza [41,42].
In order to strengthen the understanding into the molecular mechanism for R. crenulata response to extreme environment, our laboratory conducted RNA-seq firstly, and then selected the gene family of glutathione peroxidases for further research. The cDNA sequences of RcGPXs were isolated from leaves of R. crenulata, and function analysis by bioinformation. In addition, the expression levels of RcGPXs for separate organs or stressful conditions were obtained by qRT-PCR. At last, physical interaction proteins of Arabidopsis thaliana with RcGPXs were investigated by yeast two-hybrid assay.
2. Results
2.1. Identification and Alignment of GPX Genes in Rhodiola crenulata
In 2015, RNA-seq experiments were conducted for two altitudes R. crenualta with Rc5200 (located at 5237.2 m altitude) and Rc4800 (located at 4864.4 m altitude). According to the annotation information of assembly unigene database, five unigene protein sequences were presented high identify with plant phospholipid hydroperoxide glutathione peroxidases (Table S1). Based on the results with Blastp on NCBI, the potential open reading frames were predicted for RcGPXs separately. Then primers were designed according to the ORF (open reading frame) and sequences were validated by specific primer amplification products with Sanger sequencing. As the results showed, there were five unigenes annotated to GPXs, compared with eight members for Arabidopsis thaliana GPXs. The coding sequence (CDS) length of comp 28068 was 516 bp and encoded 171 amino acids which named RcGPX5 (MK084908) with protein sequence shared high identity with XP_011033684.1 (Populus euphratica GPX4) and At3g63080.1 (Arabidopsis thaliana GPX5). Meanwhile, Comp 37551 was the largest gene named RcGPX7 (MK084912) with 705 bp length encoding 234 amino acids. Comp 33811 was named RcGPX8 (MK084909) with 513 bp length encoding 170 amino acids. Comp 35764 was named RcGPX4 (MK084910) also with 513 bp length encoding 170 amino acids. Comp 36129 was named RcGPX2 (MK084911) with 606 bp length encoding 201 amino acids (Table S2).
2.2. Bioinformation for GPXs Genes in Rhodiola Crenulata
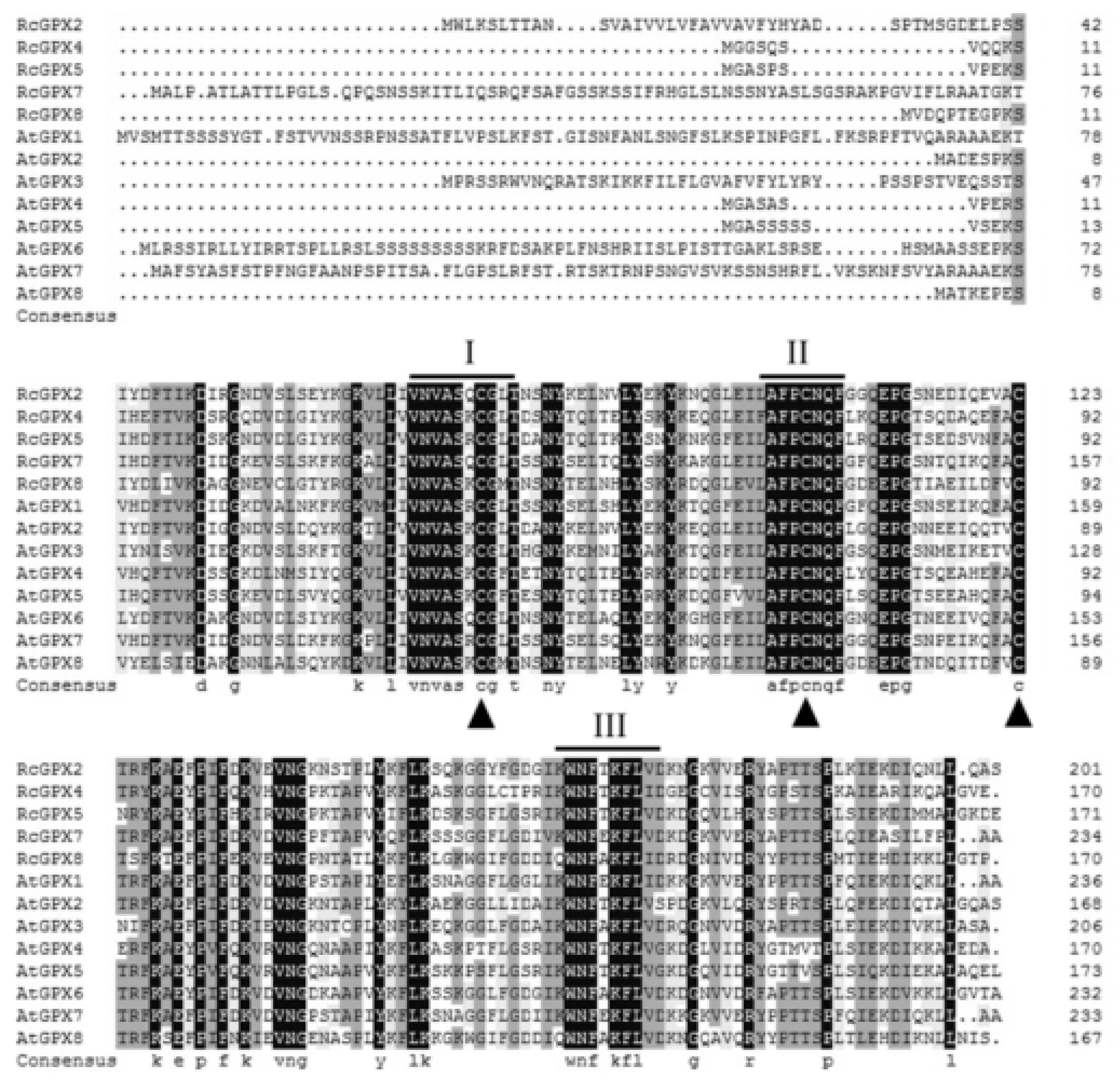
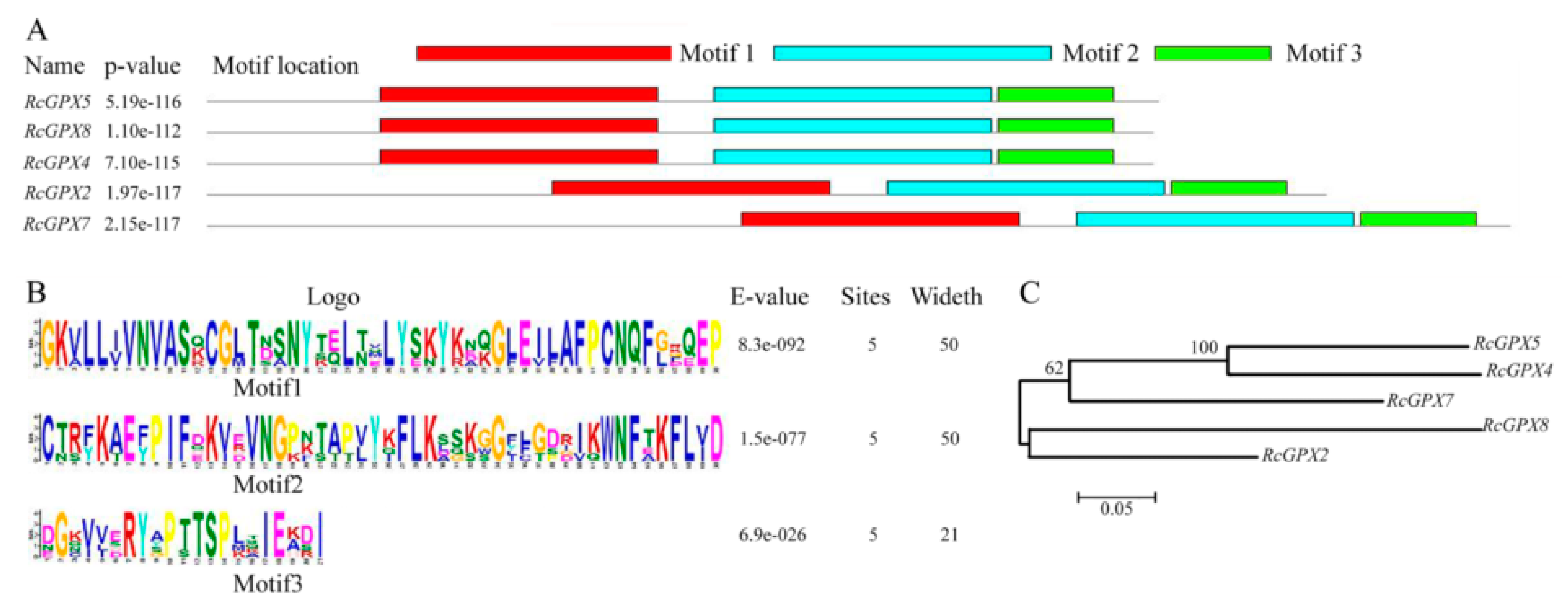
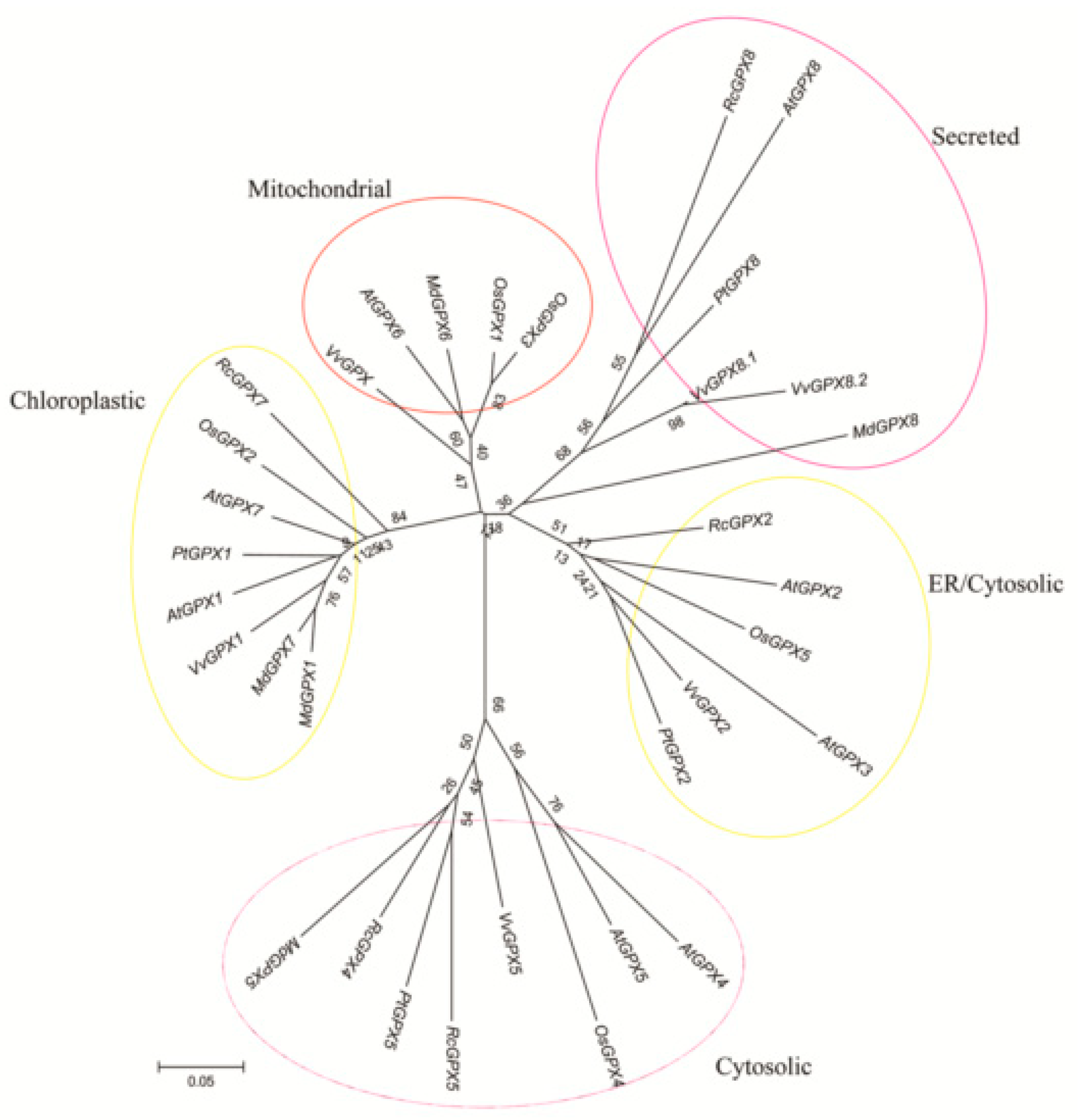
Based on the protein sequences, RcGPXs with other GPXs of diverse resource species were shared three conserved non-SeCys acted as the catalase sites (Figure 1). By MEME software, three conserved domain were searched. As the figure presented, RcGPXs included three similar conserved domains (Figure 1 and Figure 2). Two Cys sites were located in VNVAS(K/R/Q/E)CGLT and LAFPCNQF domains, another was outside domain KWNF(E/T/A/S)KFLV. By online website prediction and alignment results, five RcGPXs were located in different subcellular positions. Comparing the model species Arabidopsis thaliana GPXs, RcGPX5, and RcGPX4 were the same as AtGPX4/5 co-located in cytoplasm, RcGPX2 was likely to AtGPX2/3 located in endoplasmic reticulum or cytoplasm, RcGPX7 and AtGPX1/7 were located in chloroplast, RcGPX8 was the same with RcGPX8 located in nucleus or cytoplasm. However, there was not a GPXs member of R. crenulata similar to AtGPX6 located in mitochondria (Figure 3). The tertiary structures were predicted for RcGPXs by Swissmodel website (Figure S1). Using poplar GPX5 as a template which the crystal structure has been determined [19], RcGPXs protein tertiary structures shared similar sites and RcGPX4, RcGPX5, RcGPX7, and RcGPX8 held very similar 3D structures. In addition, four of five RcGPXs expect RcGPX2 were without transmembrane structures by TMHMM Server 2.0 (Table S2).
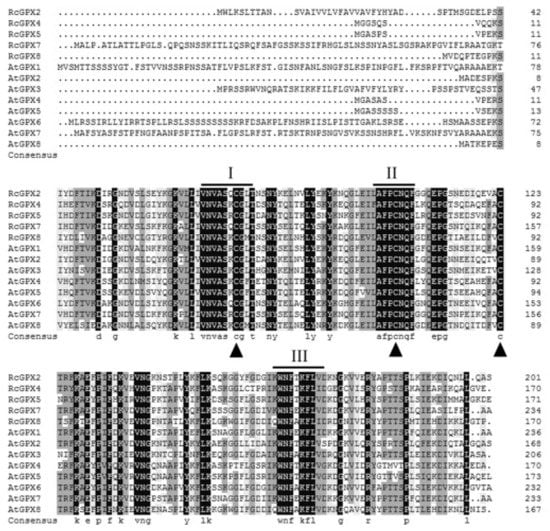
Figure 1.
Amino acid sequences alignment of plant glutathione peroxidase by DNAMAN. The highly conserved domains were presented by line and digital alphabet I, II, and III The conserved three Cys residues of plant GPX proteins are indicated by triangles. Abbreviations of plant species: Rc, Rhodiola crenualta; At, Arabidopsis thaliana. The GPX protein sequences of Arabidopsis thaliana were as followed: AtGPX1 (AT2G25080.1); AtGPX2 (AT2G31570.1); AtGPX3 (AT2G43350.1); AtGPX4 (AT2G48150.1); AtGPX5 (AT3G63080.1); AtGPX6 (AT4G11600.1); AtGPX7 (AT4G31870.1); AtGPX8 (AT1G63460.1).
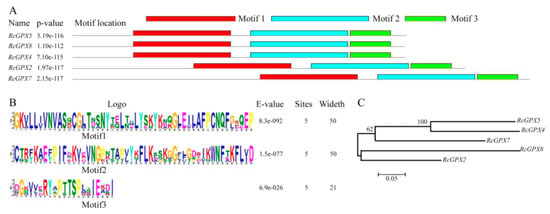
Figure 2.
Architecture and phylogenetic tree of glutathione peroxidases of Rhodiola crenulata. The structures of RcGPXs were predicted by MEME software. Distribution of conserved domains was presented by different colors (A). Hidden Markov model logos obtained using MEME (B). The phylogenetic tree of RcGPXs was constructed using the neighbor-joining method of CLUSTALW by Mega 5.0, with 1000 bootstraps, and the bar indicates 0.05 substitutions per site. Each ellipse shows a clade (C).
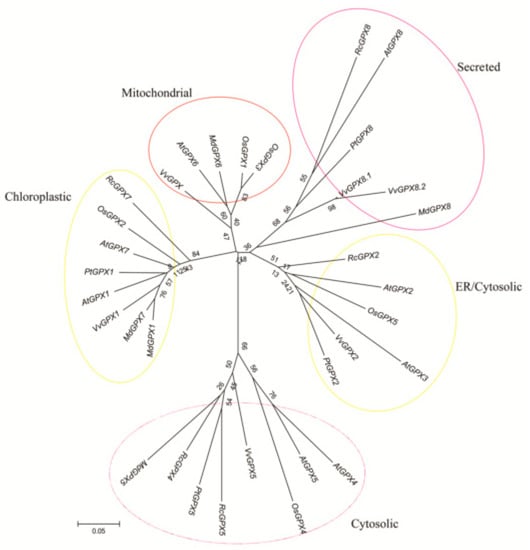
Figure 3.
Phylogenetic analyses of thirty three plant glutathione peroxidase (GPX) proteins of R. crenulata and other species. The tree was constructed using the neighbor-joining method of CLUSTALW by Mega 5.0, with 1000 bootstraps, and the bar indicates 0.05 substitutions per site. Each ellipse shows a clade. The protein sequences of GPXs were followed: AtGPX1 (AT2G25080.1); AtGPX2 (AT2G31570.1); AtGPX3 (AT2G43350.1); AtGPX4 (AT2G48150.1); AtGPX5 (AT3G63080.1); AtGPX6 (AT4G11600.1); AtGPX7 (AT4G31870.1); AtGPX8 (AT1G63460.1); VvGPX1 (XM002285528); VvGPX2 (XM002263291); VvGPX5 (XM002276220); VvGPX8.1 (XM002272900); VvGPX8.2 (XM010662496); VvGPX (XM010662503); OsGPX1 (Os04g0556300); OsGPX2 (Os06g0185900); OsGPX3 (Os02g0664000); OsGPX4 (Os03g0358100); OsGPX5 (Os11g18170); PtGPX1 (POPTR_0006s28120); PtGPX2 (POPTR_0007s02160); PtGPX5 (POPTR_0014s13490); PtGPX8 (POPTR_0001s09280); MdGPX1 (XP008379282.1); MdGPX5 (XP008355452.1); MdGPX6 (NP001280872.1); MdGPX7 (XP008347490.1); MdGPX8 (XP008384017.1). Plant species included: Rc (Rhodiola crenulata); At (Arabidopsis thaliana); Os (Oryza sativa); Pt (Populus trichocarpa); Vv (Vitis vinifera); and Md (Malus domestica).
2.3. Expression Levels for RcGPXs in Organs and Development Stages
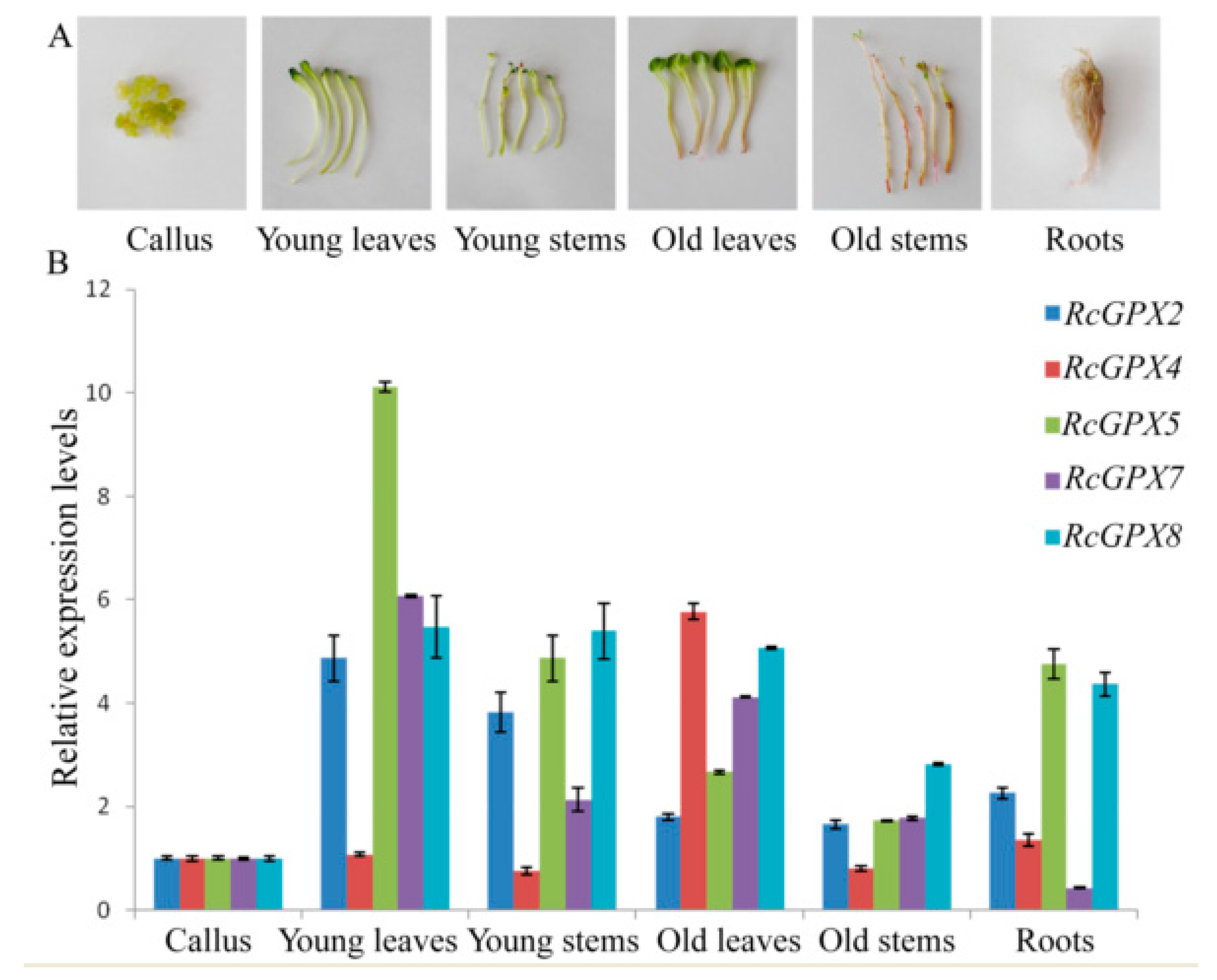
In order to analyze spatial and temporal specific expression, three organs including leaves, roots, and stems and three development stages including callus, young/old leaves, and stems were selected (Figure 4). Five RcGPXs members were expressed in all tissues and organs above with some differences. RcGPX4 presented parallel expression levels for tissues with the highest in old leaves. Other RcGPXs expression levels changed distinctly among materials with the lowest in callus and the highest in young and green tissues include leaves and stems expected RcGPX7. RcGPX7 with its chloroplast location showed higher expression levels in leaves. For the overground parts, all genes showed lower expression levels in old stems whose color was red because of the longer development time. In addition, RcGPXs presented lower expression levels in callus.
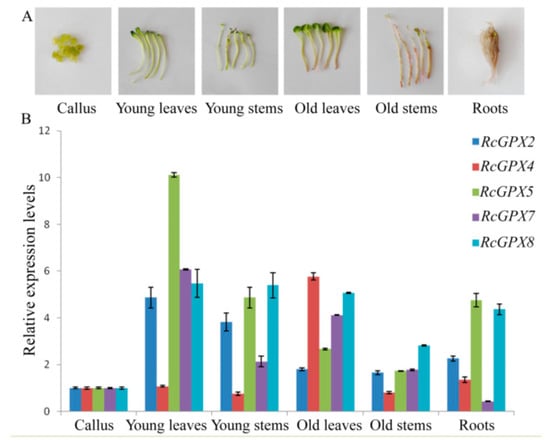
Figure 4.
Tissue-specific expression analysis of RcGPXs. Six organs and tissues were used for real-time PCR (A). The expression levels of RcGPXs under different development stages or tissues were presented by colors (B). mRNA levels were normalized with respect to RcGAPDH and are expressed relative to the values of callus tissue, which were given an arbitrary value of 1. Data represent the means ± SE of at least three replicates.
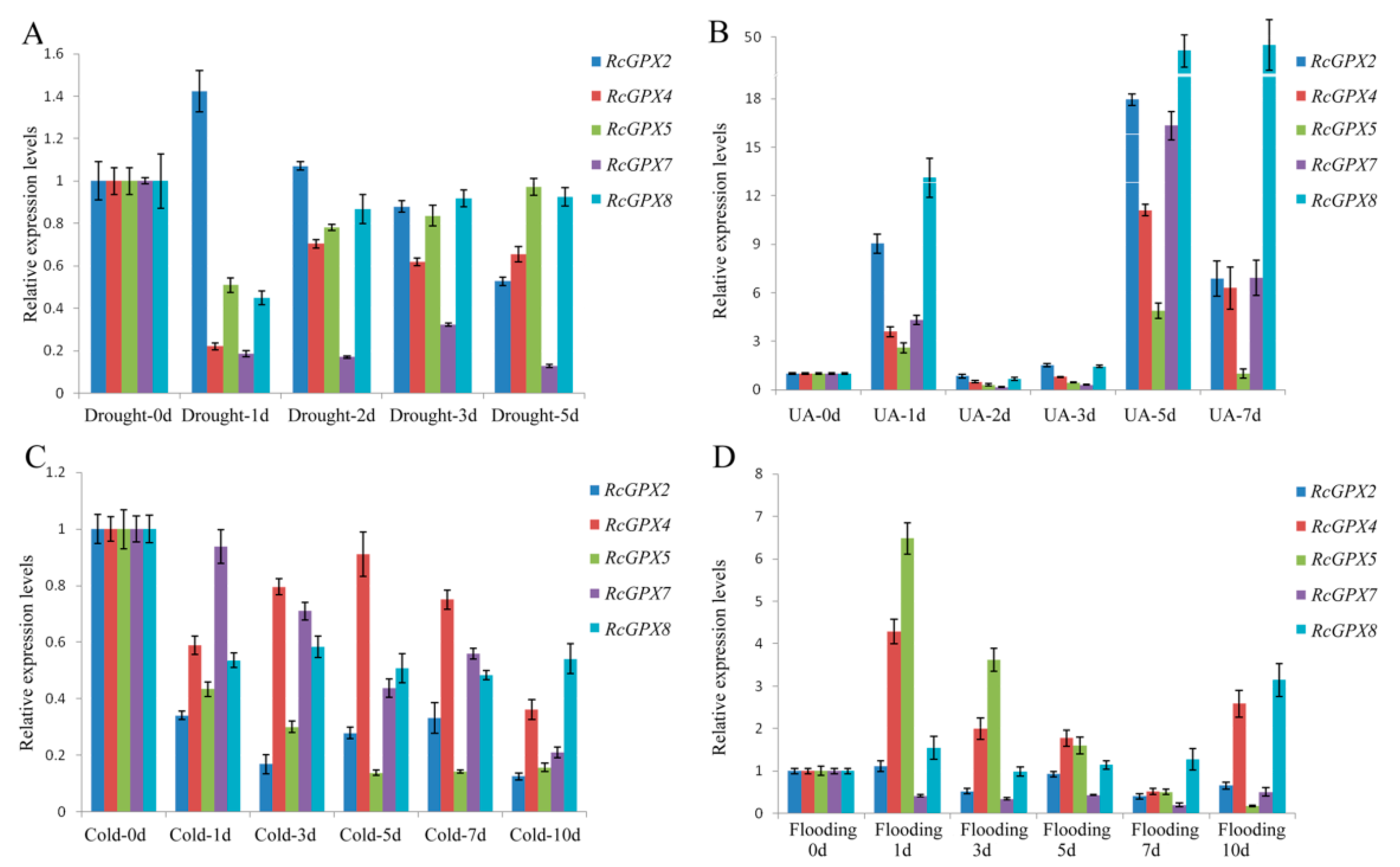
2.4. H2O2 Contents of Rhodiola Crenualta under Drought, UA, Cold and Flooding Stresses, and Expression Profiling of RcGPXs Response for These Stresses
In order to research the expression patterns of RcGPXs response to stresses, four treatment conditions include drought, UA (ultraviolet ray), cold and flooding were conducted for R. crenulata at the same developmental stage. At first, the H2O2 contents of leaves were measured to ensure the stresses were effective to plantlets (Figure S2). Compared with pre-treatment, H2O2 contents were changed distinctly with the treatment process. UA stress presented heavy damage to R. crenulata bodies with inducing the highest levels of H2O2 at the second day. Drought held similar tendency of UA stress in H2O2 contents with continuously increasing over the first days and then going down. However, the H2O2 contents of cold and flooding stress were reduced overall with a slightly rise over the following days and then dropping off finally.
Exceeding H2O2 in plants could require high enzyme activities of GPXs, which also make an important effect on the expression levels of RcGPXs. However, RcGPXs expression levels tendencies were not completely consistent with the H2O2 contents in leaves (Figure 5). Firstly, with the highest H2O2 releasing by UA stress, all member expression levels were significantly induced by UA stress especially RcGPX7 and RcGPX8. Compared with 0 day materials, RcGPXs expression levels were up-regulated at 1 day, while down-regulation at 2 days and 3 days, and abundantly expressed at last. For drought stress, RcGPXs expression levels were lower than 0 day, however up-regulated subsequently and then went to down again with the exception RcGPX2. RcGPXs response to flooding stress were very different among members. RcGPX4, RcGPX5, and RcGPX8 could be significantly increased by flooding stress, while the expression time of RcGPX8 was late for RcGPX4 and RcGPX5. RcGPX7 and RcGPX2 expression levels were down-regulated at the first two days, and up-regulated at 5 days. Cold stress could decrease the expression of RcGPXs especially for RcGPX2 and RcGPX5. RcGPX4 and RcGPX8 had persistently decreased expression levels and the others presented up at first and then down-regulation.
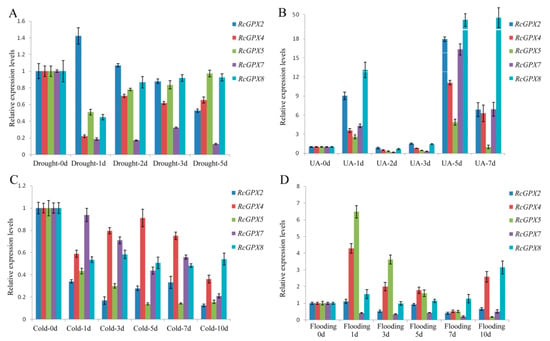
Figure 5.
Expression patterns of Rhodiola crenulata glutathione peroxidase (RcGPXs) genes for leaves under abiotic stresses. (A) Drought stress was used by PEG-4000 (20%); (B) UA stress was used the 234 nm ultraviolet ray wavelength; (C) cold stress was under ice-water environment for 0 °C; (D) flooding stress was used to build hypoxia circumstance. The total RNA was extracted from young leaves. mRNA levels were normalized with respect to RcGAPDH and were expressed relative to the values at 0 day (control), which were given an arbitrary value of 1. Data represent the means ± SE of at least three replicates.
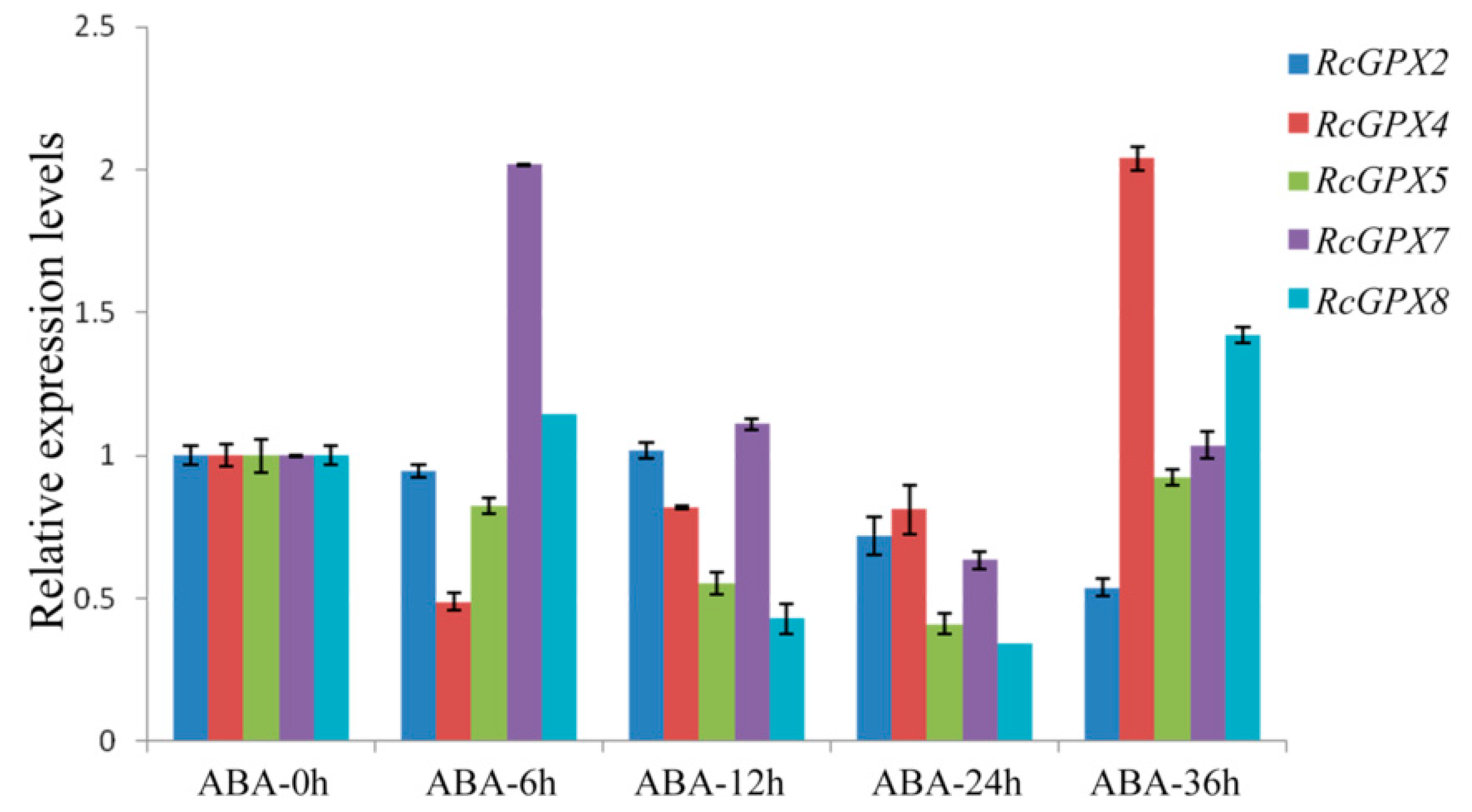
2.5. The Expression Patterns of RcGPXs for Plant Hormones
As is well known, plant hormones play a major role in plant development or response to stresses and GPXs expression levels were induced by multiple hormones [43,44]. With the extension of abscisic acid treatment, RcGPX5 and RcGPX4 were distinctly down-regulated at the early treatment stage but up-regulated at 36 h. RcGPX2 and RcGPX7 expression levels were increased at first and then went down. RcGPX8 presented a complicated tendency for abscisic acid (Figure 6). In addition, the expression patterns for RcGPXs were changed for other hormones. Exogenous MeJA and GA3 treatments could negatively regulated the expression levels of RcGPXs, exogenous IAA treatment significantly improved the expression levels of RcGPX4, RcGPX7, and RcGPX2 however reduced RcGPX5 expression and made little effect on RcGPX8 (Figure S3). These results showed that RcGPXs were widely regulated by plant hormones and involvement in the control of plant development.
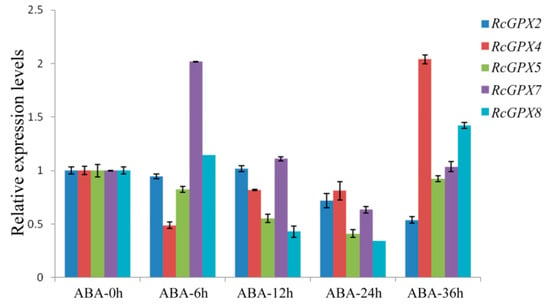
Figure 6.
Time-course expression levels of RcGPXs genes for leaves under abscisic acid treatment. Plant hormone abscisic acid (ABA) was used to treat whole seedlings of R. crenulata at 50 μM. Five time periods of conditions were selected at 0 h, 6 h, 12 h, 24 h, and 36 h. The total RNA was extracted from young leaves. mRNA levels were normalized with respect to RcGAPDH and were expressed relative to the values at 0 h (control), which were given an arbitrary value of 1. Data represent the means ± SE of at least three replicates.
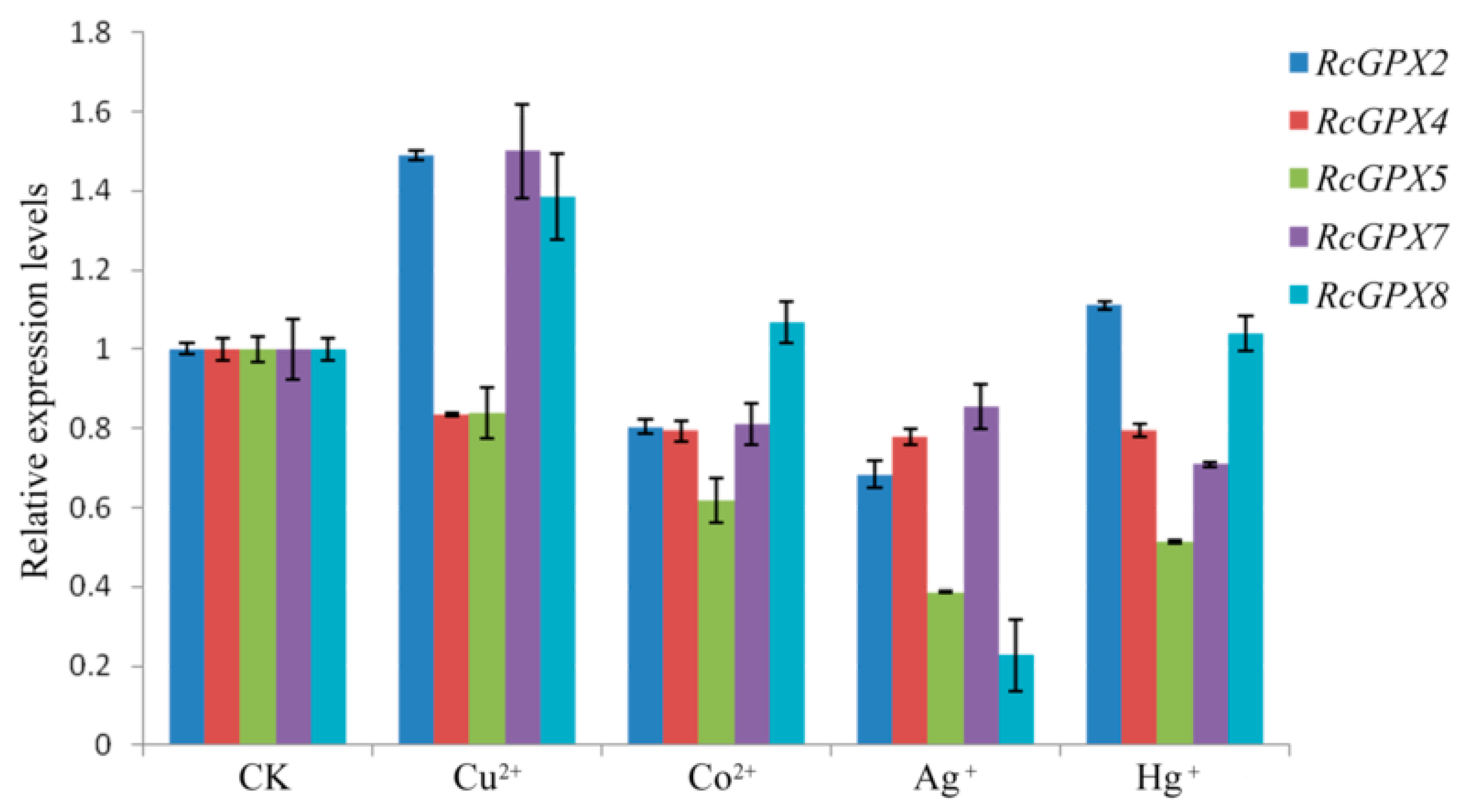
2.6. Expression Levels of RcGPXs under Heavy Metals
It is widely accepted that heavy metals could induce cell oxidative stress and irreversibly break down the protein or DNA molecules, furthermore, heavy metals also directly destroyed the antioxidant defense system with inactivation antioxidant enzymes or depletion of low molecular weight antioxidants [45,46]. So in order to investigate whether the expression levels of RcGPXs were affected by heavy metals, four ions including copper, mercury, cobalt, and silver were selected to treat the R. crenulata plantlets. As the results showed, exceeding copper (Cu) could induce RcGPX2, RcGPX7 and RcGPX8 expression with reduced RcGPX4 and RcGPX5. Cobalt (Co) could decrease four of five RcGPXs expression levels while having little effect on RcGPX8. Exogenous silver (Ag) had a negative influence on the expression of all RcGPXs. In addition, for mercury (Hg) stress, RcGPX4, RcGPX5, and RcGPX7 had decreased expression levels while RcGPXs presented slightly increasing levels compared to control materials (Figure 7).
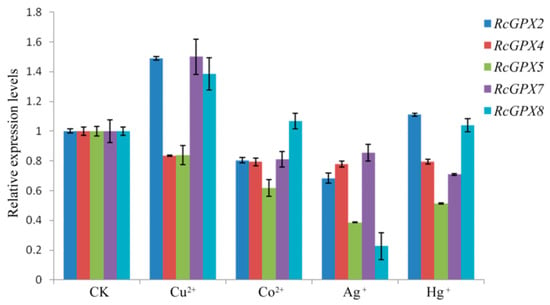
Figure 7.
Expression levels of RcGPXs genes for leaves under heavy metal conditions. Heavy metal compounds were used to treat whole seedlings of R. crenulata for 6 h. The seedlings were exposed to MS (Murashige and Skoog) media contained different heave mental ions and the concentrations were 25 μM Hg+ (HgCl), 50 μM Cu2+ (CuSO4), 50 μM Co2+ (CoCl2), and 50 μM Ag+ (AgNO4). CK indicted the negative control which seedlings were exposed for MS media for 6 h. The total RNA was extracted from young leaves. mRNA levels were normalized with respect to RcGAPDH and were expressed relative to the CK values, which were given an arbitrary value of 1. Data represent the means ± SE of at least three replicates.
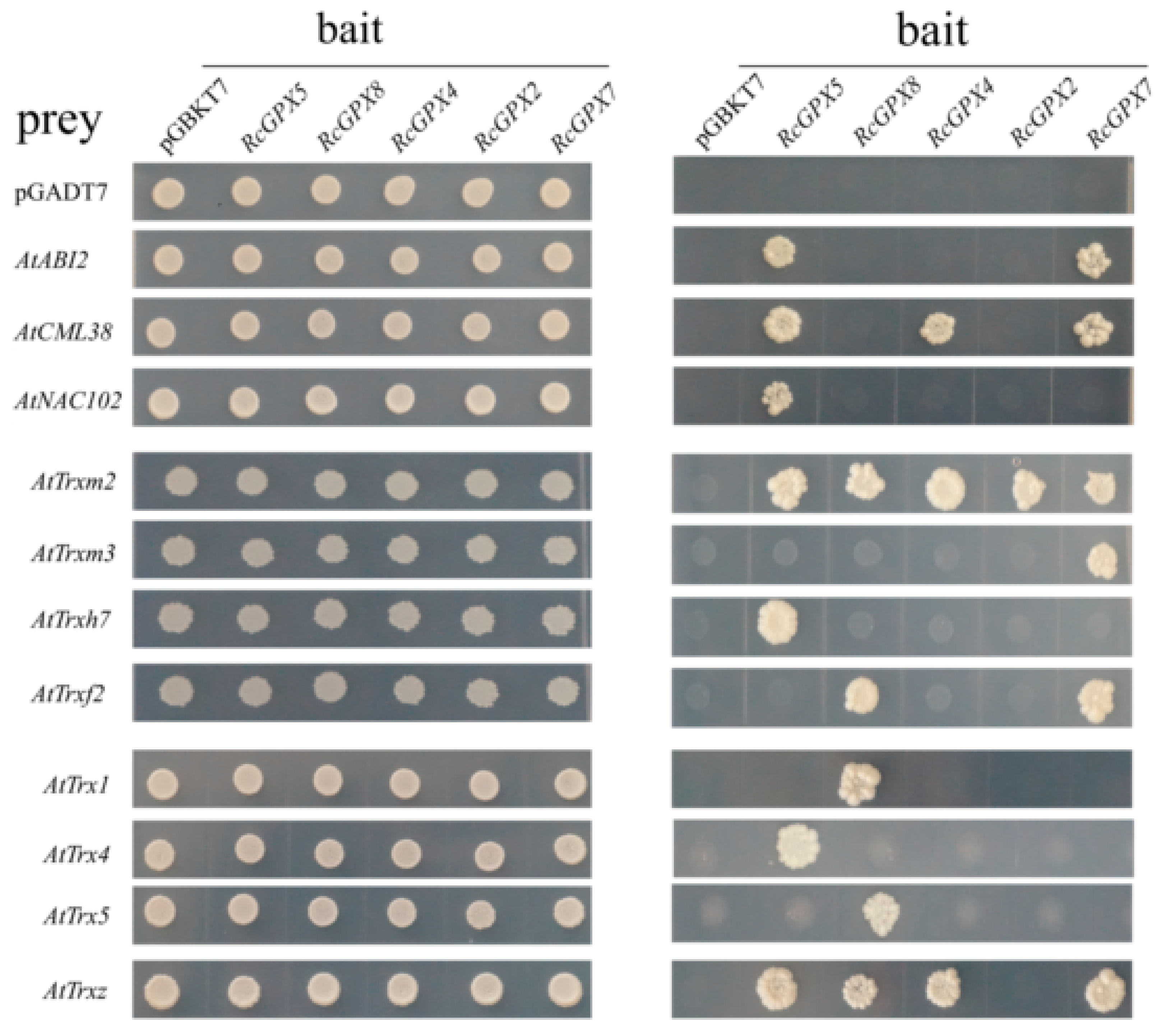
2.7. RcGPXs Physically Interaction with Proteins of Multiple Pathways
It has been reported that Arabidopsis thaliana GPX3 participated in ABA signal pathway by physically interacted with the 2C-type protein phosphatase ABA INSENSITIVE2 (ABI2) and lesser extent with ABI1 [25]. In order to analyze the function of RcGPXs further, several genes of Arabidopsis thaliana associated with transcription factors, ethylene, calcium, and ABA signal pathways were selected to detect the interaction relationship with RcGPXs by yeast two-hybrid assay. As the result showed, seven ethylene syntheses key genes (AtACS2, AtACS6, AtACS7, AtACS9, AtACO1, AtACO2, AtACO4, and AtACO5) all presented no interaction with any RcGPXs directly. For five ABA pathway genes (AtABI1, AtABI2, AtABI5, AtCYP707A2, and AtSNF4), only AtABI2 interacted with RcGPX5 and RcGPX7. In addition, a calmodulin gene AtCML38 interacted with four of five RcGPXs except RcGPX8. And a NAC (NAM/ATAF/CUC transcription factor) family member AtNAC102 was only interacted with RcGPX5, however, MYB (v-myb avian myeloblastosis viral oncogene homolog) transcription factor (AtMYB2) and alcohol dehydrogenase (AtADH) showed no interaction (Figure 8).
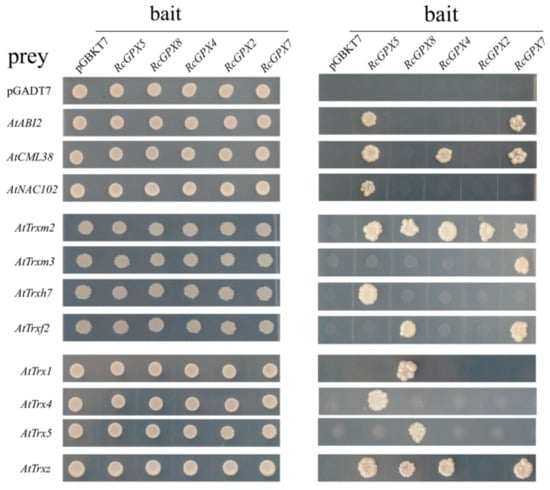
Figure 8.
The putative interaction proteins of RcGPXs were investigated by yeast two—hybrid assays. Y2H gold yeast strains containing RcGPXs as bait and Arabidopsis genes as prey were grown on SD solid media lacking Trp and Leu for 5 days (left panel) and were assayed for reporter gene His expression by growing on SD solid media lacking Trp, Leu, and His (right panel). Prey and bait empty vectors were used as negative control. The Arabidopsis genes: AtABI2 (AT5G57050.1); AtCML38 (AT1G76650); AtNAC102 (AT5G63790); AtTrxm2 (AT4G03520.1); AtTrxm3 (AT2G15570.2); AtTrxh7 (AT1G59730.1); AtTrxf2 (AT5G16400.1); AtTrx1 (AT1G76760); AtTrx4 (AT1G19730.1); AtTrx5 (AT1G45145.1); and AtTrxz (AT3G06730.1).
Thioredoxin (Trx) gene family encoding of the low weight molecules which has been confirmed to act as a substrate to provide electron donors for GPXs to catalyze H2O2 reduction and the protein model for GPXs and Trx have been calculated [19,47]. So, sixteen thioredoxin genes of Arabidopsis thaliana belonged to three subfamilies including y, m, and f type were chosen to investigate the protein’s relationship with RcGPXs. As the result showed, only half of AtTrxs genes presented physical interaction with one or more RcGPXs. For example, AtTrxm2 interacted with all RcGPXs. AtTrxz interacted with four of five RcGPXs except RcGPX2. AtTrx1 and AtTrx5 presented interaction with only RcGPX8. AtTrxh7 and AtTrx4 interacted only with RcGPX5. AtTrxf2 was interacted with RcGPX8 and RcGPX5. In addition, AtTrxm3 could only interact with RcGPX7 (Figure 8).
3. Discussion
Despite the medical values, R. crenulata with other Rhodiola species were ideal models to research the species origin and evolution of QTP and its adjacent regions [48]. In recent years, with improvement demand for R. crenulata, it is becoming severely endangered due to excessive and indiscriminate exploitation with environmental destruction, furthermore, R. crenulata and most of this genus fail to undergo artificial cultivation [49]. In order to protect and have appropriate utilization, it needs further biological and biochemical researches for R. crenulata. However, because of its specific growing environment, R. crenulata collecting and studying were notoriously different. Consequently, little was known about R. crenulata with its homogenous species on genomic and genetic information [50]. Except a report about draft genome sequence, there were only several functional genes associated with salidroside synthesis pathway and DNA barcoding sequences related to genus diversity [2,49,50,51,52]. There were no reports on the mechanism of R. crenulata adaptation to its extreme environment. In this study, the glutathione peroxidase genes were isolated and analyzed further by bioinfomation, expression patterns, and yeast two-hybrid. This paper aimed to fill the research blank and to provide insight to its relationship with the alpine environment.
Glutathione peroxidases (GPXs) were selected as an object for the important biological functions to cells and organisms. GPXs play a major role in ROS scavenging and biomembrane protection [24]. Consequently, ROS generated not only from oxidant damage of environment stimulus but also from the electron transfer chains in photosynthesis and respiration. It meant that GPXs extensively participated in the growth and development of plant. The expression levels of GPXs could change the H2O2 content and determine oxidant stress. Overexpression exogenous or endogenous glutathione peroxidases in plants or microorganism could provide tolerance to multipe stresses include salt, chilling, or H2O2, and methyl viologen [22,26,27,29,53]. However, knockdown or knockout expression levels of GPXs had a bad effect on development of the plant with compromised ROS scavenging system and accumulation of H2O2 [28,54]. For RcGPXs, our laboratory has proved overexpression RcGPX5 transgenic Arabidopsis thaliana or Salvia miltiorrhiza presented tolerance to chilling or drought (unpublished). However members of GPXs presented divergent and redundant molecular functions partly due to the subcellular locations [44]. For seven AtGPXs, single mutation for any member made little influence on the shoot phenotypes, but depletion of both AtGPX1 and AtGPX7 which co-located in chloroplast could alter the leaf cell and chloroplast morphology [18,55]. So it might be a future decision that five RcGPXs members fulfilled respective functions on different biological processes, which was needed for R. crenulata to respond to complex environmental changes.
Detection of transcription initiation is a necessary and widespread mechanism for plants utilized by controlling gene transcription or not and expression quantity [56,57]. Promoter sequences located in upstream of gene coding regions play a vital role in regulation genes expression. There are some specific DNA sequences named cis-regulation elements located in promoter regions which are recognized by transcription factor, plant hormones, and stresses. Using PlantCARE software [58] Arabidopsis thaliana and Thellungiella salsuginea GPXs have different numbers and types of cis-acting elements in the promoters [44]. For example, abscisic acid recognizing element ABRE (TACGTG) was investigated in TsGPX2/4/5/7 and AtGPX1/4/5/8. Drought inducibility element MBS (TAACTG) existed in TsGPX1/2/3/4/7 and AtGPX2/5/7 [58]. In addition, promoters also contained tissue specific elements like meristem or seed specific cis-regulatory sites which determined the positions for different GPXs members function [28]. All of these elements could explain the expression patterns of GPXs response to multiple stresses treatment. In this paper, the expression patterns were different among RcGPXs members even for RcGPX4 and RcGPX5 which were the closest genes in protein sequences and had similar subcellular location. Although we don’t obtain the promoters’ information, the expression specificity for RcGPXs was presented by the way of tissue, development time, and stressful conditions. Therefore, it needs further studies for promoter sequences of RcGPXs by genome walking and transgenic technology which provided our new sights to their functions.
As mentioned above, plant hormones or transcription factors could change the expression levels of RcGPXs by the cis-regulation elements in the promoter region, however, RcGPXs could directly interact with special proteins to participate in multiple signal pathways inversely. For example, Arabidopsis thaliana GPX3 physically interacted with the negative factors in ABA signal pathway to regulate plants response for drought stress [25,59]. In order to further research the function of RcGPXs, yeast two-hybrid assays were conducted to validate the interaction proteins from multiple pathways. Interesting, RcGPX2 was highest identity with AtGPX3 on protein sequence, subcellular location and transmembrane domain could not interact with AtABI2. However, RcGPX5 located in cytoplasm with RcGPX7 located in chloroplast presented interactions with AtABI2 instead of RcGPX4. Furthermore, a calmodulin gene AtCML38 and NAC transcription factor AtNAC102 were confirmed with important hypoxia responsive sensor were also interacted with RcGPXs specially [60,61]. However, although it was reported that the phosphatase activity of AtABI2 was highly sensitive to H2O2 and the redox state was coupled with AtGPX3, the function model for RcGPXs with interaction proteins needed further research. In addition, we also screened the interaction AtTrx protein for RcGPXs. As is well known, Trx proteins participate in glutathione cycle by acting as substrates for GPXs [19,62]. The plant Trx gene family contained multiply members and divided into h, f, m, o, x, and y types located in cytoplast, chloroplast, or mitochondria. However, there were no stable relations existed in Trxs and GPXs by which special GPX enzyme employed some type Trx proteins at the same subcellular location [47]. For sixteen Arabidopsis Trx members, eight AtTrxs interacted with RcGPXs and AtTrxm2 could interact with all RcGPXs. Besides, five of eight AtTrxs interacted with one RcGPX protein. Although the results by yeast two-hybrid were not impeccable, it also enlarged the knowledge for cellular homeostasis by RcGPXs with Trxs.
Different subcellular locations, expression patterns of tissue or stresses, and interaction proteins showed RcGPXs have extensive and complex function for R. crenulata growth and development. RcGPXs might be first in the general control of H2O2 homeostasis, and the second in linking with other pathways [25]. On the one hand, RcGPXs could change the contents of ROS or ratio of GSH/GSSG to create a different intracellular environment to mobilize other gene expressions, on the other hand, RcGPXs could participate in multiple pathways by protein-protein interaction and thiol-disulfide exchange. Consequently, RcGPXs plays an irreplaceable role for R. crenulata adapting to the treacherous weather in the highlands.
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
In July of 2015, we cultured R. crenulata wild materials around Lhasa city of Tibet. Leaves of R. crenulata from Mila Mountain were used as explants for tissue culture and regeneration seedlings were used for subsequently experiments. For tissue-specificity analysis, four organs contained callus, roots, stems, leaves and two different developmental stages of leaves or stems were collected. Young and old leaves or stems were distinguished according to the position on the plant. With development of Rhodiola crenulata seedlings, the old leaves were under the young leaves with thinker in transaction and greener in color. The old stems were all in red color which were very different from young stems. The similar size and development stage of plantlets were selected to stressful treatment. For drought, the whole plantlets were immersed in MS liquid media contained 20% PEG-4000 and slightly shaken with 80 rpm. Drought stress was treated for 0 day, 1 day, 2 day, 3 day, and 5 day. For (ultraviolet ray) UA stress, the whole plantlets were exposed in UA with wave length 234 nm for 8 h and dark for 16 h one day. The plantlets were treated for 0 day, 1 day, 2 day, 3 day, 5 day and 7 day. For cold, the whole plantlets were treated with ice-water mixture for 0 °C. For hypoxia, we used blooding treatment to build the low-oxygen circumstance. The whole plantlets were placed on the bottom of glass bottle filled with sterile water, and the liquid level was 5 cm higher than the apex of seedlings. These two stresses were treated under dark for 0 day, 1 day, 3 day, 5 day, 7 day, and 10 day. After treatments, the leaves of all materials were cut carefully and divided into two parts with one was quick-freeze by liquid nitrogen with the remained fresh leaves were used to measure physiological indices. For other stresses, the whole plantlets were submerged in MS liquid media contained different heavy metals and plant hormones (25 μM Hg+, 50 μM Cu2+, 50 μM Co2+, 50 μM Ag+, 100 μM GA3, 100 μM IAA, 100 μM MeJA, and 50 μM ABA). The sets were slightly shaken with 80 rpm for 6 hours expect ABA. For ABA, five time periods were selected at 0 h, 6 h, 12 h, 24 h, and 36 h. Finally, the young leaves were cut and quick-freeze in liquid nitrogen.
4.2. Measurement of Physiological Indices
H2O2 contents for drought, UA, cold, and hypoxia stresses were measured by 752-UV spectrophotometry. Content of H2O2 was obtained according to the method of the H2O2 Assay Kit (KeyGEN Biotech, Nanjing, China). All young leaves for these assays were come from three individuals. All data are presented as the means ± standard error (SE) of at least three replicates. The Student’s t-test was used to test the significance of differences compared to pre-treatment materials. And Asterisks (* and **) indicate a significant difference between the controls and transgenic plants at p < 0.05 and 0.01 respectively.
4.3. RNA Extraction and First Strand Synthesis
Total RNA for all treatments of materials was extracted using Easy RNA extract kit (Promega, Beijing, China). The quality and concentration of RNA were measured by agar gel-electrophoresis and Nano Drop 1000. Values of OD260/280 and OD 260/230 were kept in 1.8–2.2 and the concentration of RNA was higher than 200 ng/μL. Two μg RNA were used for the first strand synthesis by M-MLV (Takara, Japan) with 20 μL volumes. For any one of these treatments, three individuals were conducted separately and then cDNA were mixed into a pool. Finally, the cDNA pool was diluted to 10 times by ddH2O for subsequently experiments.
4.4. Primers Design, PCR Amplification and Vectors Construction of Yeast Two-Hybrid
The RcGPXs primers were designed by primer premier 5 according to the predicted coding sequences of assembly unigenes. To construct recombination vectors of yeast two-hybrid, the potential genes were obtained for Arabidopsis database (Tair: http://www.arabidopsis.org/). And primers with enzyme sites were designed according to the CDS regions. Then sequences were amplified with high fidelity thermostable DNA polymerase PrimeSTAR Max DNA Polymerase (Takara, Japan). The products were linked with pEASYT1 vectors (TransGen Biotech, Beijing, China) and sequencing by Sanger sequencing (Biotech, Shanghai, China). In addition, the recombination vectors for pGADT7 and pGBKT7 were obtained by double digestion method. Finally, the vectors were validated by PCR and sequencing.
4.5. Bioinformation Analysis
The DNA sequences of RcGPXs were validated by Sanger sequencing again. And the protein sequences were translated according to DNA sequences. Using Blastp, RcGPXs protein sequences from NCBI, the homologous genes of other species were obtained and a phylogeny tree of these genes was constructed by Mega 5.0. In addition, conserve domains were predicted by MEME (http://meme-suite.org/index.html), the protein tertiary structures were predicted by Swissmodel online website (https://swissmodel.expasy.org/interactive), and the protein sequences blast was conducted by DNAMAN. And the transmembrane structures were searched by TMHMM Server 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
4.6. Yeast Two-Hybrid Assays
The bait and prey recombination vectors were transformed into yeast strain Y2H Gold by LiTE-PEG method. Positively yeast clones grown in SD-Trp-Leu were validated by PCR and then cultured on the solid media SD-Trp-Leu-His and the well grown clones were regarded for interaction. Then the clones were cultured in liquid media SD-Trp-Leu for 16 hours to OD600 0.6–0.8 and diluted to OD600 0.01 by sterile water. At last, 6 microliters was spotted on SD-Trp-Leu-His solid media, using SD-Trp-Leu solid media as control.
4.7. Quantitative Real-Time PCR and Statistical Analysis
For gene expression level quantitative analysis, qRT-PCR was completed by iQ5 (Bio-Rad, Hercules, CA, USA). The reactions were 20 μL volumes contained 10 μL SYBR I (Roche, China), 10ng cDNA. Reference genes were RcGAPDH. Primers of RcGPXs for real-time PCR were designed by primer premier 5 (Table S3). The gene expression levels were calculated by the method 2−ΔΔCt with three biological replicates.
5. Conclusions
In conclusion, we isolated five members of the glutathione peroxidase gene family from a traditional Chinese medicinal plant Rhodiola crenulata. And RcGPXs protein sequences were analyzed by bioinformation for alignment, the conserve domain searching, sub-cellular location, and tertiary structure construction. Then the expression levels of RcGPXs under different tissues, abiotic stresses, and plant hormones were investigated by qRT-PCR. The results showed that RcGPXs were highly expressed in green and young organs, and in response to drought, cold, ultraviolet ray, and flooding stresses. Finally, several putative interaction proteins were obtained by yeast two-hybrid. RcGPXs members could present redundancy and divergence function in the plant body, which plays an important role for R. crenulata to adapt to the treacherous weather in the highlands.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/11/3329/s1.
Author Contributions
Author L.Z. designed the experiments, analyzed the data and wrote the manuscript. Author M.W. and D.Y. provided assistance on physical indices measurement, primers design, and yeast two-hybrid experiments. Y.T., T.W., and C.C. made a significant contribution for manuscript touches. Corresponding author W.S. was responsible for the overall concept, designing of problem, and experiments.
Funding
There were no funding sources relevant to this study.
Acknowledgments
We were immensely grateful to Dong-Sheng Ren, one manager of company (Tibet Rhodiola Pharmaceutical Holding Co.), who helped us gather the wild materials around Lhasa city. Secondly, we would like to thank Zhe-Hua Yang, a doctor of linguistics and applied linguistics, college of arts, Nankai University for editing the manuscript language.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kosanovic, D.; Tian, X.; Pak, O.; Lai, Y.J.; Hsieh, Y.L.; Seimetz, M.; Weissmann, N.; Schermuly, R.T.; Dahal, B.K. Rhodiola: An ordinary plant or a promising future therapy for pulmonary hypertension? a brief review. Pulm. Circ. 2013, 3, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Grech-Baran, M.; Syklowska-Baranek, K.; Pietrosiuk, A. Biotechnological approaches to enhance salidroside, rosin and its derivatives production in selected Rhodiola spp. in vitro cultures. Phytochem. Rev. 2015, 14, 657–674. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.Q.; Zhou, Y.; Zeng, Y.S.; Lin, Y.K.; Li, Y.; Zhong, Z.Q.; Chan, W.Y. Protective effects of a Rhodiola crenulata extract and salidroside on hippocampal neurogenesis against streptozotocin-induced neural injury in the rat. PLoS ONE 2012, 7, e29641. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.C.; Lin, J.G.; Pai, P.Y.; Lai, M.H.; Lin, Y.M.; Yeh, Y.L.; Cheng, S.M.; Liu, Y.; Huang, C.Y.; Lee, S.D. Effects of Rhodiola crenulata on mice hearts under severe sleep apnea. BMC Complement. Altern. Med. 2015, 15, 198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Meng, S.Y.; Rao, G.Y. Phylogeography of Rhodiola kirilowii (Crassulaceae): A story of miocene divergence and quaternary expansion. PLoS ONE 2014, 9, e112923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Meng, S.Y.; Allen, G.A.; Wen, J.; Rao, G.Y. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol. Phylogenet. Evol. 2014, 77, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Niu, F.; Lin, Z.; Luo, J.; Liu, M. Effects of local factors and climate on permafrost conditions and distribution in Beiluhe basin, Qinghai-Tibet Plateau, China. Sci. Total Environ. 2017, 581–582. [Google Scholar] [CrossRef] [PubMed]
- Barah, P.; Jayavelu, N.D.; Rasmussen, S.; Nielsen, H.B.; Mundy, J.; Bones, A.M. Genome-scale cold stress response regulatory networks in ten Arabidopsis thaliana ecotypes. BMC Genom. 2013, 14, 722. [Google Scholar] [CrossRef] [PubMed]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007, 50, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hare, P.D.; Cress, W.A.; Staden, J.V. Dissecting the roles of osmolyte accumulation during stress. Plant Cell Environ. 1998, 21, 535–553. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Anjum, S.A.; Xie, X.Y.; Wang, L.C.; Saleem, M.F.; Man, C.; Lei, W. Morphological, physiological and biochemical responses of plants to drought stress. Afr. J. Agric. Res. 2011, 6, 2026–2032. [Google Scholar] [CrossRef]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Csiszár, J.; Gallé, Á.; Horváth, E.; Dancsó, P.; Gombos, M.; Váry, Z.; Erdei, L.; Györgyey, J.; Tari, I. Different peroxidase activities and expression of abiotic stress-related peroxidases in apical root segments of wheat genotypes with different drought stress tolerance under osmotic stress. Plant Physiol. Biochem. 2012, 52, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Chang, C.C.; Slesak, I.; Jorda, L.; Sotnikov, A.; Melzer, M.; Miszalski, Z.; Mullineaux, P.M.; Parker, J.E.; Karpinska, B.; Karpinski, S. Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 2009, 150, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.S.; Didierjean, C.; Navrot, N.; Panjikar, S.; Mulliert, G.; Rouhier, N.; Jacquot, J.P.; Aubry, A.; Shawkataly, O.; Corbier, C. Crystal structures of a poplar thioredoxin peroxidase that exhibits the structure of glutathione peroxidases: Insights into redox-driven conformational changes. J. Mol. Biol. 2007, 370, 512–529. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Herbette, S.; Menn, A.L.; Rousselle, P.; Ameglio, T.; Faltin, Z.; Branlard, G.; Eshdat, Y.; Julien, J.L.; Drevet, J.R.; Roeckel-Drevet, P. Modification of photosynthetic regulation in tomato overexpressing glutathione peroxidase. Biochim. Biophys. Acta 2005, 1724, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Faltin, Z.; Holland, D.; Velcheva, M.; Tsapovetsky, M.; Roeckel-Drevet, P.; Handa, A.K.; Abu-Abied, M.; Friedman-Einat, M.; Eshdat, Y.; Perl, A. Glutathione peroxidase regulation of reactive oxygen species level is crucial for in vitro plant differentiation. Plant Cell Physiol. 2010, 51, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Maiorino, M.; Gregolin, C.; Ursini, F. Phospholipid hydroperoxide glutathione peroxidase. Int. J. Tissue React. 2007, 38, 41–48. [Google Scholar] [CrossRef]
- Gueta-Dahan, Y.; Yaniv, Z.; Zilinskas, B.A.; Ben-Hayyim, G. Salt and oxidative stress_ similar and specific responses and their relation to salt tolerance in Citrus. Planta 1997, 203, 469. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Lv, D.; Wang, P.; Wang, X.C.; Chen, J.; Miao, C.; Song, C.P. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y.; Xu, H.; Li, G.; Yu, A.; Yu, X.; Hu, W.; Zeng, X.; Li, S.; Wang, Y.; Hu, Z. Cloning a glutathione peroxidase gene from Nelumbo nucifera and enhanced salt tolerance by overexpressing in rice. Mol. Biol. Rep. 2014, 41, 4919–4927. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Miyao, K.; Gaber, A.; Takeda, T.; Kanaboshi, H.; Miyasaka, H.; Shigeoka, S. Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 2004, 37, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Passaia, G.; Fonini, L.S.; Caverzan, A.; Jardim-Messeder, D.; Christoff, A.P.; Gaeta, M.L.; de Araujo Mariath, J.E.; Margis, R.; Margis-Pinheiro, M. The mitochondrial glutathione peroxidase GPX3 is essential for H2O2 homeostasis and root and shoot development in rice. Plant Sci. 2013, 208, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Vaghchhipawala, Z.; Li, W.; Asard, H.; Dickman, M.B. Tomato phospholipid hydroperoxide glutathione peroxidase inhibits cell death induced by Bax and oxidative stresses in yeast and plants. Plant Physiol. 2004, 135, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Song, Y.; Zhang, G.; Ju, L.; Zhang, J.; Yu, Y.; Niu, N.; Wang, J.; Ma, S. De Novo assembly and transcriptome analysis of wheat with male sterility induced by the chemical hybridizing agent SQ-1. PLoS ONE 2015, 10, e0123556. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, A.; Pech, N.; Martin, J.F.; McCairns, R.J.S.; Mevy, J.P.; Chappaz, R.; Gilles, A. Challenges and advances for transcriptome assembly in non-model species. PLoS ONE 2017, 12, e0185020. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.; Zhang, R.C.; Wang, L.; Zhang, Y.Q. Researches on transcriptome sequencing in the study of traditional Chinese medicine. Evid. Based Complement. Altern. Med. 2017, 7521363. [Google Scholar] [CrossRef] [PubMed]
- Narnoliya, L.K.; Kaushal, G.; Singh, S.P.; Sangwan, R.S. De novo transcriptome analysis of rose-scented geranium provides insights into the metabolic specificity of terpene and tartaric acid biosynthesis. BMC Genom. 2017, 18, 74. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Zhang, Y.; Song, J.; Zhao, L.; Wang, Z. De novo transcriptome sequencing in Salvia miltiorrhiza to identify genes involved in the biosynthesis of active ingredients. Genomics 2011, 98, 272–279. [Google Scholar] [CrossRef]
- Shiraishi, A.; Murata, J.; Matsumoto, E.; Matsubara, S.; Ono, E.; Satake, H. De Novo transcriptomes of Forsythia koreana using a novel assembly method: Insight into tissue- and species-specific expression of lignan biosynthesis-related gene. PLoS ONE 2016, 11, e0164805. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Yamazaki, M.; Takahashi, H.; Nakamura, M.; Kojoma, M.; Suzuki, H.; Saito, K. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front Plant Sci. 2016, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Zhan, C.; Yang, Y.; Wang, X.; Yang, T.; Zhao, Z.; Zhang, Q.; Li, X.; Hu, X. The transcript profile of a traditional Chinese medicine, Atractylodes lancea, revealing its sesquiterpenoid biosynthesis of the major active components. PLoS ONE 2016, 11, e0151975. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Hou, X.; Fang, C.; An, J.; Ha, D.; Qiu, L.; Ju, Y.; Zhao, H.; Du, W.; Qi, J.; et al. Transcriptome analysis of distinct Lindera glauca tissues revealed the differences in the unigenes related to terpenoid biosynthesis. Gene 2015, 559, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ding, G.; Lin, H.; Cheng, H.; Kong, Y.; Wei, Y.; Fang, X.; Liu, R.; Wang, L.; Chen, X.; et al. Transcriptome analysis of medicinal plant Salvia miltiorrhiza and identification of genes related to tanshinone biosynthesis. PLoS ONE 2013, 8, e80464. [Google Scholar] [CrossRef]
- Wei, S.; Ma, X.; Pan, L.; Miao, J.; Fu, J.; Bai, L.; Zhang, Z.; Guan, Y.; Mo, C.; Huang, H.; et al. Transcriptome analysis of Taxillusi chinensis (DC.) danser seeds in response to water loss. PLoS ONE 2017, 12, e0169177. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Chen, H.; Wu, B.; Nelson, D.R.; Wu, K.; Liu, C. Computational identification and systematic classification of novel cytochrome P450 genes in Salvia miltiorrhiza. PLoS ONE 2014, 9, e115149. [Google Scholar] [CrossRef]
- Li, M.Y.; Tan, H.W.; Wang, F.; Jiang, Q.; Xu, Z.S.; Tian, C.; Xiong, A.S. De novo transcriptome sequence assembly and identification of AP2/ERF transcription factor related to abiotic stress in parsley (Petroselinum crispum). PLoS ONE 2014, 9, e108977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, AM. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop. Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Bela, K.; Horvath, E.; Galle, A.; Szabados, L.; Tari, I.; Csiszar, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.J.M. Accumulators and excluders—Strategies in the response of plants to heavy metals. J. Plant Nutr. 1981, 3, 643–654. [Google Scholar] [CrossRef]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Gelhaye, E.; Rouhier, N.; Gérard, J.; Jolivet, Y.; Gualberto, J.; Navrot, N.; Ohlsson, P.I.; Wingsle, G.; Hirasawa, M.; Knaff, D.B.; et al. A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc. Natl. Acad. Sci. USA 2004, 101, 14545–14550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Meng, S.Y.; Wen, J.; Rao, G.Y. DNA barcoding of Rhodiola (crassulaceae): A case study on a group of recently diversified medicinal plants from the Qinghai-Tibetan Plateau. PLoS ONE 2015, 10, e0119921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Q.; Meng, S.Y.; Wen, J.; Rao, G.Y. Phylogenetic relationships and character evolution of Rhodiola (Crassulaceae) based on nuclear ribosomal ITS and plastid trnL-F and psbA-trnH sequences. Syst. Bot. 2014, 39, 441–451. [Google Scholar] [CrossRef]
- Fu, Y.; Li, L.; Hao, S.; Guan, R.; Fan, G.; Shi, C.; Wan, H.; Chen, W.; Zhang, H.; Liu, G.; et al. Draft genome sequence of the Tibetan medicinal herb Rhodiola crenulata. Gigascience 2017, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Chang, K.; Zeng, L.; Liu, X.; Qiu, F.; Zheng, W.; Quan, H.; Liao, Z.; Chen, M.; Huang, W.; et al. Engineering salidroside biosynthetic pathway in hairy root cultures of Rhodiola crenulata based on metabolic characterization of tyrosine decarboxylase. PLoS ONE 2013, 8, e75459. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, Z.; Jaakola, L.; Neubauer, P.; Hohtola, A. Isolation and genotype-dependent, organ-specific expression analysis of a Rhodiola rosea cDNA encoding tyrosine decarboxylase. J. Plant Physiol. 2009, 166, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.Z.; Lei, Z.; Yin, L.J.; Ming, C.; Wang, Q.Y.; Li, L.C.; Xu, Z.S.; Ma, Y.Z. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis. PLoS ONE 2013, 8, e73989. [Google Scholar] [CrossRef] [PubMed]
- Gaber, A.; Ogata, T.; Maruta, T.; Yoshimura, K.; Tamoi, M.; Shigeoka, S. The involvement of Arabidopsis glutathione peroxidase 8 in the suppression of oxidative damage in the nucleus and cytosol. Plant Cell Physiol. 2012, 53, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Passaia, G.; Queval, G.; Bai, J.; Margis-Pinheiro, M.; Foyer, C.H. The effects of redox controls mediated by glutathione peroxidases on root architecture in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P. Recent advances in computational promoter analysis in understanding the transcriptional regulatory network. Biochem. Biophys. Res. Commun. 2003, 309, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, J.; Ma, T.; Li, H.; Wang, N.; Li, Z.; Zhang, Z.; Zhou, Y. The glutathione peroxidase gene family in Thellungiella salsuginea: Genome-wide identification, classification, and gene and protein expression analysis under stress conditions. Int. J. Mol. Sci. 2014, 15, 3319–3335. [Google Scholar] [CrossRef] [PubMed]
- Meinhard, M.; Rodriguez, PL.; Grill, E. The sensitivity of ABI2 to hydrogen peroxide links the abscisic acid-response regulator to redox signalling. Planta 2002, 214, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Lokdarshi, A.; Conner, W.C.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 2016, 170, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.A.; Wilson, I.W.; Llewellyn, D.J.; Dennis, E.S. The low-oxygen-induced NAC domain transcription factor ANAC102 affects viability of Arabidopsis seeds following low-oxygen treatment. Plant Physiol. 2009, 149, 1724–1738. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, A.; Yabuta, Y.; Takeda, T.; Nakano, Y.; Shigeoka, S. Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 2006, 273, 5589–5597. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).