Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells
Abstract
1. Introduction
2. Results and Discussion
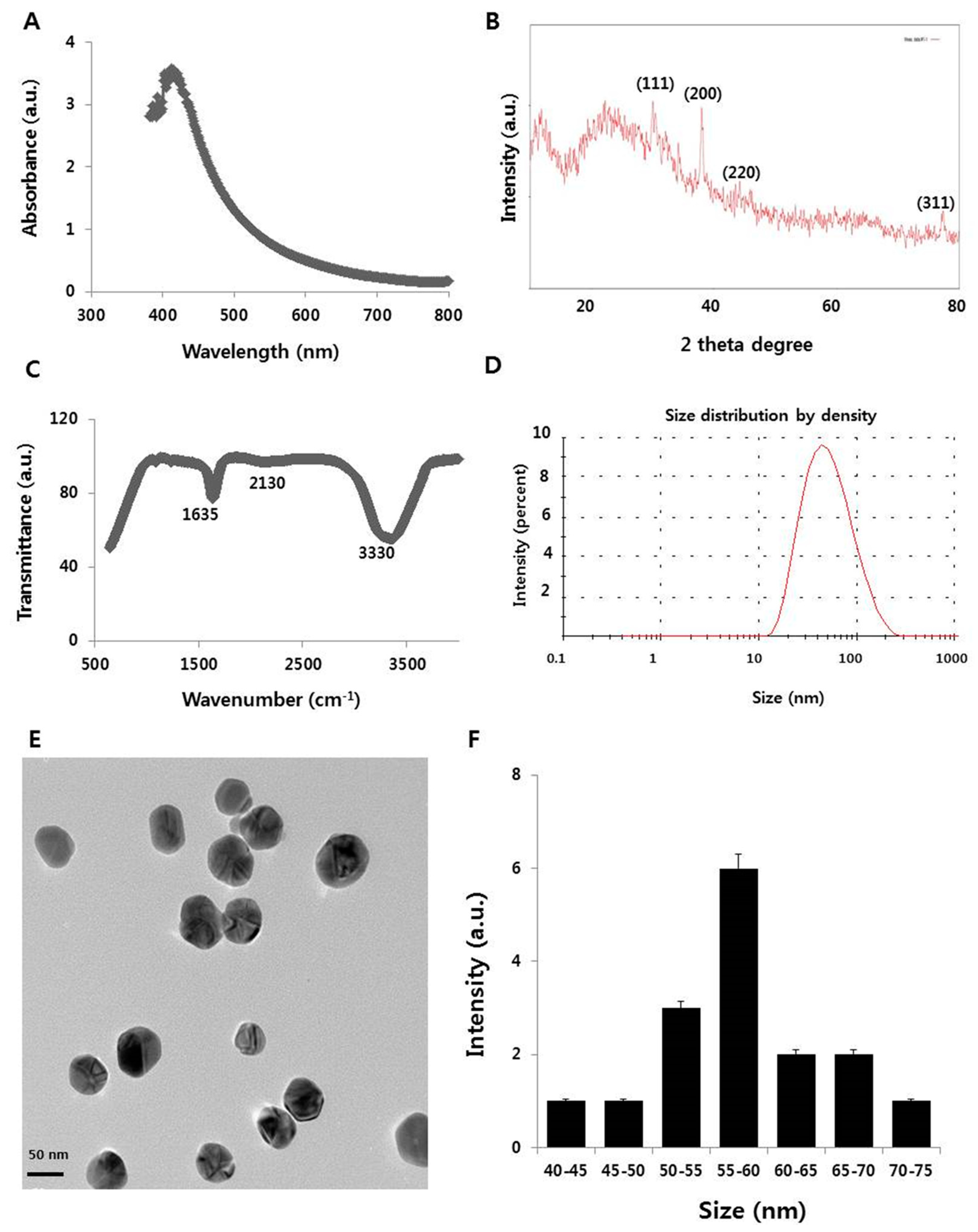
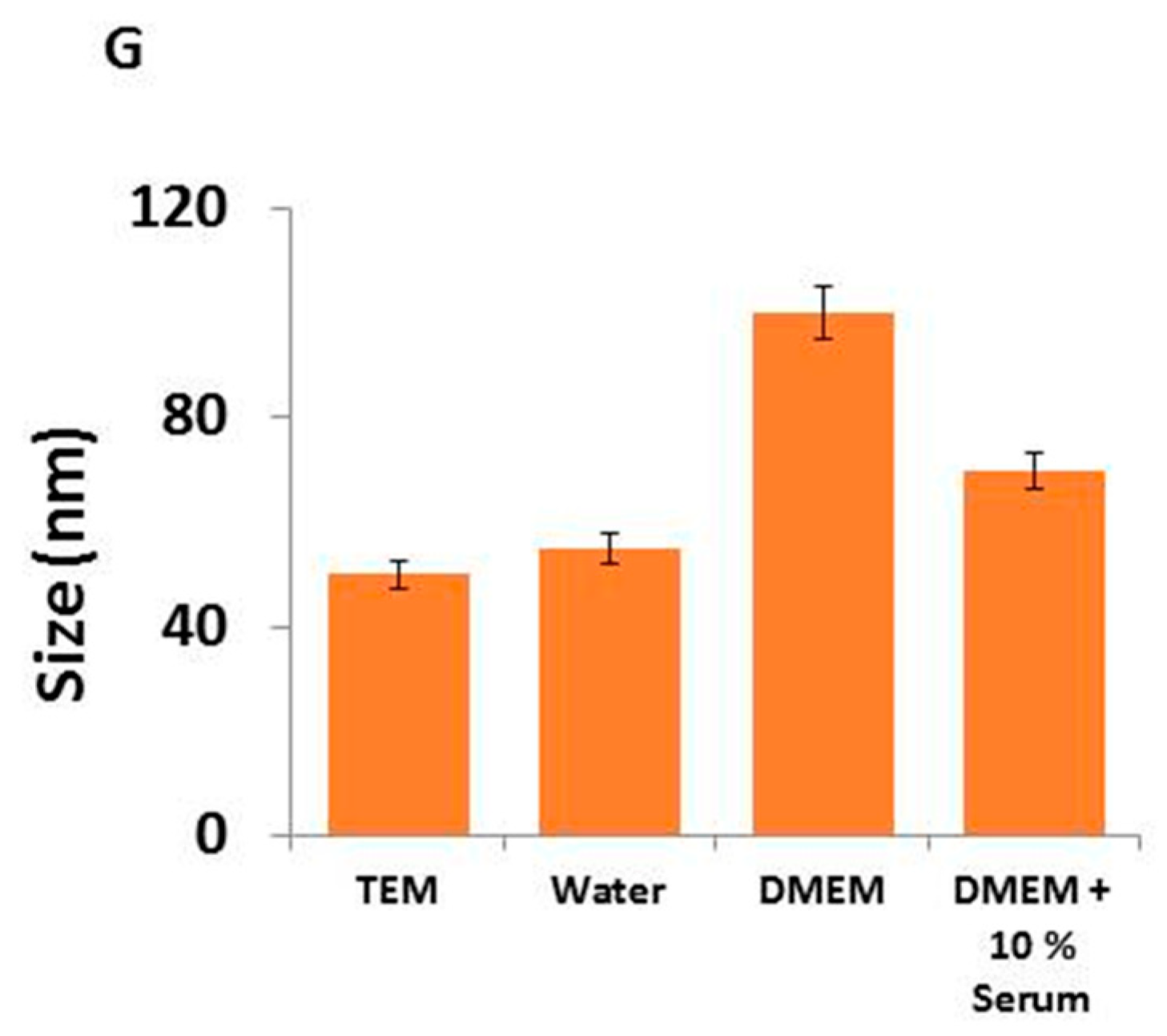
2.1. Synthesis and Characterization of Silver Nanoparticles (AgNPs) Using Myricetin
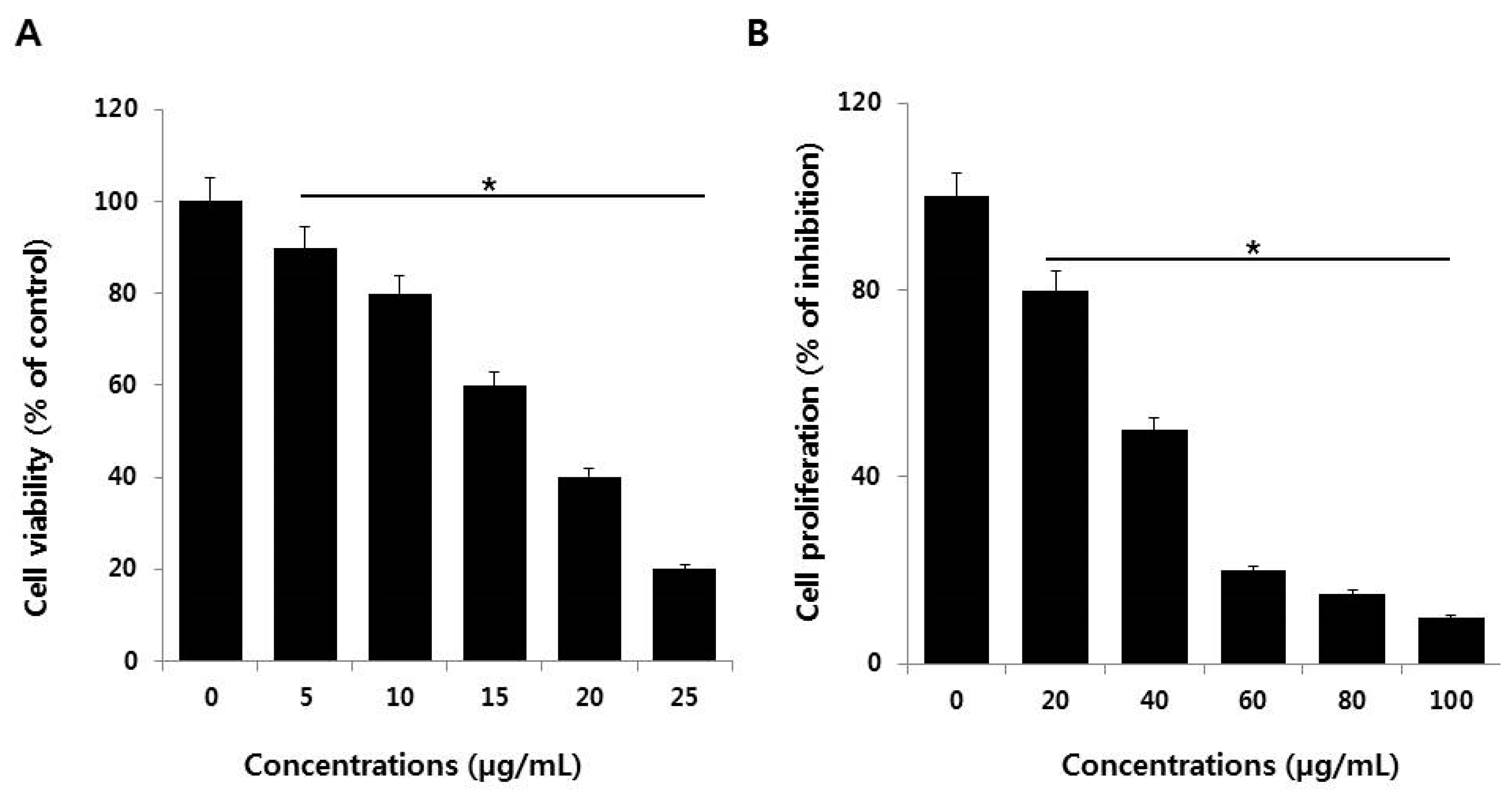
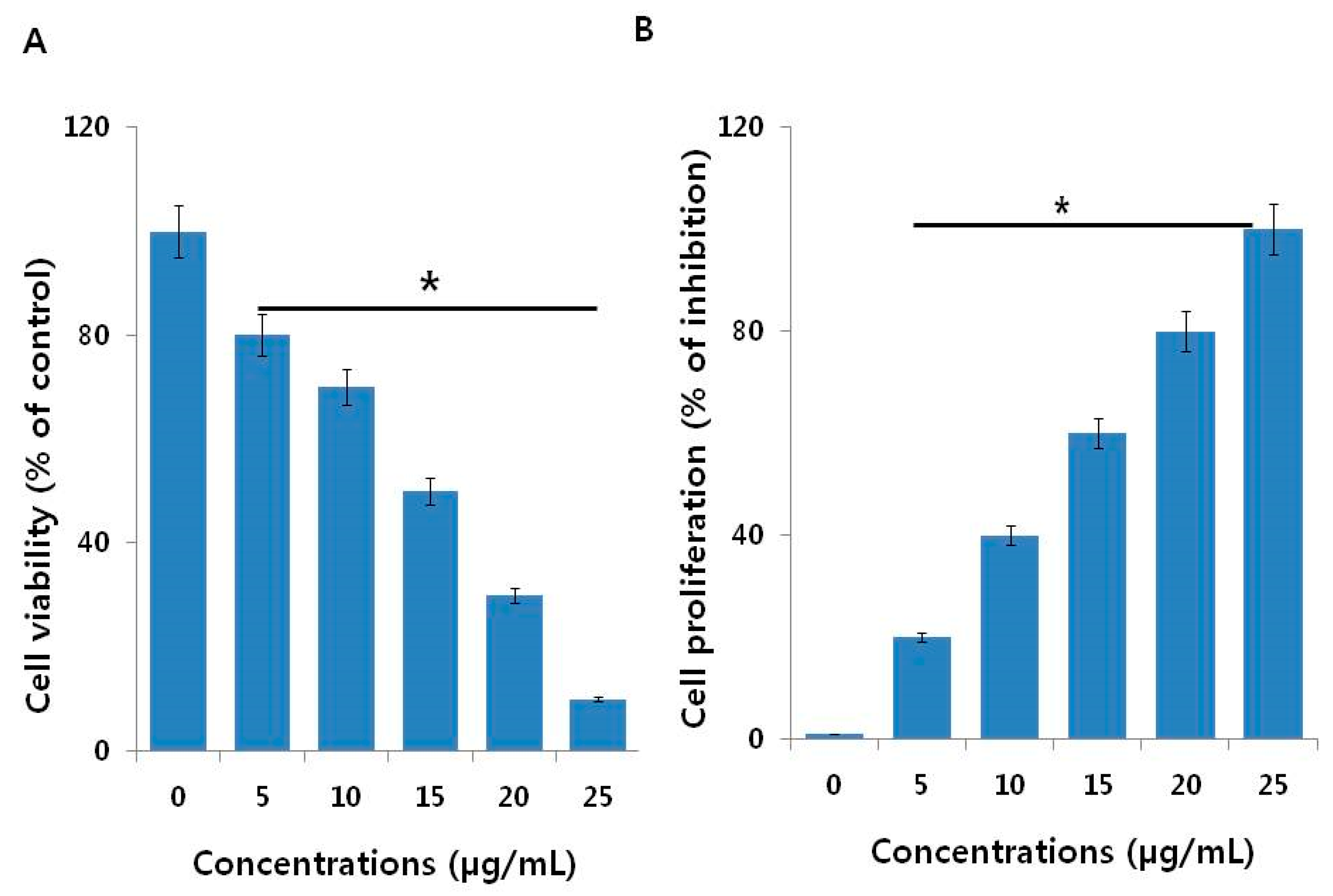
2.2. AgNPs Reduce the Viability and Cell Proliferation of NIH3T3 Cells
2.3. AgNPs Induce Cytotoxicity in NIH3T3 Cells
2.4. Cellular Uptake and Internalization of AgNPs
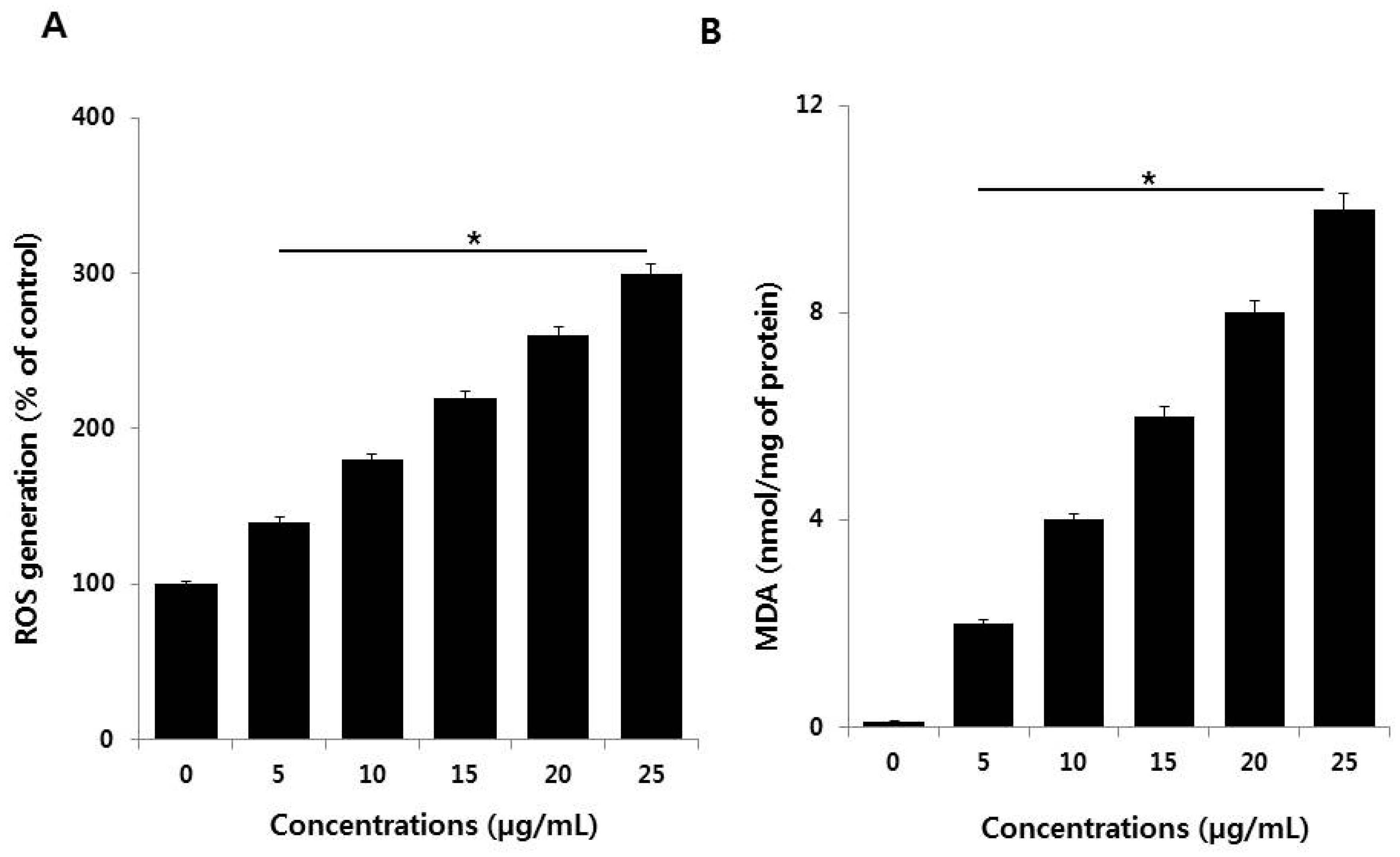
2.5. AgNPs Induce Oxidative Stress by Increased Levels of ROS and Malondialdehyde (MDA)
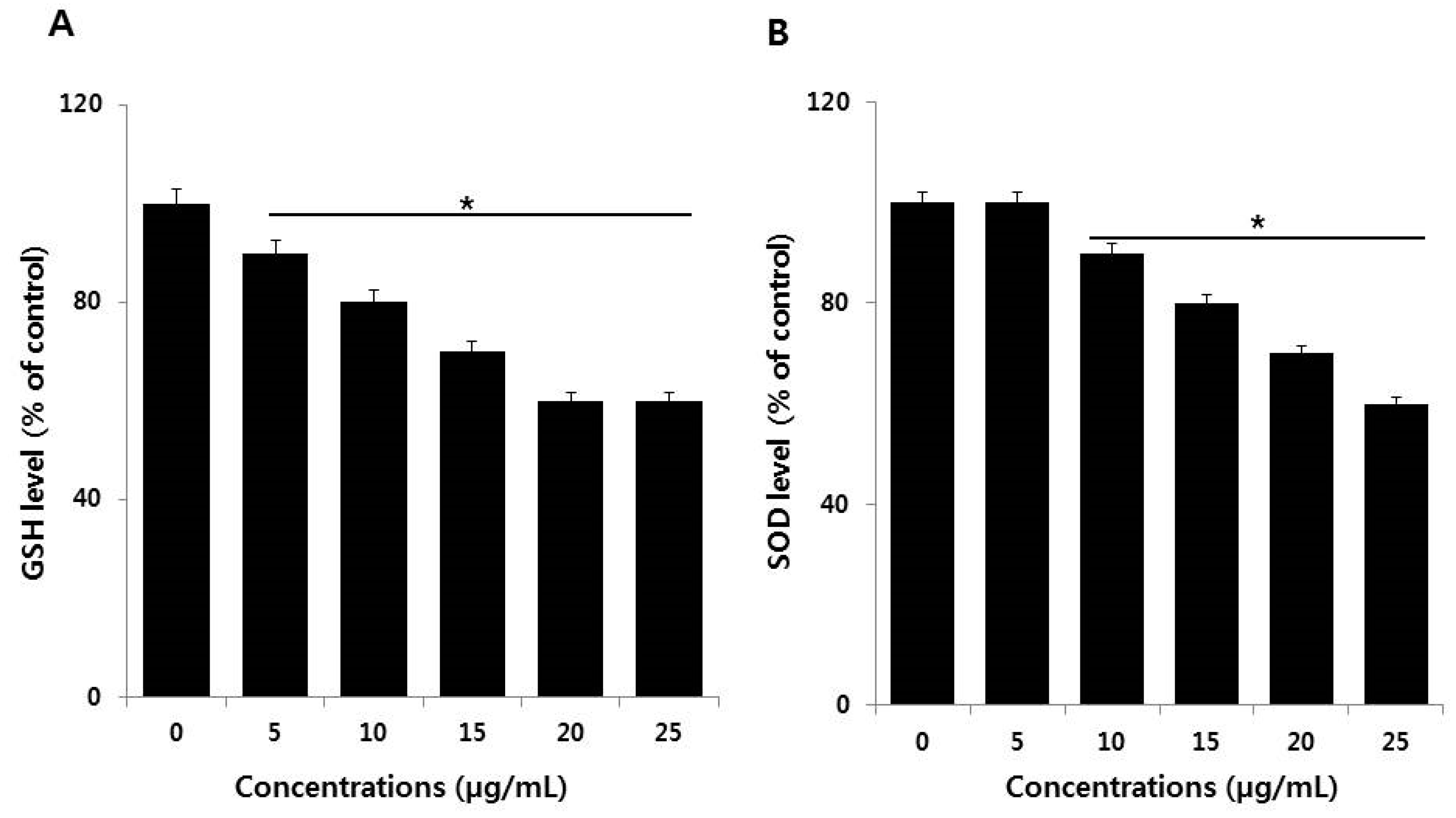
2.6. Effect of AgNPs on Antioxidants
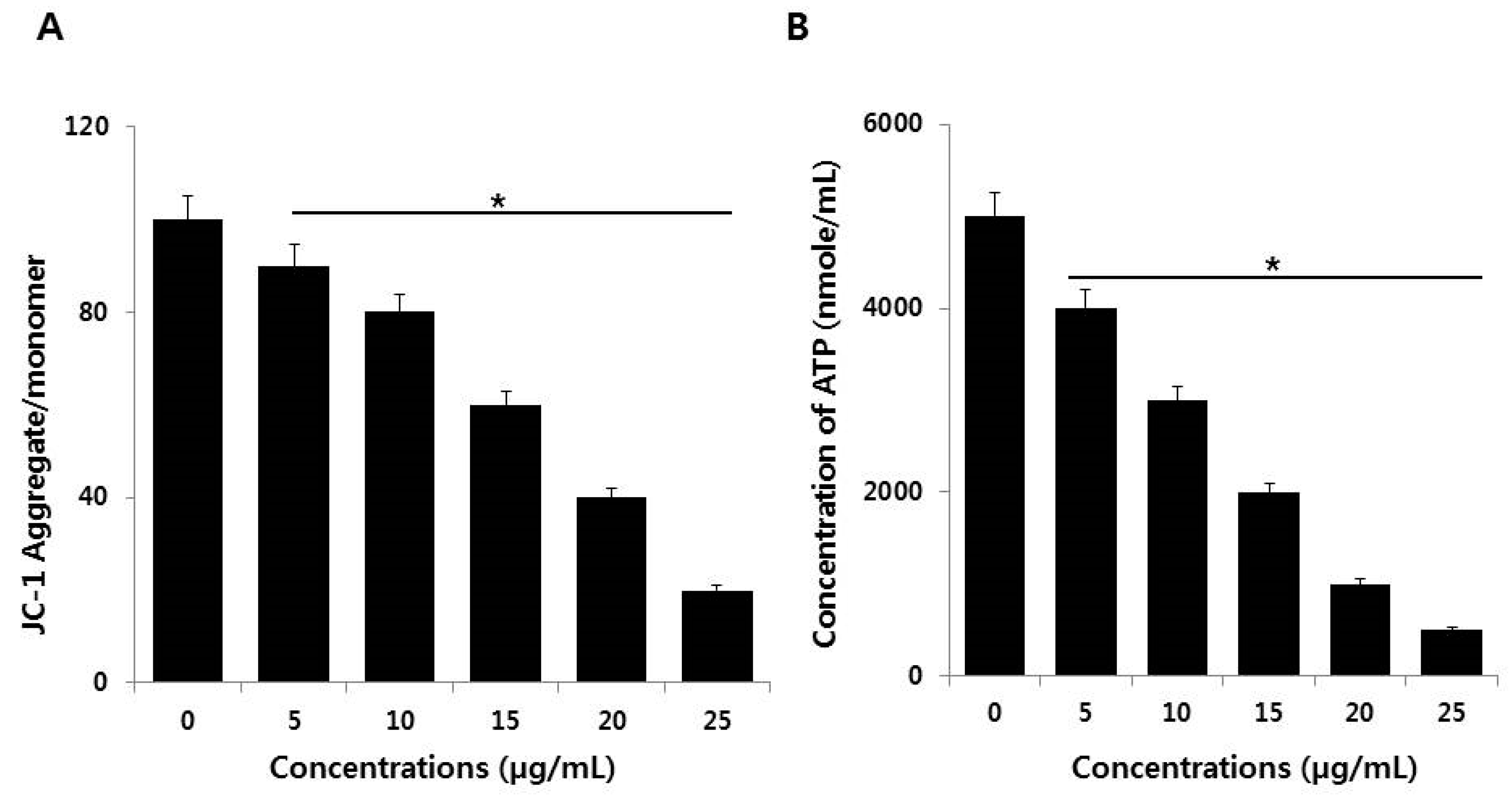
2.7. Effect of AgNPs on Mitochondrial Dysfunction and ATP Generation
2.8. AgNPs Cause DNA Damage in NIH3T3 Cells
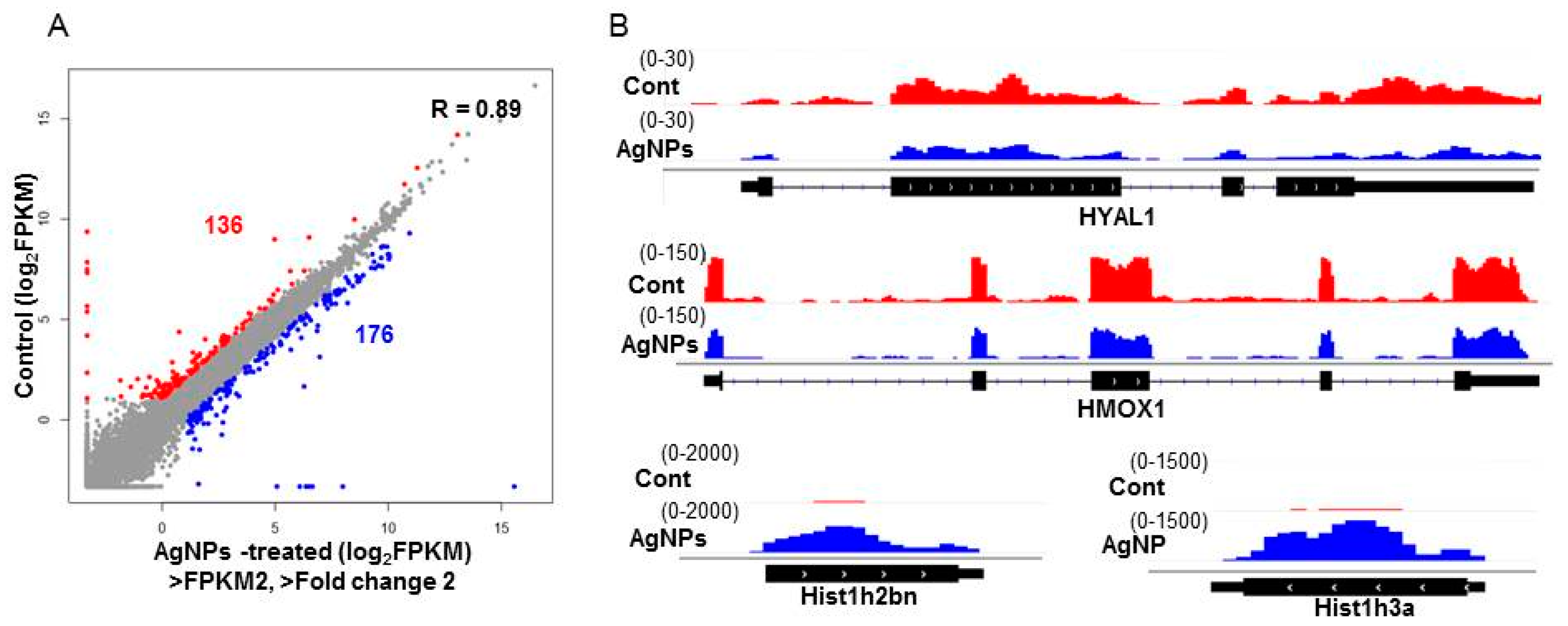
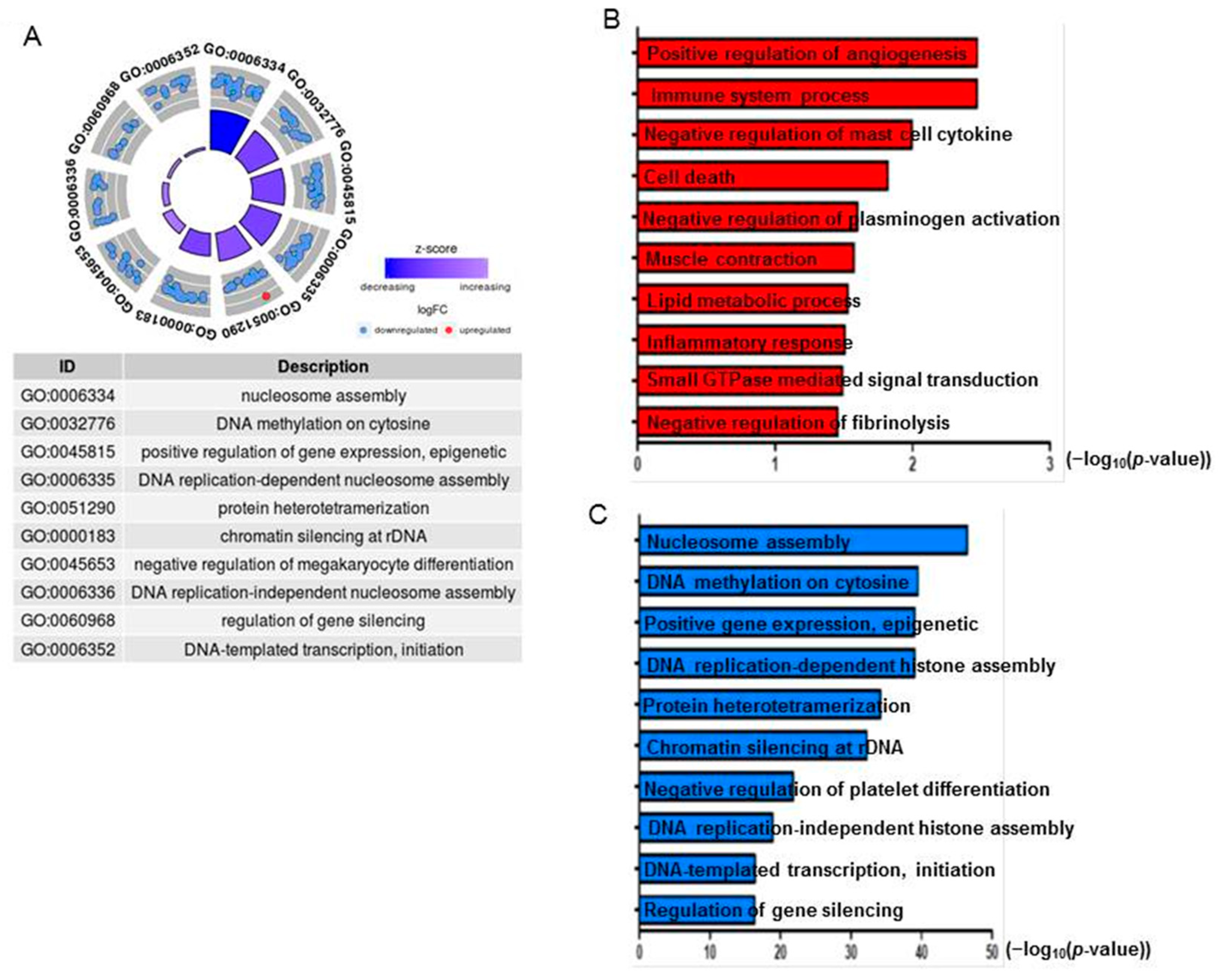
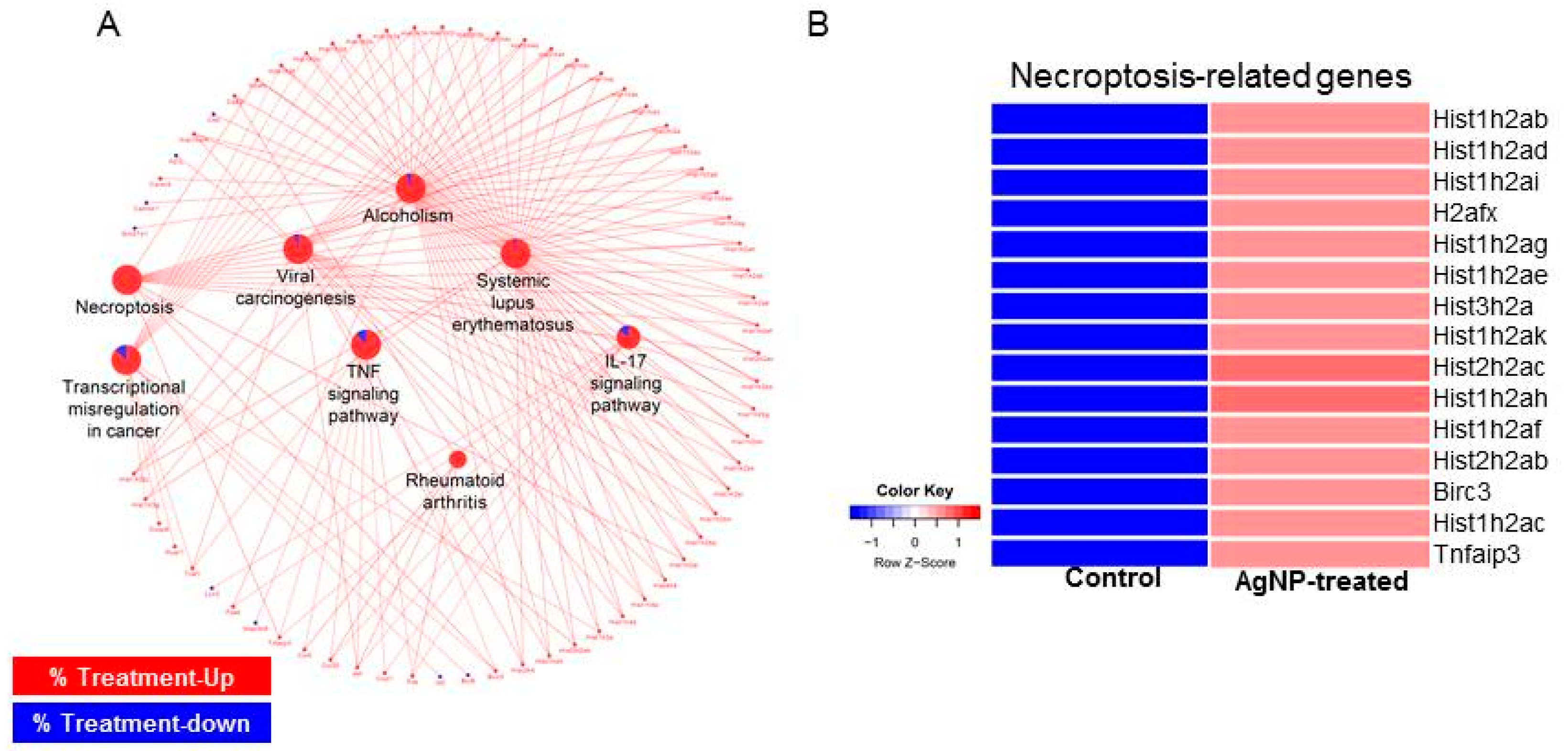
2.9. AgNPs Alter the Expression of Genes Involved in Apoptosis and Nucleosome Assembly
3. Materials and Methods
3.1. Synthesis and Characterization of AgNPs
3.2. Cell Viability and BrdU Cell Proliferation Assay
3.3. Membrane Integrity
3.4. Determination of Intracellular ROS
3.5. Measurement of Oxidative and Antioxidant Markers
3.6. JC-1 Assay and Measurement of ATP
3.7. Measurement of 8-oxo-dG
3.8. Measurement of Caspase 3 Activity
3.9. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
3.10. RNA-Seq and Downstream Bioinformatics Analysis
3.11. Statistical Analyses
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, X.; Schluesener, H.J. Nanosilver: A nanoproduct in medical application. Toxicol. Lett. 2008, 176, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Oberdörster, G.; Oberdörster, E.; Oberdörster, J. Nanotoxicology: An emerging discipline evolving from studies of ultrafine particles. Environ. Health Perspect. 2005, 113, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Lewinski, N.; Colvin, V.; Drezek, R. Cytotoxicity of nanoparticles. Small 2008, 4, 26–49. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Cheng, F.Y.; Chiu, H.W.; Tsai, J.C.; Fang, C.Y.; Chen, C.W.; Wang, Y.J. Cytotoxicity, oxidative stress, apoptosis and the autophagic effects of silver nanoparticles in mouse embryonic fibroblasts. Biomaterials 2014, 35, 4706–4715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Sai Lung, P.; Zhao, S.; Chu, Z.; Chrzanowski, W.; Li, Q. Shape dependent cytotoxicity of PLGA-PEG nanoparticles on human cells. Sci. Rep. 2017, 7, 7315. [Google Scholar] [CrossRef] [PubMed]
- Salatin, S.; Yari Khosroushahi, A. Overviews on the cellular uptake mechanism of polysaccharide colloidal nanoparticles. J. Cell Mol. Med. 2017, 21, 1668–1686. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.S.; Song, K.S.; Ryu, H.R.; Sung, J.H.; Park, J.D.; Park, H.M.; Song, N.W.; Shin, B.S.; Marshak, D.; et al. Biopersistence of silver nanoparticles in tissues from Sprague-Dawley rats. Part. Fibre Toxicol. 2013, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T.; Holbrook, N.J. Oxidants, oxidative stress and the biology of ageing. Nature 2000, 408, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Jeyaraj, M.; Kim, J.H. Cytotoxicity of biologically synthesized silver nanoparticles in MDA-MB-231 human breast cancer cells. BioMed Res. Int. 2013, 2013, 535796. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Jeong, J.K.; Han, J.W.; Zhang, X.F.; Park, J.H.; Kim, J.H. Multidimensional effects of biologically synthesized silver nanoparticles in Helicobacter pylori, Helicobacter felis, and human lung (L132) and lung carcinoma A549 cells. Nanoscale Res. Lett. 2015, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Han, J.W.; Gurunathan, S.; Jeong, J.K.; Choi, Y.J.; Kwon, D.N.; Park, J.K.; Kim, J.H. Oxidative stress mediated cytotoxicity of biologically synthesized silver nanoparticles in human lung epithelial adenocarcinoma cell line. Nanoscale Res. Lett. 2014, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Braydich-Stolle, L.; Hussain, S.; Schlager, J.J.; Hofmann, M.C. In vitro cytotoxicity of nanoparticles in mammalian germline stem cells. Toxicol. Sci. 2005, 88, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Hsin, Y.H.; Chen, C.F.; Huang, S.; Shih, T.S.; Lai, P.S.; Chueh, P.J. The apoptotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicol. Lett. 2008, 179, 130–139. [Google Scholar] [CrossRef] [PubMed]
- AshaRani, P.V.; Low Kah Mun, G.; Hande, M.P.; Valiyaveettil, S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano 2009, 3, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.K.; Gurunathan, S.; Kang, M.H.; Han, J.W.; Das, J.; Choi, Y.J.; Kwon, D.N.; Cho, S.G.; Park, C.; Seo, H.G.; et al. Hypoxia-mediated autophagic flux inhibits silver nanoparticle-triggered apoptosis in human lung cancer cells. Sci. Rep. 2016, 6, 21688. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Gurunathan, S.; Choi, Y.J.; Han, J.W.; Song, H.; Kim, J.H. Silver nanoparticles suppresses brain-derived neurotrophic factor-induced cell survival in the human neuroblastoma cell line SH-SY5Y. J. Ind. Eng. Chem. 2017, 47, 62–73. [Google Scholar] [CrossRef]
- Singhal, P.K.; Sassi, S.; Lan, L.; Au, P.; Halvorsen, S.C.; Fukumura, D.; Jain, R.K.; Seed, B. Mouse embryonic fibroblasts exhibit extensive developmental and phenotypic diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Mytych, J.; Zebrowski, J.; Lewinska, A.; Wnuk, M. Prolonged effects of silver nanoparticles on p53/p21 pathway-mediated proliferation, DNA damage response, and methylation parameters in HT22 hippocampal neuronal cells. Mol. Neurobiol. 2017, 54, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Siu, H.; Zhu, Y.; Jin, L.; Xiong, M. Implication of next-generation sequencing on association studies. BMC Genom. 2011, 12, 322. [Google Scholar] [CrossRef] [PubMed]
- Feliu, N.; Kohonen, P.; Ji, J.; Zhang, Y.; Karlsson, H.L.; Palmberg, L.; Nyström, A.; Fadeel, B. Next-generation sequencing reveals low-dose effects of cationic dendrimers in primary human bronchial epithelial cells. ACS Nano 2015, 9, 146–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guo, Y.; Ma, C.; Zhang, D.; Wang, C.; Yang, Q. Transcriptome analysis of maize resistance to Fusarium graminearum. BMC Genom. 2016, 17, 477. [Google Scholar] [CrossRef]
- Sun, Q.L.; Zhao, C.P.; Wang, T.Y.; Hao, X.B.; Wang, X.Y.; Zhang, X.; Li, Y.C. Expression profile analysis of long non-coding RNA associated with vincristine resistance in colon cancer cells by next-generation sequencing. Gene 2015, 572, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.N.; Hsu, W.L.; Berger, B. DART: A fast and accurate RNA-seq mapper with a partitioning strategy. Bioinformatics 2018, 34, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, T.; Chitale, M.; Kihara, D. Functional enrichment analyses and construction of functional similarity networks with high confidence function prediction by PFP. BMC Bioinform. 2010, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Maher, C.A.; Palanisamy, N.; Brenner, J.C.; Cao, X.; Kalyana-Sundaram, S.; Luo, S.; Khrebtukova, I.; Barrette, T.R.; Grasso, C.; Yu, J.; et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc. Natl. Acad. Sci. USA 2009, 106, 12353–12358. [Google Scholar] [CrossRef] [PubMed]
- Costa, B.M.; Smith, J.S.; Chen, Y.; Chen, J.; Phillips, H.S.; Aldape, K.D.; Zardo, G.; Nigro, J.; James, C.D.; Fridlyand, J.; et al. Reversing HOXA9 oncogene activation by PI3K inhibition: Epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010, 70, 453–462. [Google Scholar] [CrossRef] [PubMed]
- Vigneshwaran, N.; Ashtaputre, N.M.; Varadarajan, P.V.; Nachane, R.P.; Paralikar, K.M.; Balasubramanya, R.H. Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Mater. Lett. 2007, 61, 1413–1418. [Google Scholar] [CrossRef]
- Jain, S.; Mehata, M.S. Medicinal Plant Leaf Extract and Pure Flavonoid Mediated Green Synthesis of Silver Nanoparticles and their Enhanced Antibacterial Property. Sci. Rep. 2017, 7, 15867. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Lee, K.J.; Kalishwaralal, K.; Sheikpranbabu, S.; Vaidyanathan, R.; Eom, S.H. Antiangiogenic properties of silver nanoparticles. Biomaterials 2009, 30, 6341–6350. [Google Scholar] [CrossRef] [PubMed]
- Kalishwaralal, K.; BarathManiKanth, S.; Pandian, S.R.K.; Deepak, V.; Gurunathan, S. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf. B Biointerfaces 2010, 79, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S. Rapid biological synthesis of silver nanoparticles and their enhanced antibacterial effects against Escherichia fergusonii and Streptococcus mutans. Arab. J. Chem. 2014. [Google Scholar] [CrossRef]
- Fissan, H.; Ristig, S.; Kaminski, H.; Asbach, C.; Epple, M. Comparison of different characterization methods for nanoparticle dispersions before and after aerosolization. Anal. Methods 2014, 6, 7324. [Google Scholar] [CrossRef]
- Sahu, N.; Soni, D.; Chandrashekhar, B.; Satpute, D.B.; Saravanadevi, S.; Sarangi, B.K.; Pandey, R.A. Synthesis of silver nanoparticles using flavonoids: Hesperidin, naringin and diosmin, and their antibacterial effects and cytotoxicity. Int. Nano Lett. 2016, 6, 173–181. [Google Scholar] [CrossRef]
- Prathna, T.C.; Chandrasekaran, N.; Raichur, A.M.; Mukherjee, A. Biomimetic synthesis of silver nanoparticles by Citrus limon (lemon) aqueous extract and theoretical prediction of particle size. Colloids Surf. B Biointerfaces 2011, 82, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, E.; Choi, Y.J.; Kwon, D.N.; Kim, J.H. Reduced graphene oxide-silver nanoparticle nanocomposite: A potential anticancer nanotherapy. Int. J. Nanomedicine 2015, 10, 6257–6276. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Mahmood, M.; Xu, Y.; Watanabe, F.; Biris, A.S.; Hansen, D.K.; Inselman, A.; Casciano, D.; Patterson, T.A.; Paule, M.G.; et al. Effects of silver nanoparticles on human and rat embryonic neural stem cells. Front. Neurosci. 2015, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.G.; Wang, Y.H.; Xing, H.H.; Gurunathan, S. Quercetin-mediated synthesis of graphene oxide–silver nanoparticle nanocomposites: A suitable alternative nanotherapy for neuroblastoma. Int. J. Nanomed. 2017, 12, 5819–5839. [Google Scholar] [CrossRef] [PubMed]
- Dalle-Donne, I.; Rossi, R.; Giustarini, D.; Colombo, R.; Milzani, A. S-glutathionylation in protein redox regulation. Free Radic. Biol. Med. 2007, 43, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Roda, E.; Barni, S.; Milzani, A.; Dalle-Donne, I.; Colombo, G.; Coccini, T. Single silver nanoparticle instillation induced early and persisting moderate cortical damage in rat kidneys. Int. J. Mol. Sci. 2017, 18, 2115. [Google Scholar] [CrossRef] [PubMed]
- Martindale, J.L.; Holbrook, N.J. Cellular response to oxidative stress: Signaling for suicide and survival. J. Cell Physiol. 2002, 192, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Reidy, B.; Haase, A.; Luch, A.; Dawson, K.A.; Lynch, I. Mechanisms of silver nanoparticle release, transformation and toxicity: A critical review of current knowledge and recommendations for future studies and applications. Materials 2013, 6, 2295–2350. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.; Rott, S.; Mantion, A.; Graf, P.; Plendl, J.; Thünemann, A.F.; Meier, W.P.; Taubert, A.; Luch, A.; Reiser, G. Effects of silver nanoparticles on primary mixed neural cell cultures: Uptake, oxidative stress and acute calcium responses. Toxicol. Sci. 2012, 126, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.; Mantion, A.; Graf, P.; Plendl, J.; Thuenemann, A.F.; Meier, W.; Taubert, A.; Luch, A. A novel type of silver nanoparticles and their advantages in toxicity testing in cell culture systems. Arch. Toxicol. 2012, 86, 1089–1098. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, K.; Sundar, L.S.; Pereira, E.; Duarte, A.C. Graphene oxide induces cytotoxicity and oxidative stress in bluegill sunfish cells. J. Appl. Toxicol. 2018, 38, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Gliga, A.R.; di Bucchianico, S.; Lindvall, J.; Fadeel, B.; Karlsson, H.L. RNA-sequencing reveals long-term effects of silver nanoparticles on human lung cells. Sci. Rep. 2018, 8, 6668. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Park, J.H.; Han, J.W.; Kim, J.H. Comparative assessment of the apoptotic potential of silver nanoparticles synthesized by Bacillus tequilensis and Calocybe indica in MDA-MB-231 human breast cancer cells: Targeting p53 for anticancer therapy. Int. J. Nanomed. 2015, 10, 4203. [Google Scholar] [CrossRef] [PubMed]
- Mei, N.; Zhang, Y.; Chen, Y.; Guo, X.; Ding, W.; Ali, S.F.; Biris, A.S.; Rice, P.; Moore, M.M.; Chen, T. Silver nanoparticle-induced mutations and oxidative stress in mouse lymphoma cells. Environ. Mol. Mutagen. 2012, 53, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.F.; Wang, J.; Patterson, T.A.; Saini, U.T.; Robinson, B.L.; Newport, G.D.; Murdock, R.C.; Schlager, J.J.; Hussain, S.M.; Ali, S.F. Expression of genes related to oxidative stress in the mouse brain after exposure to silver-25 nanoparticles. Toxicol. Lett. 2009, 187, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.M.; Hess, K.L.; Gearhart, J.M.; Geiss, K.T.; Schlager, J.J. In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol. In Vitro 2005, 19, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Arora, M.; Kumar, A.; Kaundal, R.K.; Sharma, S.S. Amelioration of neurological and biochemical deficits by peroxynitrite decomposition catalysts in experimental diabetic neuropathy. Eur. J. Pharmacol. 2008, 596, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Piao, M.J.; Kang, K.A.; Lee, I.K.; Kim, H.S.; Kim, S.; Choi, J.Y.; Choi, J.; Hyun, J.W. Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis. Toxicol. Lett. 2011, 201, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, K.K.; Awasthi, A.; Verma, R.; Soni, I.; Awasthi, K.; John, P.J. Silver nanoparticles and carbon nanotubes induced DNA damage in mice evaluated by single cell gel electrophoresis. Macromol. Symp. 2015, 357, 210–217. [Google Scholar] [CrossRef]
- Yuan, Y.G.; Peng, Q.L.; Gurunathan, S. Combination of palladium nanoparticles and tubastatin-A potentiates apoptosis in human breast cancer cells: A novel therapeutic approach for cancer. Int. J. Nanomed. 2017, 12, 6503–6520. [Google Scholar] [CrossRef] [PubMed]
- Sakamuru, S.; Attene-Ramos, M.S.; Xia, M. Mitochondrial membrane potential assay. Methods Mol. Biol. 2016, 1473, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yin, N.; Wen, R.; Liu, W.; Jia, Y.; Hu, L.; Zhou, Q.; Jiang, G. Silver nanoparticles induced neurotoxicity through oxidative stress in rat cerebral astrocytes is distinct from the effects of silver ions. Neurotoxicology 2016, 52, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Aueviriyavit, S.; Phummiratch, D.; Maniratanachote, R. Mechanistic study on the biological effects of silver and gold nanoparticles in Caco-2 cells—Induction of the Nrf2/HO-1 pathway by high concentrations of silver nanoparticles. Toxicol. Lett. 2014, 224, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Piao, M.J.; Kim, K.C.; Kim, A.D.; Choi, J.Y.; Choi, J.; Hyun, J.W. Endoplasmic reticulum stress signaling is involved in silver nanoparticles-induced apoptosis. Int. J. Biochem. Cell Biol. 2012, 44, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, P.; Krzyzowska, M.; Zdanowski, R.; Winnicka, A.; Nowakowska, J.; Stankiewicz, W.; Tomaszewska, E.; Celichowski, G.; Grobelny, J. Assessment of in vitro cellular responses of monocytes and keratinocytes to tannic acid modified silver nanoparticles. Toxicol. In Vitro 2013, 27, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Smeitink, J.A.; Zeviani, M.; Turnbull, D.M.; Jacobs, H.T. Mitochondrial medicine: A metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006, 3, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Diebold, L.P.; Kong, H.; Schieber, M.; Huang, H.; Hensley, C.T.; Mehta, M.M.; Wang, T.; Santos, J.H.; Woychik, R.; et al. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol. Cell 2016, 61, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Bohovych, I.; Khalimonchuk, O. Sending out an SOS: Mitochondria as a signaling hub. Front. Cell Dev. Biol. 2016, 4, 109. [Google Scholar] [CrossRef] [PubMed]
- Neustadt, J.; Pieczenik, S.R. Medication-induced mitochondrial damage and disease. Mol. Nutr. Food Res. 2008, 52, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Maurer, L.L.; Meyer, J.N. A systematic review of evidence for silver nanoparticle-induced mitochondrial toxicity. Environ. Sci. Nano 2016, 3, 311–322. [Google Scholar] [CrossRef]
- Geromel, V.; Kadhom, N.; Cebalos-Picot, I.; Ouari, O.; Polidori, A.; Munnich, A.; Rötig, A.; Rustin, P. Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum. Mol. Genet. 2001, 10, 1221–1228. [Google Scholar] [CrossRef] [PubMed]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-hydroxy-2′-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health Part C 2009, 27, 120–139. [Google Scholar] [CrossRef] [PubMed]
- Sriram, M.I.; Kanth, S.B.M.; Kalishwaralal, K.; Gurunathan, S. Antitumor activity of silver nanoparticles in Dalton’s lymphoma ascites tumor model. Int. J. Nanomed. 2010, 5, 753–762. [Google Scholar] [CrossRef]
- Foldbjerg, R.; Dang, D.A.; Autrup, H. Cytotoxicity and genotoxicity of silver nanoparticles in the human lung cancer cell line, A549. Arch. Toxicol. 2011, 85, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, S.; Scherzed, A.; Kessler, M.; Hummel, S.; Technau, A.; Froelich, K.; Ginzkey, C.; Koehler, C.; Hagen, R.; Kleinsasser, N. Silver nanoparticles: Evaluation of DNA damage, toxicity and functional impairment in human mesenchymal stem cells. Toxicol. Lett. 2011, 201, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Huang, F.H.; Zhang, G.L.; Bai, D.P.; Massimo, D.F.; Huang, Y.F.; Gurunathan, S. Novel biomolecule lycopene-reduced graphene oxide-silver nanoparticle enhances apoptotic potential of trichostatin A in human ovarian cancer cells (SKOV3). Int. J. Nanomed. 2017, 12, 7551–7575. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kalishwaralal, K.; Vaidyanathan, R.; Venkataraman, D.; Pandian, S.R.K.; Muniyandi, J.; Hariharan, N.; Eom, S.H. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf. B Biointerfaces 2009, 74, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.; Liu, Q.; Liu, J.; He, B.; Cui, L.; Li, Z.; Yun, Z.; Qu, G.; Liu, S.; Zhou, Q.; et al. Silver nanoparticle exposure attenuates the viability of rat cerebellum granule cells through apoptosis coupled to oxidative stress. Small 2013, 9, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, M.; Lin, Z.; Zhao, M.; Xiao, M.; Wang, C.; Xu, T.; Chen, T.; Zhu, B. Polyethylenimine-functionalized silver nanoparticle-based co-delivery of paclitaxel to induce HepG2 cell apoptosis. Int. J. Nanomed. 2016, 11, 6693–6702. [Google Scholar] [CrossRef] [PubMed]
- Karlic, H.; Herrmann, H.; Varga, F.; Thaler, R.; Reitermaier, R.; Spitzer, S.; Ghanim, V.; Blatt, K.; Sperr, W.R.; Valent, P.; et al. The role of epigenetics in the regulation of apoptosis in myelodysplastic syndromes and acute myeloid leukemia. Crit. Rev. Oncol. Hematol. 2014, 90, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Takebe, M.; Oishi, H.; Taguchi, K.; Aoki, Y.; Takashina, M.; Tomita, K.; Yokoo, H.; Takano, Y.; Yamazaki, M.; Hattori, Y. Inhibition of histone deacetylases protects septic mice from lung and splenic apoptosis. J. Surg. Res. 2014, 187, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Banfalvi, G. Apoptotic agents inducing genotoxicity-specific chromatin changes. Apoptosis 2014, 19, 1301–1316. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zheng, H.; Huang, S.; You, N.; Xu, J.; Ye, X.; Zhu, Q.; Feng, Y.; You, Q.; Miao, H.; et al. Heme oxygenase-1 enhances autophagy in podocytes as a protective mechanism against high glucose-induced apoptosis. Exp. Cell Res. 2015, 337, 146–159. [Google Scholar] [CrossRef] [PubMed]
- McAtee, C.O.; Berkebile, A.R.; Elowsky, C.G.; Fangman, T.; Barycki, J.J.; Wahl, J.K.; Khalimonchuk, O.; Naslavsky, N.; Caplan, S.; Simpson, M.A. Hyaluronidase hyal1 increases tumor cell proliferation and motility through accelerated vesicle trafficking. J. Biol. Chem. 2015, 290, 13144–13156. [Google Scholar] [CrossRef] [PubMed]
- Sierra, M.I.; Rubio, L.; Bayón, G.F.; Cobo, I.; Menendez, P.; Morales, P.; Mangas, C.; Urdinguio, R.G.; Lopez, V.; Valdes, A.; et al. DNA methylation changes in human lung epithelia cells exposed to multi-walled carbon nanotubes. Nanotoxicology 2017, 11, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zhang, J.; Hu, Q.; Xu, M.; Chen, Y.; Hu, G.; Zhao, M.; Liu, S. Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials 2015, 70, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Toyooka, T.; Ibuki, Y. Silver nanoparticle-induced phosphorylation of histone H3 at serine 10 is due to dynamic changes in actin filaments and the activation of Aurora kinases. Toxicol. Lett. 2017, 276, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Choi, Y.J.; Kim, J.H. Antibacterial efficacy of silver nanoparticles on endometritis caused by Prevotella melaninogenica and Arcanobacterum pyogenes in dairy cattle. Int. J. Mol. Sci. 2018, 19, 1210. [Google Scholar] [CrossRef] [PubMed]




| Solvent | Zeta Potential |
|---|---|
| Water | −25.2 ± 0.1 |
| DMEM | 20.5 ± 1.5 |
| DMEM + 10% serum | 11.5 ± 2.2 |
| Gene | Primer |
|---|---|
| p53 | F: AGAGACCGTACAGAAGA |
| R: CTGTAGCATGGGATCCTTT | |
| p21 | F: GTTGCTGTCCGGACTACCG |
| R: AAAAACAATGCCACCACTCC | |
| PARP-1 | F: CTCCATCCTGGCCTCGCTGT |
| R: GCTGTCACCTT CACCGTTCC | |
| CDK2 | F: GCTAGCAGACTTTGGACTAGCCAG |
| R: AGCTCGGTACCACAGGGTCA | |
| CDK4 | F: CTGGTGTTTGAGCATGTAGACC |
| R: AAACTGGCGCATCAGATCCTT | |
| GAPP45A | F: TGCTCAGCAAAGCCCTGAGT |
| R: GCTTGGCCGCTTCGTACA | |
| GAPDH | F: TGCACCACCAACTGCTTAGC |
| R: GGCATGGACTGTGGTCATGAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Choi, D.Y.; Song, H.; Park, C.; Kim, J.-H.; Hong, K. Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells. Int. J. Mol. Sci. 2018, 19, 3618. https://doi.org/10.3390/ijms19113618
Gurunathan S, Qasim M, Park C, Yoo H, Choi DY, Song H, Park C, Kim J-H, Hong K. Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells. International Journal of Molecular Sciences. 2018; 19(11):3618. https://doi.org/10.3390/ijms19113618
Chicago/Turabian StyleGurunathan, Sangiliyandi, Muhammad Qasim, Chanhyeok Park, Hyunjin Yoo, Dong Yoon Choi, Hyuk Song, Chankyu Park, Jin-Hoi Kim, and Kwonho Hong. 2018. "Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells" International Journal of Molecular Sciences 19, no. 11: 3618. https://doi.org/10.3390/ijms19113618
APA StyleGurunathan, S., Qasim, M., Park, C., Yoo, H., Choi, D. Y., Song, H., Park, C., Kim, J.-H., & Hong, K. (2018). Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells. International Journal of Molecular Sciences, 19(11), 3618. https://doi.org/10.3390/ijms19113618






