Newly Identified Essential Amino Acids Affecting Chlorella ellipsoidea DGAT1 Function Revealed by Site-Directed Mutagenesis
Abstract
:1. Introduction
2. Results
2.1. Sequence Conservation among DGAT1s from Different Species and the Selection of Mutagenesis Sites
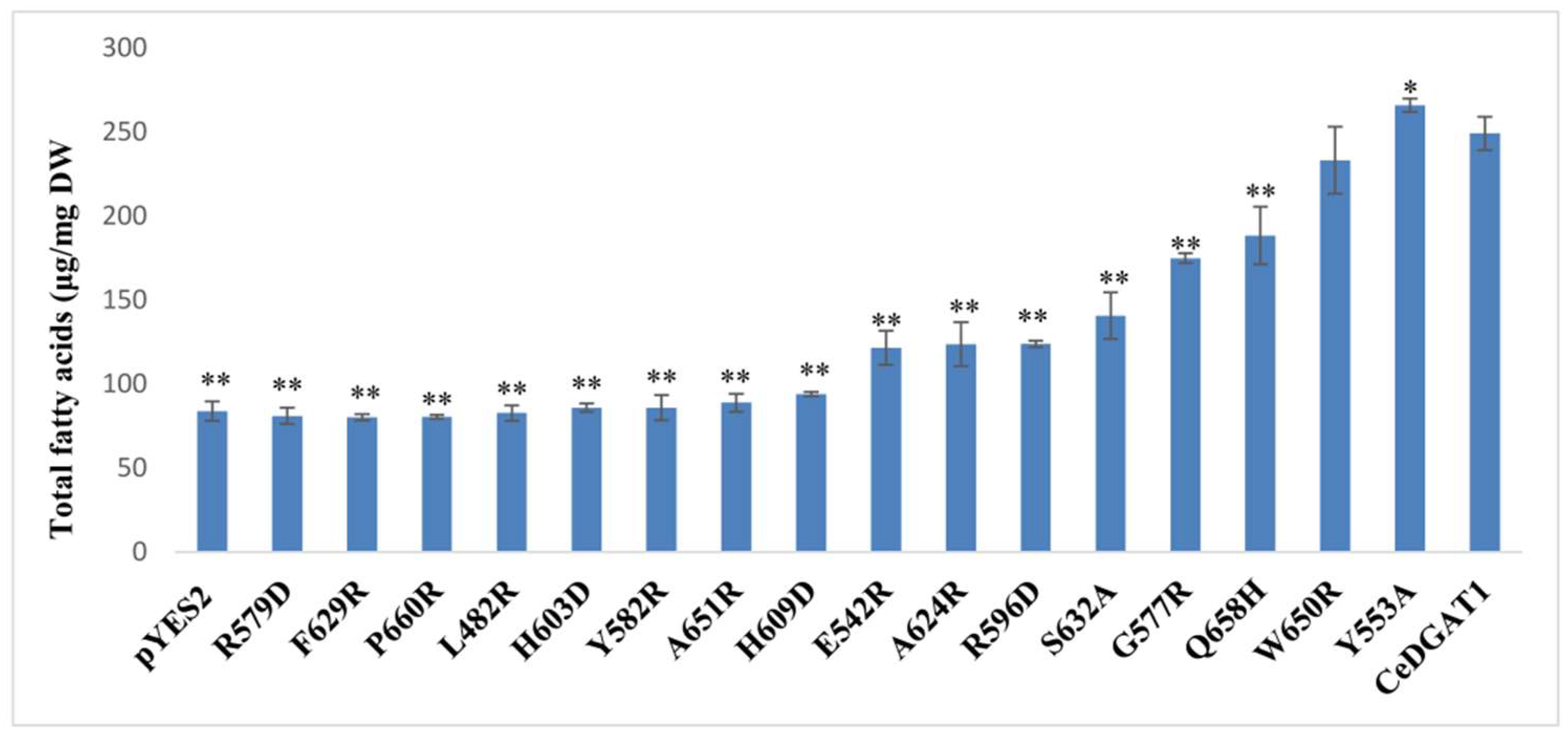
2.2. Analysis of Mutants Function by GC–MS
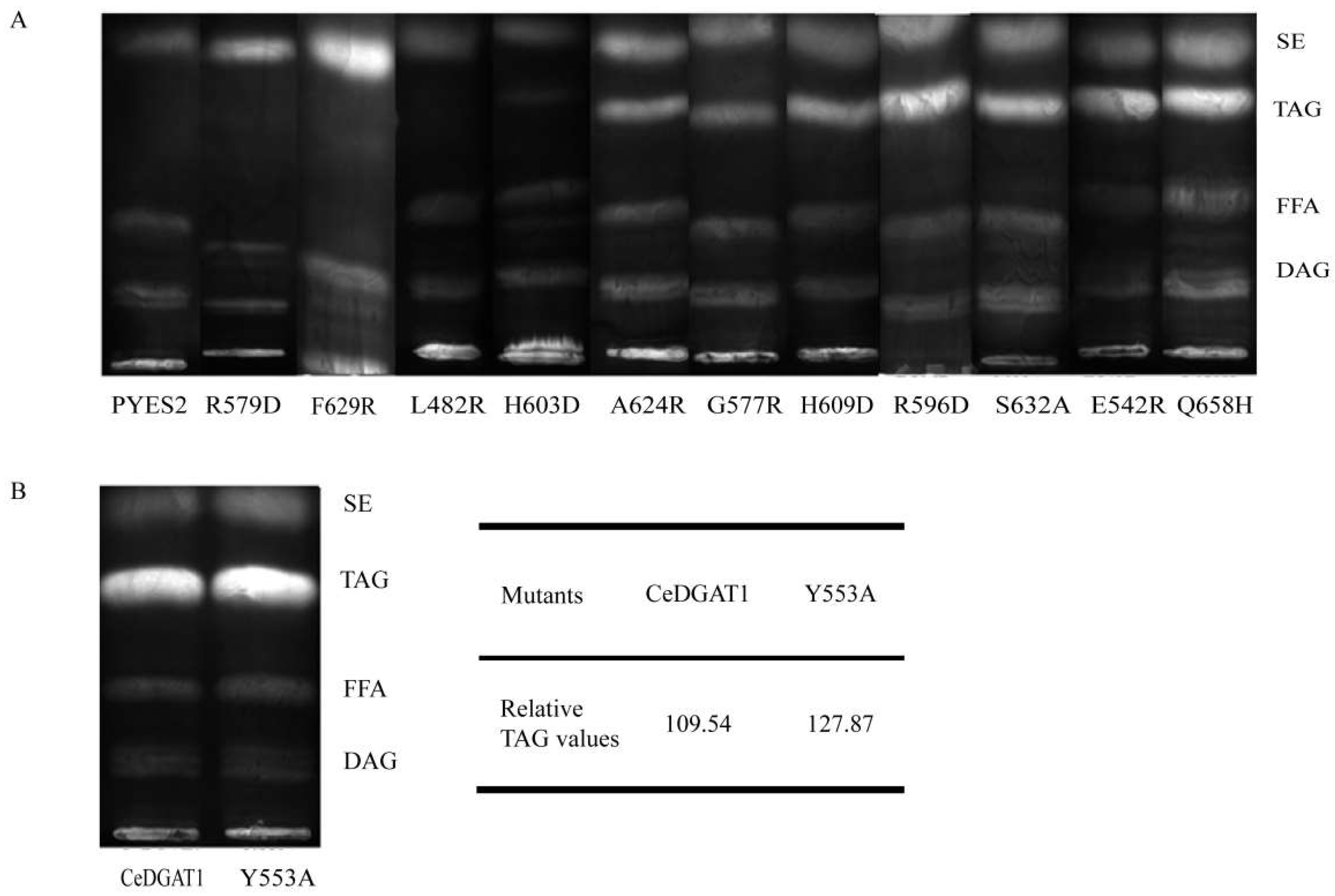
2.3. Mutant Functional Analysis by TLC
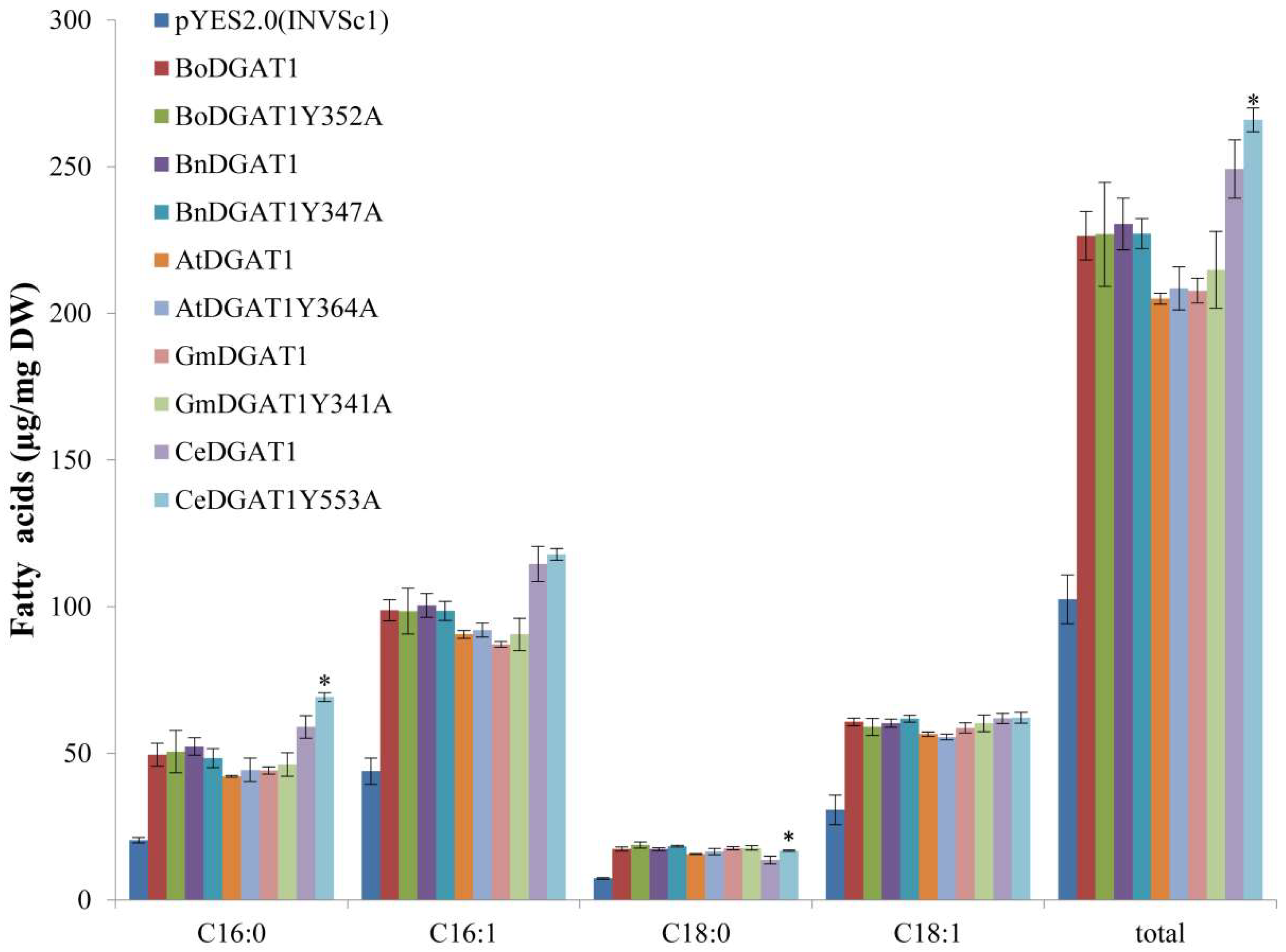
2.4. Mutation of DGAT1 from Different Species Corresponding to Position 553 of CeDGAT1
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Plasmids
4.2. CeDGAT1 Site-Directed Mutagenesis and Yeast Transformation
4.3. RT-PCR Analysis of Yeast Transformed with CeDGAT1 and Mutants
4.4. Lipid Analysis by GC–MS and TLC
4.5. Alignment and Three-Dimensional Structure and Topology Model Prediction
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DGAT | Diacylglycerol acyltransferase |
| TAG | Triacylglycerol |
| TMD | Transmembrane domain |
| ER | Endoplasmic reticulum |
References
- Turchetto-Zolet, A.C.; Maraschin, F.S.; de Morais, G.L.; Cagliari, A.; Andrade, C.M.B.; Margis-Pinheiro, M.; Margis, R. Evolutionary view of acyl-CoA diacylglycerol acyltransferase (DGAT), a key enzyme in neutral lipid biosynthesis. BMC Evol. Biol. 2011, 11, 263. [Google Scholar] [CrossRef] [PubMed]
- Shockey, J.M.; Gidda, S.K.; Chapital, D.C.; Kuan, J.C.; Dhanoa, P.K.; Bland, J.M.; Rothstein, S.J.; Mullen, R.T.; Dyer, J.M. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 2006, 18, 2294–2313. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Enugutti, B.; Rajakumari, S.; Rajasekharan, R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006, 141, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.H.; Krishna, T.H.A.; Saha, S.; Negi, A.S.; Rajasekharan, R. Defective in Cuticular Ridges (DCR) of Arabidopsis thaliana, a gene associated with surface cutin formation, encodes a soluble diacylglycerol acyltransferase. J. Biol. Chem. 2010, 285, 38337–38347. [Google Scholar] [CrossRef] [PubMed]
- Cases, S.; Novak, S.; Zheng, Y.W.; Myers, H.M.; Lear, S.R.; Sande, E.; Welch, C.B.; Lusis, A.J.; Spencer, T.A.; Krause, B.R.; et al. ACAT-2, a second mammalian acyl-CoA : Cholesterol acyltransferase-its cloning, expression, and characterization. J. Biol. Chem. 1998, 273, 26755–26764. [Google Scholar] [CrossRef] [PubMed]
- Guihéneuf, F.; Leu, S.; Zarka, A.; Khozin-Goldberg, I.; Khalilov, I.; Boussiba, S. Cloning and molecular characterization of a novel acyl-CoA:diacylglycerol acyltransferase 1-like gene (PtDGAT1) from the diatom Phaeodactylum tricornutum. FEBS J. 2011, 278, 3651–3666. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, D.H.; Lu, C.; Hills, M.J. Cloning of a cDNA encoding diacylglycerol acyltransferase from Arabidopsis thaliana and its functional expression. FEBS Lett. 1999, 452, 145–149. [Google Scholar] [CrossRef]
- Bouvier-Nave, P.; Benveniste, P.; Oelkers, P.; Sturley, S.L.; Schaller, H. Expression in yeast and tobacco of plant cDNAs encoding acyl CoA: Diacylglycerol acyltransferase. Eur. J. Biochem. 2000, 267, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Nykiforuk, C.L.; Furukawa-Stoffer, T.L.; Huff, P.W.; Sarna, M.; Laroche, A.; Moloney, M.M.; Weselake, R.J. Characterization of cDNAs encoding diacylglycerol acyltransferase from cultures of Brassica napus and sucrose-mediated induction of enzyme biosynthesis. Biochim. Biophys. Acta 2002, 1580, 95–109. [Google Scholar] [CrossRef]
- He, X.; Turner, C.; Chen, G.Q.; Lin, J.T.; McKeon, T.A. Cloning and characterization of a cDNA encoding diacylglycerol acyltransferase from castor bean. Lipids 2004, 39, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Milcamps, A.; Tumaney, A.W.; Paddock, T.; Pan, D.A.; Ohlrogge, J.; Pollard, M. Isolation of a gene encoding a 1,2-diacylglycerol-sn-acetyl-CoA acetyltransferase from developing seeds of Euonymus alatus. J. Biol. Chem. 2005, 280, 5370–5377. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Zhang, J.S.; Gai, J.Y.; Chen, S.Y. Cloning and comparative analysis of the gene encoding diacylglycerol acyltransferase from wild type and cultivated soybean. Theor. Appl. Genet. 2006, 112, 1086–1097. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Li, R.; Hatanaka, T.; Hildebrand, D. Cloning and functional analysis of two type 1 diacylglycerol acyltransferases from Vernonia galamensis. Phytochemistry 2008, 69, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Misra, A.; Khan, K.; Niranjan, A.; Nath, P.; Sane, V.A. Over-expression of JcDGAT1 from Jatropha curcas increases seed oil levels and alters oil quality in transgenic Arabidopsis thaliana. Phytochemistry 2013, 96, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.K.; Huang, W.J.; Chang, J.M.; Sebastian, A.; Li, Y.G.; Li, H.Y.; Wu, X.X.; Zhang, B.B.; Meng, F.L.; Li, W.B. Overexpression of SiDGAT1, a gene encoding acyl-CoA:diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ 2014, 119, 399–410. [Google Scholar] [CrossRef]
- Kirchner, L.; Wirshing, A.; Kurt, L.; Reinard, T.; Glick, J.; Cram, E.J.; Jacobsen, H.-J.; Lee-Parsons, C.W.T. Identification, characterization, and expression of diacylgylcerol acyltransferase type-1 from Chlorella vulgaris. Algal Res. 2016, 13, 167–181. [Google Scholar] [CrossRef]
- Lung, S.C.; Weselake, R.J. Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids 2006, 41, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Jako, C.; Kumar, A.; Wei, Y.; Zou, J.; Barton, D.L.; Giblin, E.M.; Covello, P.S.; Taylor, D.C. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001, 126, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Roesler, K.; Shen, B.; Bermudez, E.; Li, C.J.; Hunt, J.; Damude, H.G.; Ripp, K.G.; Everard, J.D.; Booth, J.R.; Castaneda, L.; et al. An improved variant of soybean type 1 diacylglycerol acyltransferase increases the oil content and decreases the soluble carbohydrate content of soybeans. Plant Physiol. 2016, 171, 878–893. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Francis, T.; Mietkiewska, E.; Giblin, E.M.; Barton, D.L.; Zhang, Y.; Zhang, M.; Taylor, D.C. Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 2008, 6, 799–818. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K. A superfamily of membrane-bound O-acyltransferases with implications for Wnt signaling. Trends Biochem. Sci. 2000, 25. [Google Scholar] [CrossRef]
- Guo, Z.Y.; Lin, S.; Heinen, J.A.; Chang, C.C.; Chang, T.Y. The active site His-460 of human acyl-coenzyme A: Cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. J. Biol. Chem. 2005, 280, 37814–37826. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Lu, X.; Chang, C.C.; Chang, T.Y. Human acyl-coenzyme A: Cholesterol acyltransferase expressed in Chinese hamster ovary cells: Membrane topology and active site location. Mol. Biol. Cell 2003, 14, 2447–2460. [Google Scholar] [CrossRef] [PubMed]
- McFie, P.J.; Banman, S.L.; Kary, S.; Stone, S.J. Murine Diacylglycerol Acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. J. Biol. Chem. 2011, 286, 28235–28246. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Zhu, L.; Schwaneberg, U. Directed evolution 2.0: Improving and deciphering enzyme properties. Chem. Commun. 2015, 51, 9760–9772. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xu, Y.; Siloto, R.M.P.; Caldo, K.M.P.; Vanhercke, T.; Tahchy, A.E.; Niesner, N.; Chen, Y.; Mietkiewska, E.; Weselake, R.J. High-performance variants of plant diacylglycerol acyltransferase 1 generated by directed evolution provide insights into structure function. Plant J. 2017, 92, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Siloto, R.M.P.; Lehner, R.; Stone, S.J.; Weselake, R.J. Acyl-CoA:diacylglycerol acyltransferase: Molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 2012, 51, 350–377. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Allen, W.B.; Roesler, K.; Williams, M.E.; Zhang, S.; Li, J.; Glassman, K.; Ranch, J.; Nubel, D.; Solawetz, W.; et al. A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat. Genet. 2008, 40, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Fan, C.; Chen, Y.; Wang, J.; Yin, W.; Wang, R.R.; Hu, Z. Identification and characterization of an efficient acyl-CoA: Diacylglycerol acyltransferase 1 (DGAT1) gene from the microalga Chlorella ellipsoidea. BMC Plant Biol. 2017, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Omasits, U.; Ahrens, C.H.; Muller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sandager, L.; Gustavsson, M.H.; Stahl, U.; Dahlqvist, A.; Wiberg, E.; Banas, A.; Lenman, M.; Ronne, H.; Stymne, S. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 2002, 277, 6478–6482. [Google Scholar] [CrossRef] [PubMed]
- Elble, R. A simple and efficient procedure for transformation of yeasts. Biotechniques 1992, 13, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]


| Mutants | Primers: Mutagenic Oligonucleotide Sequence (5′-3′, Sense) |
|---|---|
| L482R | TGGTGGTGCCGGCCCGGGTGTACCAGACCAC |
| E542R | TGGCGGGCATTGTGCGGCGCTTGTTGAAGCT |
| Y553A | CGTTCCTACTTTGGCCGGCTGGCTCATCA |
| G577R | GATCACGTACTTCCGGGACCGCGAGTTCT |
| R579D | GTACTTCGGGGACGACGAGTTCTACAAGG |
| Y582R | GGACCGCGAGTTCCGCAAGGACTGGTGGA |
| R596D | TGGGGACTACTGGGAGCTGTGGAATGTGC |
| H603D | GAATGTGCCGGTCGACAAGTGGCTGCTGC |
| H609D | AGTGGCTGCTGCGGGATGTGTACTACCCCTG |
| A624R | TCCGAAGACGGTTCGGGCAGTGGCGGTGT |
| F629R | GGCAGTGGCGGTGCGCTTCGTGAGTGCCG |
| S632A | GGTGTTCTTCGTGGCTGCCGTGTTCCACG |
| W650R | CATGGTGCGCTTCCGGGCATTCTGGGGCC |
| A651R | GGTGCGCTTCTGGCGATTCTGGGGCCTCA |
| Q658H | GGGCCTCATGTTCCACGTGCCTCTGCTGT |
| P660R | CATGTTCCAGGTGCGTCTGCTGTTCTTGA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Guo, X.; Fan, C.; Chen, Y.; Wang, J.; Hu, Z. Newly Identified Essential Amino Acids Affecting Chlorella ellipsoidea DGAT1 Function Revealed by Site-Directed Mutagenesis. Int. J. Mol. Sci. 2018, 19, 3462. https://doi.org/10.3390/ijms19113462
Sun B, Guo X, Fan C, Chen Y, Wang J, Hu Z. Newly Identified Essential Amino Acids Affecting Chlorella ellipsoidea DGAT1 Function Revealed by Site-Directed Mutagenesis. International Journal of Molecular Sciences. 2018; 19(11):3462. https://doi.org/10.3390/ijms19113462
Chicago/Turabian StyleSun, Baocheng, Xuejie Guo, Chengming Fan, Yuhong Chen, Jingqiao Wang, and Zanmin Hu. 2018. "Newly Identified Essential Amino Acids Affecting Chlorella ellipsoidea DGAT1 Function Revealed by Site-Directed Mutagenesis" International Journal of Molecular Sciences 19, no. 11: 3462. https://doi.org/10.3390/ijms19113462





