iTRAQ-Based Comparative Proteomic Analysis of the Roots of TWO Winter Turnip Rapes (Brassica rapa L.) with Different Freezing-Tolerance
Abstract
:1. Introduction
2. Results
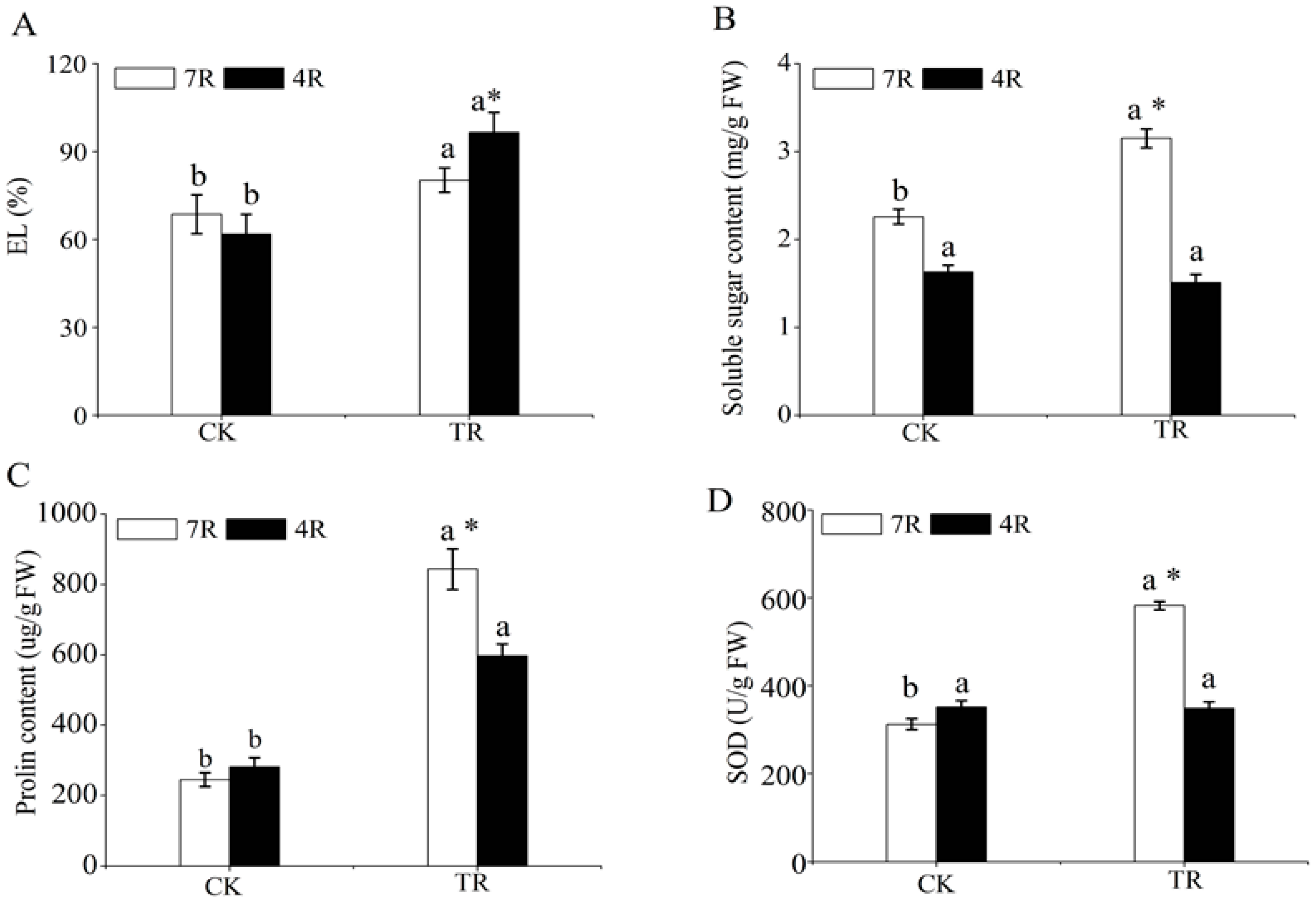
2.1. Analysis of Physiological Parameters under Freezing Stress
2.2. Primary Data Analysis and Protein Identification by iTRAQ
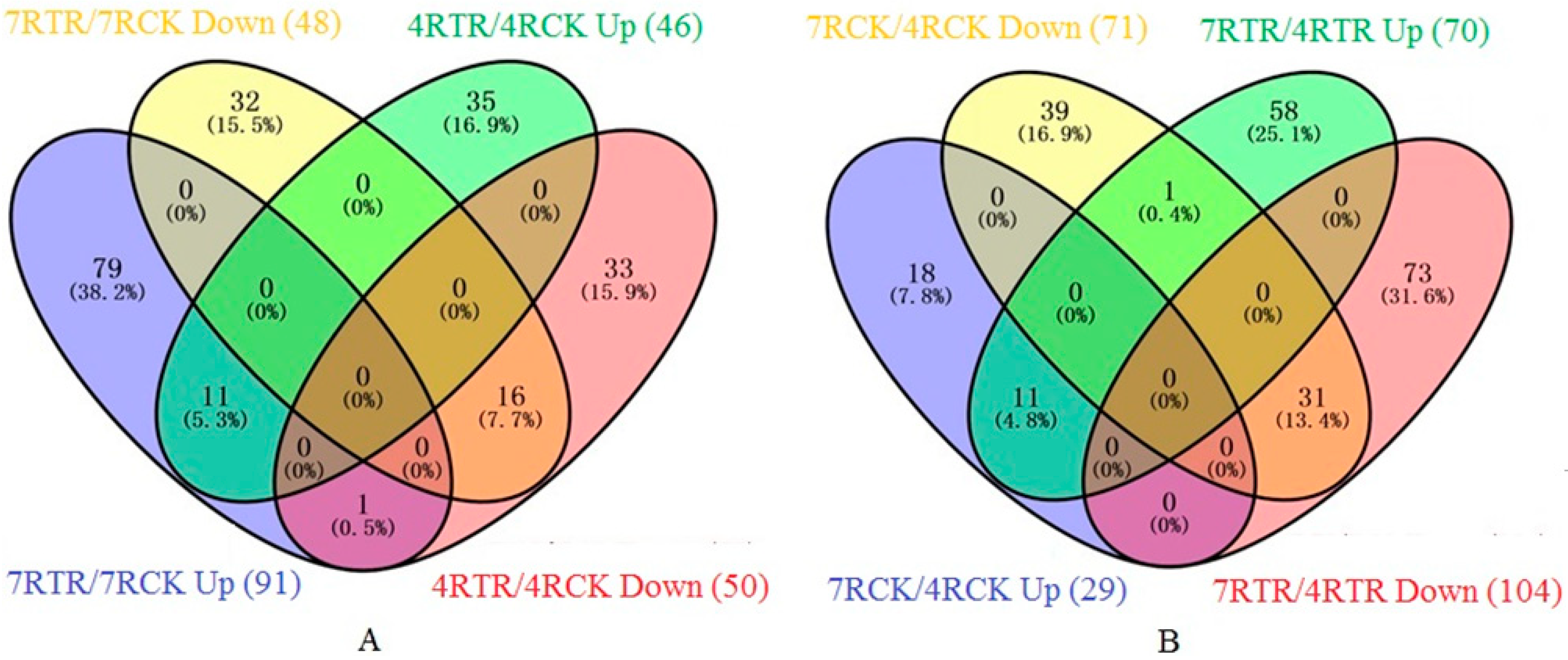
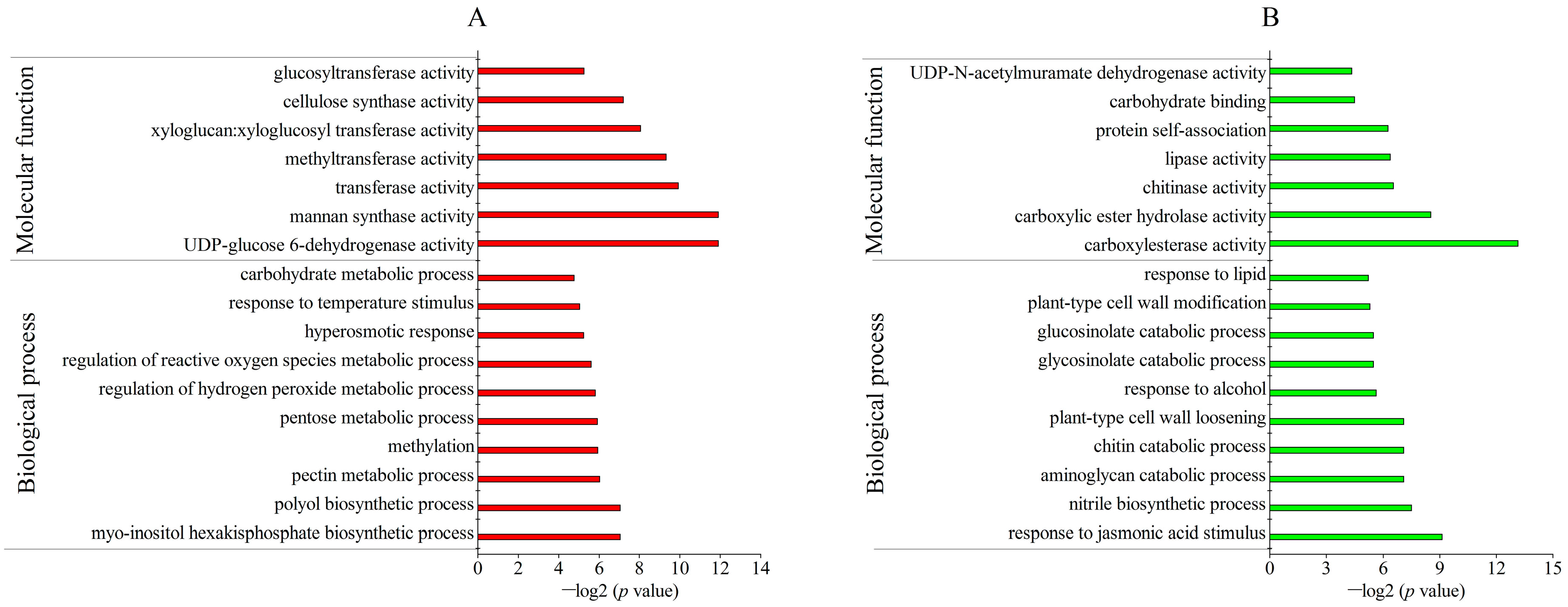
2.3. Identification and Gene Ontology (GO) Annotation of Differentially Abundant Proteins (DAPs) under Freezing Stress
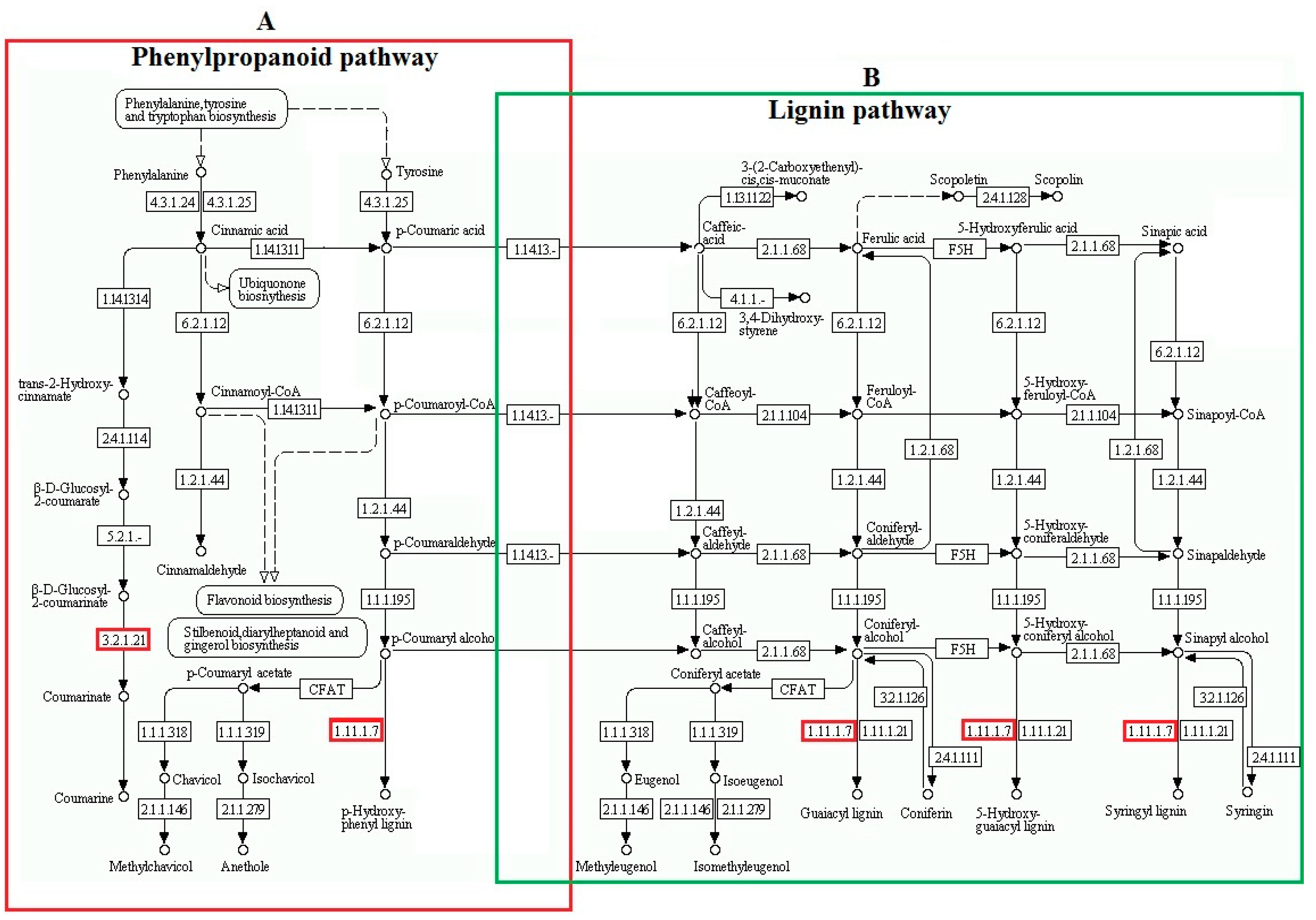
2.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis of DAPs between Two Freezing-Stressed Varieties
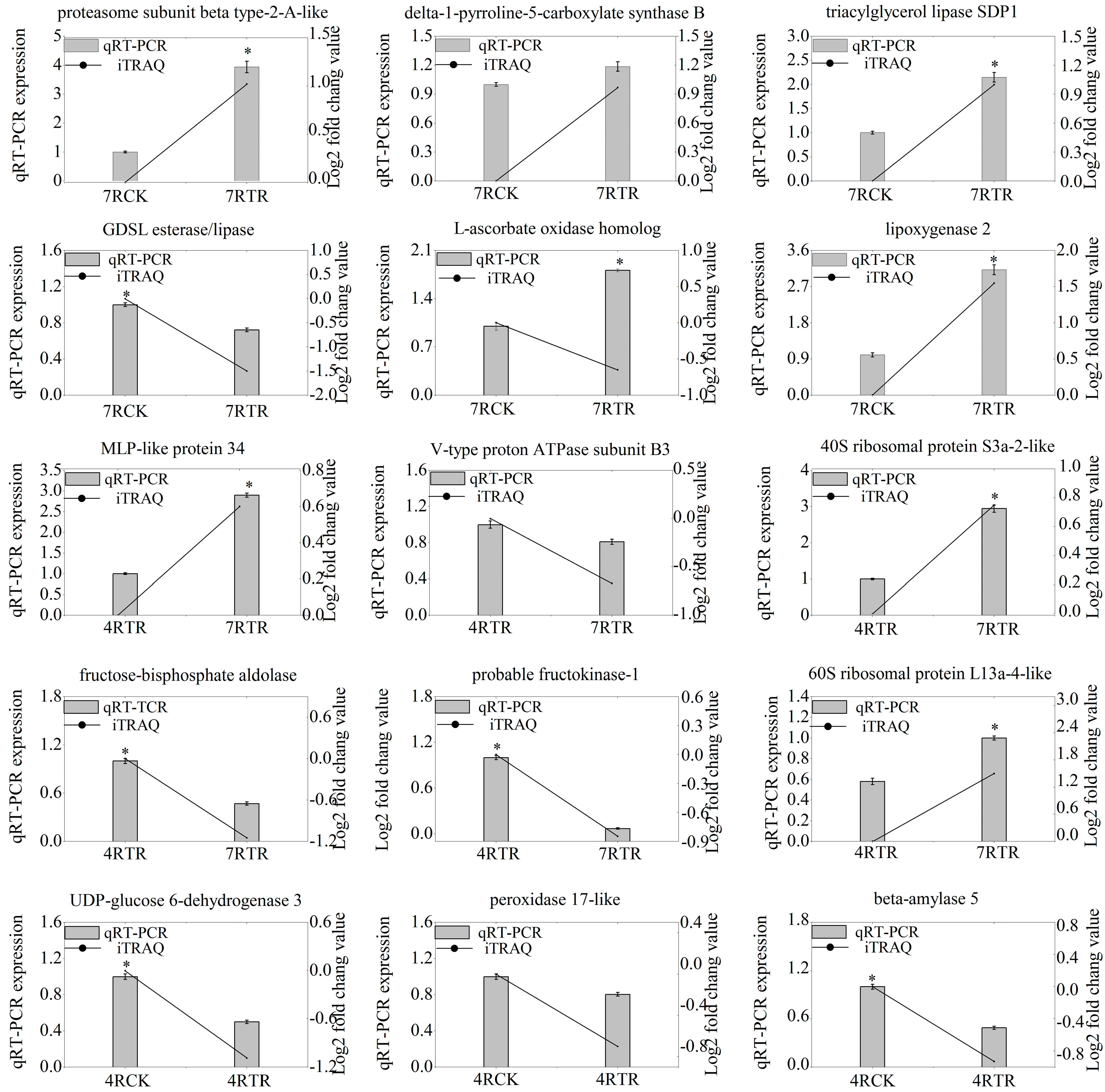
2.5. Validation of Genes-Encoding DAPs by qRT-PCR
3. Discussion
3.1. Increased Abundance of Ribosome-Related Proteins under Freezing Stress
3.2. Increased Abundance of Proteins Involved in the Biosynthesis of Secondary Metabolites
3.3. Decreased Abundance of Energy and Carbohydrate Metabolism-Related Proteins under Freezing Stress
3.4. Decreased Abundance of Alpha-linolenic Acid Metabolism-Related Proteins under Cold Stress
3.5. Decreased Abundance of Ascorbate and Aldarate Metabolism-Related Proteins under Freezing Stress
3.6. Carbon Fixation-Related Proteins in Freezing-Stressed Winter Rapes
4. Materials and Methods
4.1. Plant Materials and Freezing Stress Treatment
4.2. Analysis of Physiological and Biochemical Parameters
4.3. Protein Extraction and iTRAQ Labeling
4.4. Separation of Peptides and LC-MS/MS Analysis
4.5. iTRAQ Protein Identification and Quantification
4.6. RNA Extraction and qPCR Analysis of Gene Expression
4.7. Data Treatment and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| iTRAQ | isobaric tags for relative and absolute quantification |
| 2DE | two-dimensional electrophoresis |
| EL | electrolyte leakage |
| SOD | superoxide dismutase |
| APX | ascorbateperoxidase |
| TEAB | triethylamonium bicarbonate |
| CK | control |
| TR | treatment |
| DAPs | differently accumulated proteins |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| ROS | reactive oxygen species |
| FBPA | fructose-bisphosphate aldolase |
| cPGM | probable phosphoglucomutase cytoplasmic |
| FRK | fructokinase |
| POX | peroxidase |
| GELP | GDSL esterase/lipase-like protein |
| V-ATPase | V-type proton ATPase subunit proteins |
| P5CS | delta-1-pyrroline-5-carboxylate synthase |
| JA | jasmonic acid |
| AOC | allene oxide cyclase |
| MFP | glyoxysomal fatty acid beta-oxidation multifunctional protein |
| WAT1 | protein walls are thin 1 |
| OPR | 12-oxophytodienoate reductase |
| MDAR | monodehydroascorbate reductase |
| MLP | major latex-like protein |
| GR | glutathione reductase |
| RuBisCO | ribulose bisphosphate carboxylase large chain |
References
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Wilen, R.W.; Wu, G.H.; Robertson, A.J.; Gusta, L.V. Dehydrin gene expression and leaf water potential differs between spring and winter cereals during cold acclimation. J. Plant Phys. 2000, 156, 394–400. [Google Scholar] [CrossRef]
- Xu, W.; Li, R.; Zhang, N.; Ma, F.; Jiao, Y.; Wang, Z. Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol. Biol. 2014, 86, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Pradet-Balade, B.; Boulmé, F.; Beug, H.; Müllner, E.W.; Garcia-Sanz, J.A. Translation control: Bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001, 26, 225–229. [Google Scholar] [CrossRef]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.K.M. Proteomics and a future generation of plant molecular biologists. Plant Mol. Biol. 2002, 48, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.S.; Chong, P.K.; Pham, T.K.; Wright, P.C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 2007, 6, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Zieske, L.R. A perspective on the use of iTRAQ TM reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006, 57, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Men, J.L.; Chang, M.C.; Feng, C.P.; Yuan, L.G. ITRAQ-based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress. J. Proteomics 2017, 156, 75–84. [Google Scholar] [CrossRef]
- Hu, Z.; Liu, A.; Bi, A.; Amombo, E.; Gitau, M.M.; Huang, X.; Chen, L.; Fu, J. Identification of differentially expressed proteins in bermudagrass response to cold stress in the presence of ethylene. Environ. Exp. Bot. 2017, 139, 67–78. [Google Scholar] [CrossRef]
- Wang, X.; Shan, X.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Han, J.; Xue, C.; Yuan, Y. iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways responding to chilling stress in maize seedlings. J. Proteomics 2016, 146, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.C.; Wu, J.Y.; Fang, Y.; Liu, Q.; Yang, R.Y.; Ma, W.G.; Li, X.C.; Zhang, J.J.; Zhang, P.F.; Lei, J.M. Growth and development characteristics of winter rapeseed northern-extended from the cold and arid regions in China. Acta Agron. Sin. 2010, 36, 2124–2134. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Q.; Sun, W.C.; Liu, Z.G.; Wang, Z.J.; Fang, Y.; Wu, J.Y.; Li, X.C.; Fang, Y. Safe wintering and economic and ecological benefit of winter rapeseed in dry and cold areas of northern China. Chin. J. Appl. Ecol. 2015, 26, 3035–3044. [Google Scholar]
- Yang, N.N.; Sun, W.C.; Liu, Z.G.; Shi, P.H.; Fang, Y.; Wu, J.Y.; Zeng, X.C.; Kong, D.J.; Lu, M.H.; Wang, Y. Morphological characters and physiological mechanisms of cold resistance of winter rapeseed in northern China. Sci. Agric. Sin. 2014, 47, 452–461. [Google Scholar]
- Yoshida, M.; Abe, J.; Moriyama, M.; Kuwabara, T. Carbohydrate levels among winter wheat cultivars varying in freezing tolerance and snow mold resistance during autumn and winter. Physiol. Plantarum. 1998, 103, 8–16. [Google Scholar] [CrossRef]
- Pastori, G.; Foyer, C.H.; Mullineaux, P. Low temperature-induced changes in the distribution of H2O2 and antioxidants between the bundle sheath and mesophyll cells of maize leaves. J. Exp. Bot. 2000, 51, 107–113. [Google Scholar] [CrossRef]
- Premachandra, G.S.; Saneoka, H.; Fujita, K.; Ogata, S. Leaf water relations, osmotic adjustment, cell membrane stability, epicuticular wax load and growth as affected by increasing water deficits in sorghum. J. Exp. Bot. 1992, 43, 1569–1576. [Google Scholar] [CrossRef]
- Wu, S.; Hu, C.; Tan, Q.; Nie, Z.; Sun, X. Effects of molybdenum on water utilization, antioxidative defense system and osmotic-adjustment ability in winter wheat (Triticum aestivum) under drought stress. Plant Physiol. Biochem. 2014, 83, 365–374. [Google Scholar] [CrossRef]
- Uemura, M.; Steponkus, P. Modification of the intracellular sugar content alters the incidence of freeze-induced membrane lesions of protoplasts isolated from Arabidopsis thaliana leaves. Plant Cell Environ. 2003, 26, 1083–1096. [Google Scholar] [CrossRef]
- Hong, Z.; Lakkineni, K.; Zhang, Z.; Verma, D.P.S. Removal of feedback inhibition of 1-pyroline-5-carboxilare synthetase results in increased proline accumulation and protection of plant from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Gossett, D.R.; Millhollon, E.P.; Lucas, M.C.; Banks, S.W.; Marney, M.M. The effects of NaCl on antioxidant enzyme activities in callus tissue of salt-tolerant and salt-sensitive cotton cultivars (Gossypium hirsutum L.). Plant Cell Rep. 1994, 13, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.L. Advances of the studies on the mechanism of plant cold hardiness. Chin. Bull. Bot. 1992, 3, 17–22. [Google Scholar]
- Carroll, A.J. The Arabidopsis cytosolic ribosomal proteome: From form to function. Front. Plant Sci. 2013, 4, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, B.L.; Edrada-Ebel, R.; Da Costa, F.B. Effect of the environment on the secondary metabolic profile of Tithonia diversifolia: A model for environmental metabolomics of plants. Sci. Rep. 2016, 6, 29265. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, L.; Shang, H.; Liu, S.; Peng, J.; Gong, W.; Shi, Y.; Zhang, S.; Li, J.; Gong, J.; et al. iTRAQ-based quantitative proteomic analysis of cotton roots and leaves reveals pathways associated with salt stress. PLoS ONE 2016, 11, e0148487. [Google Scholar] [CrossRef]
- Rogers, L.A.; Campbell, M.M. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004, 164, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Lange, B.M.; Lapierre, C.; Sandermann, H., Jr. Elicitor-induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 1995, 108, 1277–1287. [Google Scholar] [CrossRef]
- Hong, J.K.; Choi, H.W.; Hwang, I.S.; Kim, D.S.; Kim, N.H.; Choi, D.S.; Kim, Y.J.; Hwang, B.K. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 2008, 227, 539–558. [Google Scholar] [CrossRef]
- Clauss, K.; Baumert, A.; Nimtz, M.; Milkowski, C.; Strack, D. Role of a GDSL lipase-like protein as sinapine esterase in Brassicaceae. Plant J. 2008, 53, 802–813. [Google Scholar] [CrossRef] [Green Version]
- Quiroga, M.; Guerrero, C.; Botella, M.A.; Barceló, A.; Amaya, I.; Medina, M.I.; Alonso, F.J.; de Forchetti, S.M.; Tigier, H.; Valpuesta, V. A tomato peroxidase involved in the synthesis of lignin and suberin. Plant Physiol. 2000, 122, 1119. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratge-Faillie, C.; Novak, O.; Morreel, K.; Lacombe, B.; Martinez, Y.P.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef] [PubMed]
- Ranocha, P.; Denance, N.; Vanholme, R.; Freydier, A.; Martinez, Y.; Hoffmann, L.; Kohler, L.; Pouzet, C.; Renou, J.P.; Sundberg, B.; et al. Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010, 63, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Rajashekar, C.B.; Lafta, A. Cell-wall changes and cell tension in response to cold acclimation and exogenous abscisic acid in leaves and cell cultures. Plant Physiol. 1996, 111, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Rutter, W.J. Evolution of aldolase. Fed. Proc. 1964, 23, 1248–1257. [Google Scholar] [PubMed]
- Mo, Y.; Guo, Z. The effects of temperature stress on photosynthesis and nitrogen reduction of alfalfa and guyanastylosanthes. Acta Agrestia Sin. 2008, 16, 100–102. [Google Scholar]
- Egli, B.; Kölling, K.; Köhler, C.; Zeeman, S.C.; Streb, S. Loss of cytosolic phosphoglucomutase compromises gametophyte development in Arabidopsis. Plant Physiol. 2010, 154, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Malinova, I.; Kunz, H.H.; Alseekh, S.; Herbst, K.; Fernie, A.R.; Gierth, M.; Fettke, J. Reduction of the cytosolic phosphoglucomutase in Arabidopsis reveals impact on plant growth, seed and root development, and carbohydrate partitioning. PLoS ONE 2014, 9, e112468. [Google Scholar] [CrossRef]
- Klimov, S.V.; Popov, V.N.; Dubinina, I.M.; Burakhanova, E.A.; Trunova, T.I. The decreased cold-resistance of chilling-sensitive plants is related to suppressed CO2 assimilation in leaves and sugar accumulation in roots. Russ. J. Plant Physiol. 2002, 49, 776–781. [Google Scholar] [CrossRef]
- Granot, D. Role of tomato hexose kinases. Funct. Plant Biol. 2007, 34, 564–570. [Google Scholar] [CrossRef]
- Schaffer, A.A.; Petreikov, M. Sucrose-to-Starch Metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol. 1997, 113, 739–746. [Google Scholar] [CrossRef]
- Wu, X.; Yuan., J.; Luo, A.; Chen, Y.; Fan, Y. Drought stress and re-watering increase secondary metabolites and enzyme activity in dendrobium moniliforme. Ind. Crop. Prod. 2016, 94, 385–393. [Google Scholar] [CrossRef]
- Ragan, C.I.; Galante, Y.M.; Hatefi, Y.; Ohnishi, T. Resolution of mitochondrial NADH dehydrogenase and isolation of two iron-sulfur proteins. Biochemistry 1982, 21, 590–594. [Google Scholar] [CrossRef]
- Chen, J.S.; Zhu, R.F.; Zhang, Y.X.; Cao, G.; Di, G.L. Yieds of alfalfa varieties with different fall dormancy levels in northeast China. Pak. J. Bot. 2014, 46, 167–172. [Google Scholar]
- Mckenzie, J.S.; Paquin, R.; Duke, S.H. Cold and heat tolerance. Alfalfa Alfalfa Improv. 1988, 29, 259–302. [Google Scholar]
- Guo, M.L.; Gao, W.X.; Li, L.; Li, H.; Xu, Y.L.; Zhou, C.X. Proteomic and phosphoproteomic analyses of NaCl stress-responsive proteins in Arabidopsis roots. J. Plant Interact. 2014, 9, 396–401. [Google Scholar] [CrossRef]
- Schulze, W.X.; Schneider, T.; Starck, S.; Martinoia, E.; Trentmann, O. Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 2012, 69, 529–541. [Google Scholar] [CrossRef]
- Uemura, M.; Steponkus, P.L. Cold acclimation in plants: Relationship between the lipid composition and the cryostability of the plasma membrane. J. Plant Res. 1999, 112, 245–254. [Google Scholar] [CrossRef]
- HernaÂndez, M.L.; Sicardo, M.D.; MartõÂnez-Rivas, J.M. Differential contribution of endoplasmic reticulum and chloroplast ω-3 fatty acid desaturase genes to the linolenic acid content of olive (Olea europaea) fruit. Plant Cell Physiol. 2016, 57, 138–151. [Google Scholar] [CrossRef]
- Ohlrogge, J.; Browse, J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [CrossRef]
- Kuiper, P.J. Lipids in alfalfa leaves in relation to cold hardiness. Plant Physiol. 1970, 45, 684–686. [Google Scholar] [CrossRef]
- Jiang, N.; Tan, X.F.; Zhang, L.; Zeng, Y.L. Gene Analysis of α-Linolenic acid metabolism of Camellia oleifera seeds based on RNA-Seq. Sci. Silvae Sin. 2014, 50, 68–75. [Google Scholar]
- Leterrier, M.; Corpas, F.J.; Barroso, J.B.; Sandalio, L.M.; delRío, L.A. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiol. 2005, 138, 2111–2123. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Oidaira, H.; Sano, S.; Koshiba, T.; Ushimaru, T. Enhancement of antioxidative enzyme activities in chilled rice seedlings. J. Plant Physiol. 2000, 156, 811–813. [Google Scholar] [CrossRef]
- Tao, D.L.; Oquist, G.; Wingsle, G. Active oxygen scavengers during cold acclimation of Scots pine seedlings in relation to freezing tolerance. Cryobiology 1998, 37, 38–45. [Google Scholar] [CrossRef]
- Kubo, A.; Aono, M.; Nakajima, N.; Saji, H.; Tanaka, K.; Kondo, N. Differential responses in activity of antioxidant enzymes to different environmental stresses in Arabidopsis thaliana. J. Plant Res. 1999, 112, 279–290. [Google Scholar] [CrossRef]
- Jiménez, A.; Hernández, J.A.; Pastori, G.; del Río, L.A.; Sevilla, F. Role of the ascorbate-glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiol. 1998, 118, 1327–1335. [Google Scholar] [CrossRef]
- Bareven, A.; Noor, E.; Lewis, N.E.; Miloa, R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. USA 2010, 107, 8889–8894. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.H.; Perez-Losada, J.; Wu, D.; Delrosario, R.; Tsunematsu, R.; Nakayama, K.I.; Brown, K.; Bryson, S.; Balmain, A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 2004, 432, 775–779. [Google Scholar] [CrossRef]
- Huang, X.B.; Shi, H.Y.; Hu, Z.R.; Liu, A.; Amombo, E.; Chen, L.; Fu, J.M. ABA is involved in regulation of cold stress response in bermudagrass. Front. Plant Sci. 2017, 8, 1613. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Deng, Y.M.; Chen, S.M.; Chen, F.D.; Cheng, X.; Zhang, F. The embryo rescue derived intergeneric hybrid between chrysanthemum and Ajania przewalskii shows enhanced cold tolerance. Plant Cell Rep. 2011, 30, 2177–2186. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Yang, L.T.; Qi, Y.P.; Lu, Y.B.; Guo, P.; Sang, W.; Feng, H.; Zhang, H.X.; Chen, L.S. iTRAQ protein profile analysis of Citrus sinensis roots in response to long-term boron-deficiency. J. Proteomics 2013, 93, 179–206. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wang, J.C.; Yao, L.R.; Li, B.C.; Meng, Y.X.; Ma, X.L.; Yong, L.; Si, E.; Ren, P.R.; Yang, K.; Shang, X.W.; et al. Comparative proteomic analysis of cultured suspension cells of the Halophyte halogeton glomeratus by iTRAQ provides insights into response mechanisms to salt stress. Front. Plant Sci. 2016, 7, 110. [Google Scholar] [CrossRef]
- The Brassica Database (BRAD). Available online: http://brassicadb.org/brad/datasets/pub/BrassicaceaeGenome/Brassica_rapa/V2.0/Scaffold2.0/BrapaV2.0_evm.out.pep.fa.gz (accessed on 8 October 2016).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Uniprot ID | Protein | Fold Changes a | ||
|---|---|---|---|---|
| 7RTR/7RCK Fold Change | 4RTR/4RCK Fold Change | 7RTR/4RTR Fold Change | ||
| PSB2A_ARATH | Proteasome subunit beta type-2-A | 2.028884928 | - | 1.716545467 |
| ECH2_ARATH | Enoyl-CoA hydratase 2, peroxisomal | 0.636897776 | - | - |
| SPD1_ARATH | Spermidine synthase 1 | 1.736926897 | - | - |
| NOP56_SCHPO | Nucleolar protein 56 | 1.801418945 | - | - |
| LOX2_ARATH | Lipoxygenase 2, chloroplastic | 2.921783613 | - | - |
| P5CS2_ARATH | Delta-1-pyrroline-5-carboxylate synthase B | 1.957524834 | - | - |
| CBF5_ARATH | H/ACA ribonucleoprotein complex subunit 4 | 1.995205069 | - | - |
| SAHH1_ARATH | Adenosylhomocysteinase 1 | - | 1.75364388 | - |
| BAM5_ARATH | Beta-amylase 5 | - | 0.52337023 | 2.646139818 |
| PGIP1_ARATH | Polygalacturonase inhibitor 1 | - | 0.61557221 | - |
| PER17_ARATH | Peroxidase 17 | - | 0.61557221 | - |
| UGDH3_ARATH | UDP-glucose 6-dehydrogenase 3 | - | 1.54961483 | - |
| GDL20_ARATH | GDSL esterase/lipase | - | 0.46826289 | 1.695003118 |
| VATB3_ARATH | V-type proton ATPase subunit B3 | - | - | 0.553787078 |
| ALF_CICAR | Fructose-bisphosphate aldolase | - | - | 0.502403227 |
| MDAR3_ARATH | Probable monodehydroascorbate reductase | - | - | 0.478651486 |
| NDUA9_ARATH | Nicotinamide adenine dinucleotide (NADH) dehydrogenase [ubiquinone] 1 alpha subcomplex | - | - | 0.608297667 |
| NDUS1_ARATH | NADH dehydrogenase [ubiquinone] iron-sulfur protein 1 | - | - | 0.551175594 |
| SCRK1_ARATH | Probable fructokinase-1 | - | 1.55896 | 0.629339177 |
| RS3A2_ARATH | 40S ribosomal protein S3a-2 | - | - | 1.624504793 |
| PERA2_ARMRU | Peroxidase A2 | - | - | 1.990433075 |
| AOC4_ARATH | Allene oxide cyclase 4 | - | - | 0.566248831 |
| RBL_BRAOL | Ribulose bisphosphate carboxylase large chain | - | 0.55295901 | 0.510849202 |
| PGMC1_ARATH | Probable phosphoglucomutase, cytoplasmic 1 | 0.662925032 | - | 0.545940075 |
| PER3_ARMRU | Peroxidase C3 | 1.622786225 | - | 2.427233592 |
| R13A4_ARATH | 60S ribosomal protein L13a-4 | - | - | 2.828427125 |
| RS3A_BRACM | 40S ribosomal protein S3a | - | - | 1.686837421 |
| RS262_ARATH | 40S ribosomal protein S26-2 | - | - | 1.688842594 |
| WAT1_ARATH | Protein WALLS ARE THIN 1 | - | - | 1.684822612 |
| MFPA_BRANA | Glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a | - | - | 0.543328591 |
| MLP34_ARATH | MLP-like protein 34 | - | - | 1.512082689 |
| MDAR3_ARATH | Monodehydroascorbate reductase | - | - | 0.478651486 |
| OPR1_ARATH | 12-oxophytodienoate reductase 1 | - | - | 0.509956713 |
| ALF_CICAR | Fructose-bisphosphate aldolase | - | - | 0.451437041 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. iTRAQ-Based Comparative Proteomic Analysis of the Roots of TWO Winter Turnip Rapes (Brassica rapa L.) with Different Freezing-Tolerance. Int. J. Mol. Sci. 2018, 19, 4077. https://doi.org/10.3390/ijms19124077
Zeng X, Xu Y, Jiang J, Zhang F, Ma L, Wu D, Wang Y, Sun W. iTRAQ-Based Comparative Proteomic Analysis of the Roots of TWO Winter Turnip Rapes (Brassica rapa L.) with Different Freezing-Tolerance. International Journal of Molecular Sciences. 2018; 19(12):4077. https://doi.org/10.3390/ijms19124077
Chicago/Turabian StyleZeng, Xiucun, Yaozhao Xu, Jinjin Jiang, Fenqin Zhang, Li Ma, Dewei Wu, Youping Wang, and Wancang Sun. 2018. "iTRAQ-Based Comparative Proteomic Analysis of the Roots of TWO Winter Turnip Rapes (Brassica rapa L.) with Different Freezing-Tolerance" International Journal of Molecular Sciences 19, no. 12: 4077. https://doi.org/10.3390/ijms19124077
APA StyleZeng, X., Xu, Y., Jiang, J., Zhang, F., Ma, L., Wu, D., Wang, Y., & Sun, W. (2018). iTRAQ-Based Comparative Proteomic Analysis of the Roots of TWO Winter Turnip Rapes (Brassica rapa L.) with Different Freezing-Tolerance. International Journal of Molecular Sciences, 19(12), 4077. https://doi.org/10.3390/ijms19124077





