Dissection of the Structural Features of a Fungicidal Antibody-Derived Peptide
Abstract
:1. Introduction
2. Results
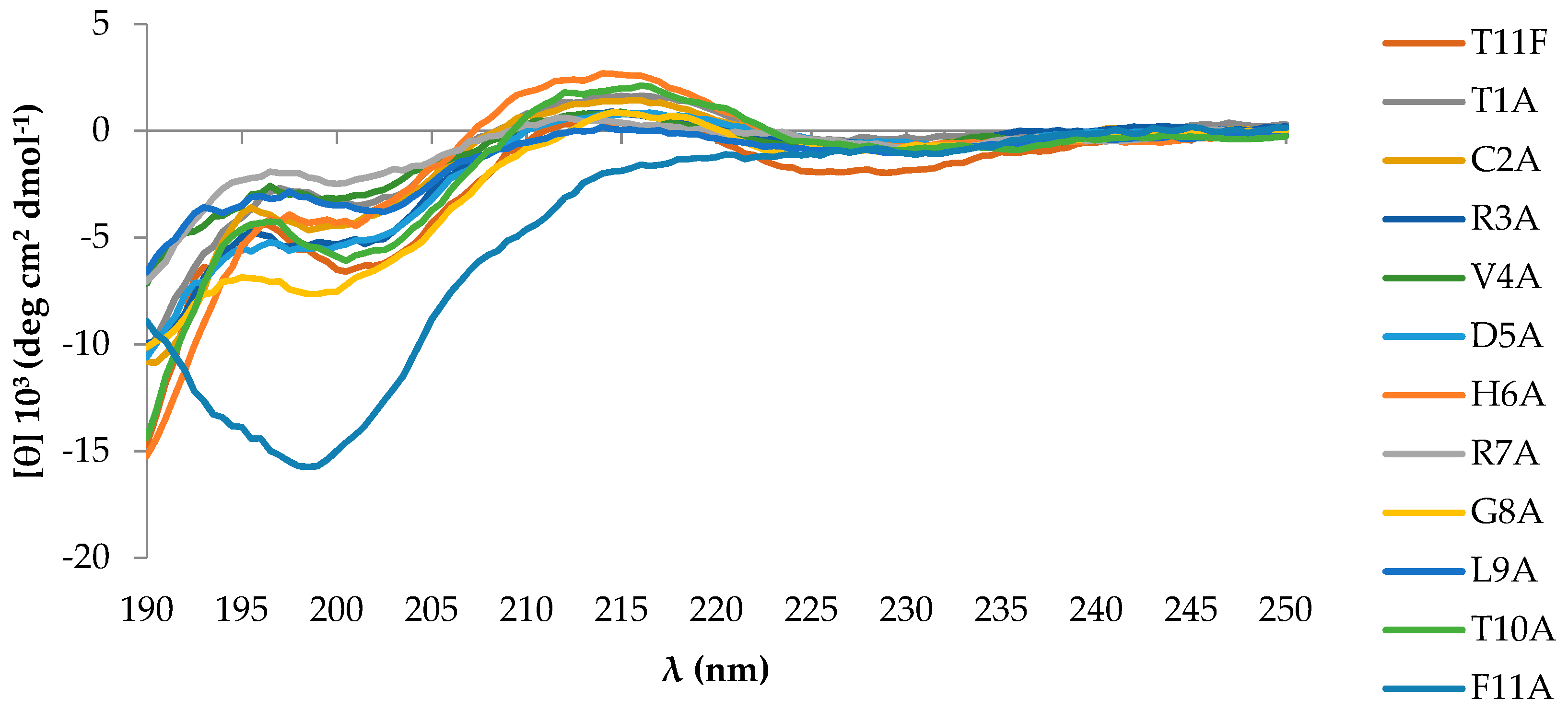
2.1. Peptides Conformational State
2.2. NMR Spectroscopy Characterisation
2.3. Time Kinetics of C. albicans Killing
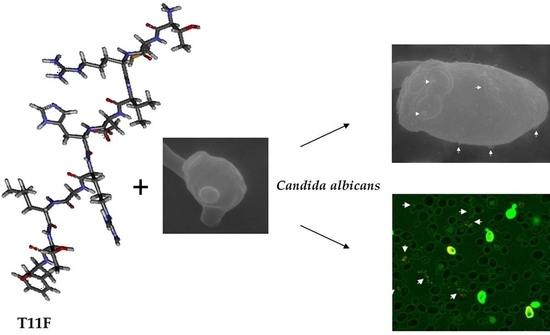
2.4. Peptide–Yeast Cells Interaction
2.4.1. Confocal Microscopy Studies
2.4.2. Scanning Electron Microscopy (SEM) Studies
2.5. Evaluation of Apoptosis in C. albicans Cells after Treatment with Selected Peptides
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. CD Spectroscopy
4.3. NMR Spectroscopy
4.4. Time Kinetics of Peptide-Mediated Killing of C. albicans
4.5. Confocal Microscopy Studies
4.6. SEM Studies
4.7. Evaluation of Apoptosis Induction Profiles in C. albicans after Treatment with Selected Peptides
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | circular dichroism |
| CFU | colony forming unit |
| FITC | fluorescein isothiocyanate |
| HPLC | high-performance liquid chromatography |
| NMR | nuclear magnetic resonance |
| PI | propidium iodide |
| SEM | scanning electron microscopy |
| TMRM | tetramethylrhodamine methyl ester |
References
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.M.B.; Dias, S.C.; Franco, O.L. Antimicrobial Peptides and Nanotechnology, Recent Advances and Challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Augusto, M.T.; Felicio, M.R.; Hollmann, A.; Franco, O.L.; Goncalves, S.; Santos, N.C. Designing improved active peptides for therapeutic approaches against infectious diseases. Biotechnol. Adv. 2018, 36, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Kokel, A.; Torok, M. Recent Advances in the Development of Antimicrobial Peptides (AMPs): Attempts for Sustainable Medicine? Curr. Med. Chem. 2018, 25, 2503–2519. [Google Scholar] [CrossRef] [PubMed]
- Magliani, W.; Giovati, L.; Ciociola, T.; Sperinde, M.; Santinoli, C.; Conti, G.; Conti, S.; Polonelli, L. Antibodies as a source of anti-infective peptides: An update. Future Microbiol. 2015, 10, 1163–1175. [Google Scholar] [CrossRef] [PubMed]
- Polonelli, L.; Ciociola, T.; Magliani, W.; Zanello, P.P.; D’Adda, T.; Galati, S.; De Bernardis, F.; Arancia, S.; Gabrielli, E.; Pericolini, E.; et al. Peptides of the constant region of antibodies display fungicidal activity. PLoS ONE 2012, 7, e34105. [Google Scholar] [CrossRef]
- Shi, Z.; Olson, C.A.; Rose, G.D.; Baldwin, R.L.; Kallenbach, N.R. Polyproline II structure in a sequence of seven alanine residues. Proc. Natl. Acad. Sci. USA 2002, 99, 9190–9195. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.; Chen, K.; Liu, Z.; Ng, A.; Bracken, W.C.; Kallenbach, N.R. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with beta-sheet scales. Proc. Natl. Acad. Sci. USA 2005, 102, 17964–17968. [Google Scholar] [CrossRef]
- Chen, K.; Liu, Z.; Zhou, C.; Shi, Z.; Kallenbach, N.R. Neighbor effect on PPII conformation in alanine peptides. J. Am. Chem. Soc. 2005, 127, 10146–10147. [Google Scholar] [CrossRef]
- Morrison, K.L.; Weiss, G.A. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001, 5, 302–307. [Google Scholar] [CrossRef]
- Carvajal-Rondanelli, P.; Arostica, M.; Alvarez, C.A.; Ojeda, C.; Albericio, F.; Aguilar, L.F.; Marshall, S.H.; Guzman, F. Understanding the antimicrobial properties/activity of an 11-residue Lys homopeptide by alanine and proline scan. Amino Acids 2018, 50, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Jarvet, J.; Damberg, P.; Danielsson, J.; Johansson, I.; Eriksson, L.E.G.; Gräslund, A. A left-handed 31 helical conformation in the Alzheimer Abeta(12-28) peptide. FEBS Lett. 2003, 555, 371–374. [Google Scholar] [CrossRef]
- Chellgren, B.W.; Creamer, T.P. Short sequences of non-proline residues can adopt the polyproline II helical conformation. Biochemistry 2004, 43, 5864–5869. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Woody, R.W.; Kallenbach, N.R. Is polyproline II a major backbone conformation in unfolded proteins? Adv. Protein Chem. 2002, 62, 163–240. [Google Scholar] [PubMed]
- Foroutan, A.; Lazarova, T.; Padros, E. Study of membrane-induced conformations of substance P: Detection of extended polyproline II helix conformation. J. Phys. Chem. B 2011, 115, 3622–3631. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, A.A.; Sternberg, M.J.; Makarov, A.A. Polyproline-II helix in proteins: Structure and function. J. Mol. Biol. 2013, 425, 2100–2132. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.S.; Cutro, A.C.; Frias, M.A.; Disalvo, E.A. Interaction of Phenylalanine with DPPC Model Membranes: More Than a Hydrophobic Interaction. J. Phys. Chem. B 2015, 119, 15844–15847. [Google Scholar] [CrossRef]
- Shahmiri, M.; Cornell, B.; Mechler, A. Phenylalanine residues act as membrane anchors in the antimicrobial action of Aurein 1.2. Biointerphases 2017, 12, 05G605. [Google Scholar] [CrossRef]
- Ciociola, T.; Giovati, L.; Conti, S.; Magliani, W.; Santinoli, C.; Polonelli, L. Natural and synthetic peptides with antifungal activity. Future Med. Chem. 2016, 8, 1413–1433. [Google Scholar] [CrossRef]
- Bondaryk, M.; Staniszewska, M.; Zielinska, P.; Urbanczyk-Lipkowska, Z. Natural Antimicrobial Peptides as Inspiration for Design of a New Generation Antifungal Compounds. J. Fungi. (Basel) 2017, 3, 46. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Naseem, M.; Khan, R.S.; Najeeb, S. Human Oral Defensins Antimicrobial Peptides: A Future Promising Antimicrobial Drug. Curr. Pharm Des. 2018, 24, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- Ciociola, T.; Pertinhez, T.A.; Giovati, L.; Sperinde, M.; Magliani, W.; Ferrari, E.; Gatti, R.; D’Adda, T.; Spisni, A.; Conti, S.; et al. Dissecting the Structure-Function Relationship of a Fungicidal Peptide Derived from the Constant Region of Human Immunoglobulins. Antimicrob. Agents Chemother. 2016, 60, 2435–2442. [Google Scholar] [CrossRef] [Green Version]
- Machado, A.; Sforca, M.L.; Miranda, A.; Daffre, S.; Pertinhez, T.A.; Spisni, A.; Miranda, M.T.M. Truncation of amidated fragment 33-61 of bovine alpha-hemoglobin: Effects on the structure and anticandidal activity. Biopolymers 2007, 88, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Polonelli, L.; Beninati, C.; Teti, G.; Felici, F.; Ciociola, T.; Giovati, L.; Sperinde, M.; Lo Passo, C.; Pernice, I.; Domina, M.; et al. Yeast killer toxin-like candidacidal Ab6 antibodies elicited through the manipulation of the idiotypic cascade. PLoS ONE 2014, 9, e105727. [Google Scholar] [CrossRef] [PubMed]
- Polonelli, L.; Ciociola, T.; Sperinde, M.; Giovati, L.; D’Adda, T.; Galati, S.; Travassos, L.R.; Magliani, W.; Conti, S. Fungicidal activity of peptides encoded by immunoglobulin genes. Sci. Rep. 2017, 7, 10896. [Google Scholar] [CrossRef] [PubMed]










| Residue | 3JHN-α T11F | 3JHN-α * (PPII) | 3JHN-α * (β-sheet) | %PPII |
|---|---|---|---|---|
| Thr1 | − | 6.06 | 9.62 | − |
| Cys2 | 6.92 | 5.93 | 9.70 | 74 |
| Arg3 | 6.87 | 5.89 | 9.51 | 73 |
| Val4 | 8.00 | 6.09 | 9.82 | 48 |
| Asp5 | 7.16 | 5.72 | 9.69 | 64 |
| His6 | 7.67 | 5.65 | 9.41 | 46 |
| Arg7 | 6.92 | 5.89 | 9.51 | 71 |
| Gly8 | nd | − | − | nd |
| Leu9 | 7.13 | 5.60 | 9.24 | 58 |
| Thr10 | 8.93 | 6.06 | 9.62 | 19 |
| Phe11 | 8.19 | 5.34 | 9.86 | 37 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pertinhez, T.A.; Ciociola, T.; Giovati, L.; Magliani, W.; Belletti, S.; Polonelli, L.; Conti, S.; Spisni, A. Dissection of the Structural Features of a Fungicidal Antibody-Derived Peptide. Int. J. Mol. Sci. 2018, 19, 3792. https://doi.org/10.3390/ijms19123792
Pertinhez TA, Ciociola T, Giovati L, Magliani W, Belletti S, Polonelli L, Conti S, Spisni A. Dissection of the Structural Features of a Fungicidal Antibody-Derived Peptide. International Journal of Molecular Sciences. 2018; 19(12):3792. https://doi.org/10.3390/ijms19123792
Chicago/Turabian StylePertinhez, Thelma A., Tecla Ciociola, Laura Giovati, Walter Magliani, Silvana Belletti, Luciano Polonelli, Stefania Conti, and Alberto Spisni. 2018. "Dissection of the Structural Features of a Fungicidal Antibody-Derived Peptide" International Journal of Molecular Sciences 19, no. 12: 3792. https://doi.org/10.3390/ijms19123792
APA StylePertinhez, T. A., Ciociola, T., Giovati, L., Magliani, W., Belletti, S., Polonelli, L., Conti, S., & Spisni, A. (2018). Dissection of the Structural Features of a Fungicidal Antibody-Derived Peptide. International Journal of Molecular Sciences, 19(12), 3792. https://doi.org/10.3390/ijms19123792