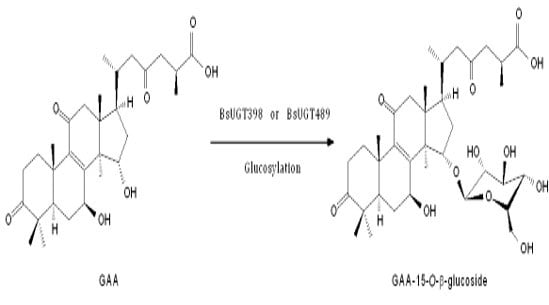
Uridine Diphosphate-Dependent Glycosyltransferases from Bacillus subtilis ATCC 6633 Catalyze the 15-O-Glycosylation of Ganoderic Acid A
Abstract
1. Introduction
2. Results
2.1. Confirming Biotransformation of GAA by Bacillus Subtilis ATCC 6633
2.2. Phylogenetic Analysis of GTs from B. subtilis ATCC 6633
2.3. Cloning and Overexpression of GT1 from B. subtilis ATCC 6633 in E. coli
2.4. Activity Assays of GT1 from B. subtilis ATCC 6633 Toward GAA
2.5. Optimal Catalyzing Conditions for BsUGT398 and BsUGT489
2.6. Isolation and Identification of the Biotransformation Metabolite
3. Discussion
4. Materials and Methods
4.1. Microorganism and Chemicals
4.2. Identification of Bacteria B. subtilis ATCC 6633 with Biotransformation Activity
4.3. UPLC
4.4. Expression and Purification of GT1 of B. subtilis ATCC 6633
4.5. In Vitro Biotransformation Assay
4.6. Scale-Up, Isolation, and Identification of the Biotransformation Product
4.7. Phylogenetic Analysis of GTs from B. subtilis ATCC 6633
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GAA | Ganoderic acid A |
| GT | Glycosyltransferase |
| GT1 | Glycosyltransferase family 1 |
| UDP | Uridine diphosphate |
| UGT | Uridine diphosphate-dependent glycosyltransferases |
References
- Shah, S.A.A.; Tan, H.L.; Sultan, S.; Faridz, M.A.B.M.; Shah, M.A.B.M.; Nurfazilah, S.; Hussain, M. Microbial-catalyzed biotransformation of multifunctional triterpenoids derived from phytonutrients. Int. J. Mol. Sci. 2014, 15, 12027–12060. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Saify, Z.S. Enzymatic biotransformation of terpenes as bioactive agents. J. Enzym. Inhib. Med. Chem. 2013, 28, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Leipold, D.; Scheller, M.C.; Haas, C.; Steingroewer, J.; Bley, T.; Neuhaus, H.E.; Mirata, M.A.; Schrader, J.; Ulber, R. Biotransformation of triterpenes. Process Biochem. 2011, 46, 1–15. [Google Scholar] [CrossRef]
- Parra, A.; Rivas, F.; Garcia-Granados, A.; Martinez, A. Microbial transformation of triterpenoids. Mini-Rev. Org. Chem. 2009, 6, 307–320. [Google Scholar] [CrossRef]
- Wu, J.W.; Zhao, W.; Zhong, J.J. Biotechnological production and application of ganoderic acids. Appl. Microbiol. Biotechnol. 2010, 87, 457–466. [Google Scholar]
- Xia, Q.; Zhang, H.; Sun, X.; Zhao, H.; Wu, L.; Zhu, D.; Yang, G.; Shao, Y.; Zhang, X.; Mao, X.; et al. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Asaka, Y. Structures of ganoderic acid A and B, two new lanostane type bitter triterpenes from Ganoderma lucidum (FR.) Karst. Helv. Chim. Acta 1982, 65, 611–619. [Google Scholar] [CrossRef]
- Jiang, J.; Grieb, B.; Thyagarajan, A.; Sliva, D. Ganoderic acids suppress growth and invasive behavior of breast cancer cells by modulating AP-1 and NF-kB signaling. Int. J. Mol. Med. 2008, 21, 577–584. [Google Scholar] [PubMed]
- Yao, X.; Li, G.; Xu, H.; Lu, C. Inhibition of the JAK-STAT3 signaling pathway by ganoderic acid A enhances chemosensitivity of HepG2 cells to cisplatin. Planta Med. 2012, 78, 1740–1748. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, D.; Tai, J.; Wang, L. Ganoderic acid A inhibits proliferation and invasion, and promotes apoptosis in human hepatocellular carcinoma cells. Mol. Med. Rep. 2017, 16, 3894–3900. [Google Scholar] [CrossRef] [PubMed]
- Akihisa, T.; Nakamura, Y.; Tagata, M.; Tokuba, H.; Yasukawa, K.; Uchiyama, E.; Suzukli, T.; Kimura, Y. Anti-inflammatory and anti-tumor-promoting effects of triterpene acids and sterols from the fungus Ganoderma lucidum. Chem. Biod. 2007, 4, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Koyama, K.; Imaizumi, T.; Akiba, M.; Kinoshita, K.; Takahashi, K.; Suzuki, A.; Yano, S.; Horie, S.; Watanabe, K.; Naoi, Y. Antinociceptive components of Ganoderma lucidum. Planta Med. 1997, 63, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chang, Q.; Wong, L.K.; Chong, F.S.; Li, R.C. Triterpene antioxidants from Ganoderma lucidum. Phytother. Res. 1999, 13, 529–531. [Google Scholar] [CrossRef]
- Chang, T.S.; Chiang, C.M.; Siao, Y.Y.; Wu, J.Y. Sequential biotransformation of antcin K by Bacillus subtilis ATCC 6633. Catalysts 2018, 8, 349. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, NS. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 716–739. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Yang, S.M.; Kim, S.Y.; Cha, M.N.; Ahn, J.H. Biosynthesis and production of glycosylated flavonoids in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Hofer, B. Recent developments in the enzymatic O-glycosylation of flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 4269–4281. [Google Scholar] [CrossRef] [PubMed]
- CAZy Database. Available online: www.cazy.org (accessed on 10 October 2018).
- Dai, L.; Li, J.; Yang, J.; Zhu, Y.; Men, Y.; Zeng, Y.; Cai, Y.; Dong, C.; Dai, Z.; Zhang, X.; et al. Use of a promiscuous glycosyltransferase from Bacillus subtilis 168 for the enzymatic synthesis of novel protopanaxtriol-type ginsenosides. J. Agric. Food Chem. 2017, 66, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Li, J.; Yao, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Yang, J.; Sun, Y. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 2017, 248, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Hu, Z.; Zhang, T.; Gong, T.; Chen, J.; Zhu, P.; Li, Y.; Yang, J. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab. Eng. 2017, 44, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Jin, Y.; Wang, C.; Kim, Y.J.; Perez, J.E.J.; Baek, N.I.; Mathiyalagan, R.; Markus, J.; Yang, D.C. Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: Synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J. Ginseng Res. 2018, 42, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Adachi, J.; Hasegawa, M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 1996, 42, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.M.; Wang, T.Y.; Yang, S.Y.; Wu, J.Y.; Chang, T.S. Production of new isoflavone glucosides from glycosylation of 8-hydroxydaidzein by glycosyltransferase from Bacillus subtilis ATCC 6633. Catalysts 2018, 8, 349. [Google Scholar] [CrossRef]
- Ünligil, U.; Rini, J.M. Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol. 2000, 10, 510–517. [Google Scholar] [CrossRef]
- Lariviere, L.; Gueguen-Chaignon, V.; Morera, S. Crystal structures of the T4 phage b-glucosyltransferase and the D100A mutant in complex with UDP-glucose: Glucose binding and identification of the catalytic base for a direct displacement mechanism. J. Mol. Biol. 2003, 330, 1077–1086. [Google Scholar] [CrossRef]
- Chang, T.S.; Chiang, C.M.; Wang, T.Y.; Lee, C.H.; Lee, Y.W.; Wu, J.Y. New triterpenoid from novel triterpenoid 15-O-glycosylation on ganoderic acid A by intestinal bacteria of zebrafish. Molecules 2018, 23, 2345. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Xu, S.H.; Zhao, Y.Z.; Zhang, C.; Zhang, Y.Y.; Yu, B.Y.; Zhang, J. Microbial hydroxylation and glycosylation of pentacyclic triterpenes as inhibitors on tissue factor procoagulant activity. Bioorg. Med. Chem. Lett. 2017, 27, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Moulis, C.; Andre, I.; Remaud-Simeon, M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell. Mol. Life Sci. 2016, 73, 2661–2679. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, T.-S.; Wu, J.-Y.; Wang, T.-Y.; Wu, K.-Y.; Chiang, C.-M. Uridine Diphosphate-Dependent Glycosyltransferases from Bacillus subtilis ATCC 6633 Catalyze the 15-O-Glycosylation of Ganoderic Acid A. Int. J. Mol. Sci. 2018, 19, 3469. https://doi.org/10.3390/ijms19113469
Chang T-S, Wu J-Y, Wang T-Y, Wu K-Y, Chiang C-M. Uridine Diphosphate-Dependent Glycosyltransferases from Bacillus subtilis ATCC 6633 Catalyze the 15-O-Glycosylation of Ganoderic Acid A. International Journal of Molecular Sciences. 2018; 19(11):3469. https://doi.org/10.3390/ijms19113469
Chicago/Turabian StyleChang, Te-Sheng, Jiumn-Yih Wu, Tzi-Yuan Wang, Kun-Yuan Wu, and Chien-Min Chiang. 2018. "Uridine Diphosphate-Dependent Glycosyltransferases from Bacillus subtilis ATCC 6633 Catalyze the 15-O-Glycosylation of Ganoderic Acid A" International Journal of Molecular Sciences 19, no. 11: 3469. https://doi.org/10.3390/ijms19113469
APA StyleChang, T.-S., Wu, J.-Y., Wang, T.-Y., Wu, K.-Y., & Chiang, C.-M. (2018). Uridine Diphosphate-Dependent Glycosyltransferases from Bacillus subtilis ATCC 6633 Catalyze the 15-O-Glycosylation of Ganoderic Acid A. International Journal of Molecular Sciences, 19(11), 3469. https://doi.org/10.3390/ijms19113469