Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’)
Abstract
:1. Introduction
2. Results
2.1. Morphological Characteristics and Anatomical Observation
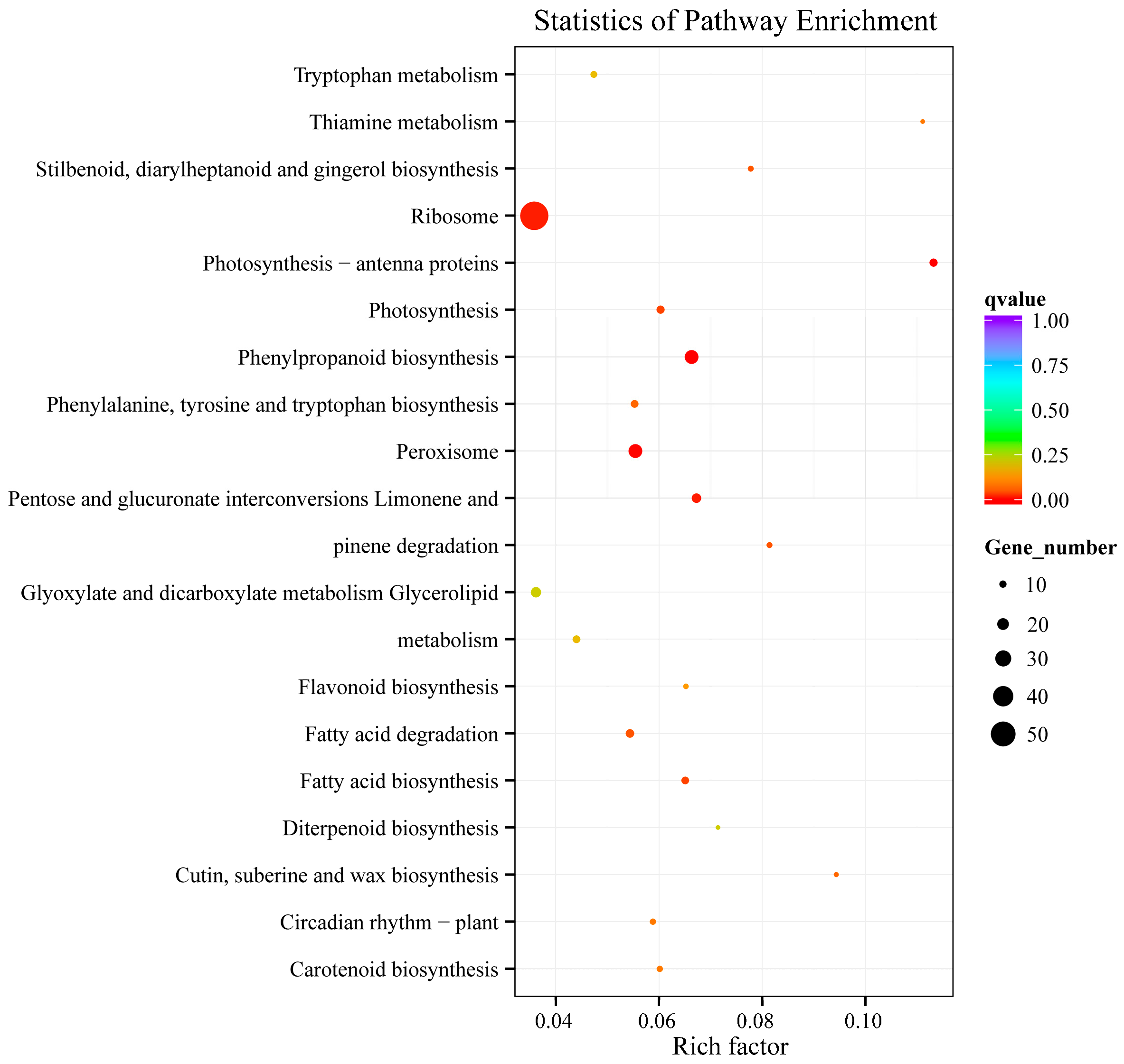
2.2. Overview of the RNA Sequencing and Classification of Enriched GO and KEGG Terms
2.3. Analysis of Differentially-Expressed Genes
2.4. qRT-PCR Validation of DEGs
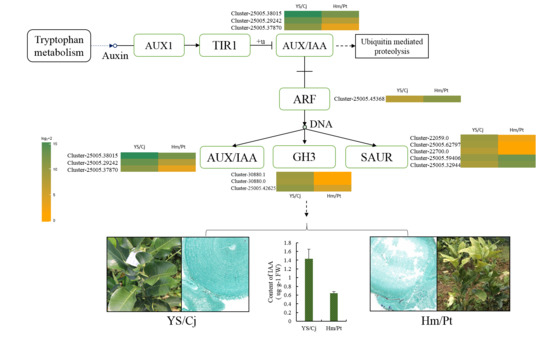
2.5. Auxin Signal Pathway and Response of Auxin Level
3. Discussion
3.1. Formation of the Stock/Scion Union
3.2. Metabolic Pathways
3.3. The Role of Hormones
4. Materials and Methods
4.1. Plant Materials
4.2. Anatomical Structure Observation
4.3. RNA Extraction, Library Construction, and Sequencing
4.4. De Novo Assembly and Gene Expression Analysis
4.5. Gene Annotation
4.6. qRT-PCR Validation
4.7. Determination of the Chlorophyll Content; Photosynthetic Characteristics
4.8. Determination of the Endogenous Phytohormones
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gong, Y.Q.; Qi, Y.H.; Fu, X.K.; Rui, Y.; Chen, T.; Tang, H.R.; Wang, X.R. Graft compatibility evaluation of different rootstock-scion combinations of honey pomelo at seedling stage. J. Trop. Subtrop. Bot. 2016, 24, 287–295. [Google Scholar]
- Melnyk, C.W.; Meyerowitz, E.M. Plant grafting. Curr. Biol. 2015, 25, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, domestication, and impacts on shoot phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Irisarri, P.; Zhebentyayeva, T.; Errea, P.; Pina, A. Differential expression of phenylalanine ammonia lyase (PAL) genes implies distinct roles in development of graft incompatibility symptoms in Prunus. Sci. Hortic. (Amsterdam) 2016, 204, 16–24. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, J.T.; Hu, F.C.; Qin, Y.H.; Wang, X.H.; Hu, G.B. Transcriptome changes between compatible and incompatible graft combination of Litchi chinensis by digital gene expression profile. Sci. Rep. 2017, 7, 3954. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, J.T.; Qin, Y.H.; Hu, G.B. Study on the graft compatibility between ‘Jingganghongnuo’ and other litchi cultivars. Sci. Hortic. (Amsterdam) 2016, 199, 56–62. [Google Scholar] [CrossRef]
- Qiu, L.L.; Jiang, B.; Fang, J.; Shen, Y.K.; Fang, Z.X.; Saravana, K.R.M.; Yi, K.K.; Shen, C.J.; Yan, D.L.; Zheng, B.S. Analysis of transcriptome in hickory (Carya cathayensis), and uncover the dynamics in the hormonal signaling pathway during graft process. BMC Genomics 2016, 17, 935. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.W.; Guo, L.X.; Liu, Y.Z.; Jin, L.F.; Hussain, S.B.; Du, W.; Deng, Z.; Peng, S.A. Transcriptome changes associated with boron deficiency in leaves of two citrus scion-rootstock combinations. Front. Plant Sci. 2017, 8, 317. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, B.; Achard, P. Long-distance transport of phytohormones through the plant vascular system. Curr. Opin. Plant Biol. 2016, 34, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, W.C. Plant grafting: Insights into tissue regeneration. Regeneration 2016, 4, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Kumar, J.; Kumar, A.; Chaudhury, A.; Singh, S.P. Grafting triggers differential responses between scion and rootstock. PLoS ONE 2015, 10, e0124438. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, L.B.; Wu, R.L. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2016, 214, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Corso, M.; Vannozzi, A.; Ziliotto, F.; Zouine, M.; Maza, E.; Nicolato, T.; Vitulo, N.; Meggio, F.; Valle, G.; Bouzayen, M.; et al. Grapevine rootstocks differentially affect the rate of ripening and modulate auxin-related genes in cabernet sauvignon berries. Front. Plant Sci. 2016, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J. The Trinity Package. Available online: https://github.com/trinityrnaseq/trinityrnaseq/wiki/Transcriptome-Contig-Nx-and-ExN50-stats (accessed on 27 September 2016).
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-Seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Yu, Y.; Tang, X.Y.; Chen, H.M.; Xiao, J.Y.; Su, X.D. Transcriptome analyses of insect cells to facilitate baculovirus-insect expression. Protein Cell 2016, 7, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Olmstead, M.A.; Lang, N.S.; Ewers, F.W.; Owens, S.A. Xylem vessel anatomy of sweet cherries grafted onto dwarfing and nondwarfing rootstocks. J. Am. Soc. Hortic. Sci. 2006, 131, 577–585. [Google Scholar]
- Cohen, R.; Burger, Y.; Horev, C.; Koren, A.; Edelstein, M. Introducing grafted cucurbits to modern agriculture: The Israeli experience. Plant Dis. 2007, 91, 916–923. [Google Scholar] [CrossRef]
- Aloni, B.; Cohen, R.; Karni, L.; Aktas, H.; Edelstein, M. Hormonal signaling in rootstock–scion interactions. Sci. Hortic. (Amsterdam) 2010, 127, 119–126. [Google Scholar] [CrossRef]
- Pina, A.; Errea, P. A review of new advances in mechanism of graft compatibility–incompatibility. Sci. Hortic. (Amsterdam) 2005, 106, 1–11. [Google Scholar] [CrossRef]
- Yin, H.; Yan, B.; Sun, J.; Jia, P.F.; Zhang, Z.J.; Yan, X.S.; Chai, J.; Ren, Z.Z.; Zheng, G.C.; Liu, H. Graft-union development: A delicate process that involves cell—Cell communication between scion and stock for local auxin accumulation. J. Exp. Bot. 2012, 63, 4219–4232. [Google Scholar] [CrossRef] [PubMed]
- Koepke, T.; Dhingra, A. Rootstock scion somatogenetic interactions in perennial composite plants. Plant Cell Rep. 2013, 32, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Dengler, N.G. Regulation of vascular development. J. Plant Growth Regul. 2001, 20, 1–13. [Google Scholar] [CrossRef]
- Pina, A.; Zhebentyayeva, T.; Errea, P.; Abbott, A. Isolation and molecular characterization of cinnamate 4-hydroxylase from apricot and plum. Biol. Plant. 2012, 56, 441–450. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Guillén-Climent, M.L.; Hernández-Clemente, R.; Catalina, A.; González, M.R.; Martín, P. Estimating leaf carotenoid content in vineyards using high resolution hyperspectral imagery acquired from an unmanned aerial vehicle (UAV). Agric. For. Meteorol. 2013, 171–172, 281–294. [Google Scholar] [CrossRef]
- Young, A.; Britton, G. Carotenoids in photosynthesis. Photochem. Photobiol. 1996, 63, 257–264. [Google Scholar]
- Ritz, T.; Damjanović, A.; Schulten, K.; Zhang, J.P.; Koyama, Y. Efficient light harvesting through carotenoids. Photosynth. Res. 2000, 66, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yan, T.; Fu, L.; Zhou, G.F.; Liu, Y.Z.; Peng, S.A. Differences in boron distribution and forms in four citrus scion—Rootstock combinations with contrasting boron efficiency under boron-deficient conditions. Trees 2014, 28, 1589–1598. [Google Scholar] [CrossRef]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem—Mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, C.W.; Schuster, C.; Leyser, O.; Meyerowitz, E.M. A developmental framework for graft formation and vascular reconnection in Arabidopsis thaliana. Curr. Biol. 2015, 25, 1306–1318. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Jaina, M.; Mitchell, A.L.; Potter, S.C.; Marco, P.; Matloob, Q.; Amaia, S.V. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, R7. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, X.M.; Jin, L.F.; Shi, C.Y.; Liu, Y.Z.; Peng, S.A. Identification and transcript analysis of two glutamate decarboxylase genes, CsGAD 1 and CsGAD 2, reveal the strong relationship between CsGAD 1 and citrate utilization in citrus fruit. Mol. Biol. Rep. 2014, 41, 6253–6262. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, E.; Koga, H.; Mori, M.; Mori, M. Auxin production by the rice blast fungus and its localization in host tissue. J. Phytopathol. 2011, 159, 522–530. [Google Scholar] [CrossRef]




| Sample ID | Raw Reads | Clean Reads | Clean Bases (G) | GC Content (%) | Q30 Ratio (%) | Mapped Ratio (%) |
|---|---|---|---|---|---|---|
| YS/Cj_1 | 50,901,804 | 49,522,214 | 7.43 | 43.81 | 92.77 | 83.91 |
| YS/Cj_2 | 54,064,700 | 51,584,962 | 7.74 | 43.29 | 92.94 | 79.9 |
| YS/Cj_3 | 45,805,274 | 44,227,228 | 6.63 | 43.7 | 90.04 | 81.72 |
| Hm/Pt_1 | 52,696,450 | 51,269,554 | 7.69 | 43.86 | 92.91 | 83.29 |
| Hm/Pt_2 | 44,024,194 | 41,907,724 | 6.29 | 43.17 | 93.42 | 79.18 |
| Hm/Pt_3 | 42,844,292 | 40,428,286 | 6.06 | 43.49 | 91.52 | 81.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, W.; Wang, Y.; Chen, Q.; Sun, B.; Tang, H.-R.; Pan, D.-M.; Wang, X.-R. Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’). Int. J. Mol. Sci. 2018, 19, 505. https://doi.org/10.3390/ijms19020505
He W, Wang Y, Chen Q, Sun B, Tang H-R, Pan D-M, Wang X-R. Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’). International Journal of Molecular Sciences. 2018; 19(2):505. https://doi.org/10.3390/ijms19020505
Chicago/Turabian StyleHe, Wen, Yan Wang, Qing Chen, Bo Sun, Hao-Ru Tang, Dong-Ming Pan, and Xiao-Rong Wang. 2018. "Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’)" International Journal of Molecular Sciences 19, no. 2: 505. https://doi.org/10.3390/ijms19020505
APA StyleHe, W., Wang, Y., Chen, Q., Sun, B., Tang, H.-R., Pan, D.-M., & Wang, X.-R. (2018). Dissection of the Mechanism for Compatible and Incompatible Graft Combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’). International Journal of Molecular Sciences, 19(2), 505. https://doi.org/10.3390/ijms19020505