The Effect of N-Terminal Domain Removal towards the Biochemical and Structural Features of a Thermotolerant Lipase from an Antarctic Pseudomonas sp. Strain AMS3
Abstract
:1. Introduction
2. Results and Discussion
2.1. Homology Modelling and Molecular Dynamic Simulation of AMS3 Lipase
Homology Modelling and Molecular Dynamic Simulation of Truncated AMS3 Lipase
2.2. Modification of AMS3 Lipase
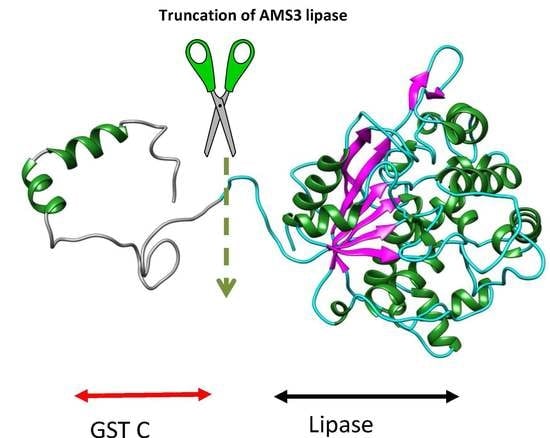
Deletion of N-Terminal Domain
2.3. Purification of Truncated AMS3 Lipase
2.4. Biochemical Characteristics of Truncated AMS3 Lipase
2.4.1. Effect of Temperature
2.4.2. Effect of pH
2.4.3. Effect of Metal Ions
2.4.4. Specific Activity of the Truncated Lipases to Different Substrates
2.4.5. Effect of Organic Solvents
3. Materials and Methods
3.1. Predicted Structure and Validation of AMS3 Lipase (Native)
3.2. Molecular Dynamic Simulation of Truncated AMS3 Lipase
3.3. Deletion of N-Terminal Domain of AMS3 Lipase
Sub Cloning and Primer Design without N-Terminal Domain
3.4. Purification of Truncated AMS3 Lipase
3.5. SDS-PAGE and Native PAGE Analysis
3.6. Lipase Activity Assay
3.7. Characterization of Truncated AMS3 Lipases
3.7.1. Effect of Temperature and the Stability on Truncated AMS3 Lipase
3.7.2. Effect of pH and Stability on Truncated AMS3 Lipase
3.7.3. Effect of Metal Ions on Truncated AMS3 Lipase
3.7.4. Effect of Substrate Specificity on Truncated AMS3 Lipase Activity
3.7.5. Effect of Natural Oil on Truncated AMS3 Lipase
3.7.6. Solvent Tolerant Profile of Truncated AMS3 Lipase
3.7.7. Predicted Structure and Validation of AMS3 Lipase (Native)
3.7.8. Molecular Dynamic Simulation of Truncated AMS3 Lipase
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Verma, N.; Thakur, S.; Bhatt, A.K. Microbial lipases: Industrial Applications and properties. Int. J. Biol. Sci. 2012, 1, 88–92. [Google Scholar]
- Singh, R.; Kumar, M.; Mittal, A.; Kumar, P. Microbial enzymes: Industrial progress in 21st century. 3 Biotech 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Sangeetha, R.; Arul Pandi, I.; Geetha, A. Bacterial lipases as potential industrial biocatalysts: An overview. Res. J. Microbiol. 2011, 6, 1–24. [Google Scholar] [CrossRef]
- Ali, M.S.; Ganasen, M.; Rahman, R.N.; Chor, A.L.; Salleh, A.B.; Basri, M. Cold-adapted RTX lipase from antarctic Pseudomonas sp. strain AMS8: Isolation, molecular modeling and heterologous expression. Protein J. 2013, 32, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baharum, S.N.; Abdul Rahman, R.N.Z.; Basri, M.; Salleh, A.B. Chaperone-dependent gene expression of organic solvent-tolerant lipase from Pseudomonas aeruginosa strain S5. Process Biochem. 2010, 45, 346–354. [Google Scholar] [CrossRef]
- Hvidsten, T.R.; Laegreid, A.; Kryshtafovych, A.; Andersson, G.; Fidelis, K.; Komorowski, J. A Comprehensive analysis of the structure-function relationship in proteins based on local structure similarity. PLoS ONE 2009, 4, e6266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.M.; Englander, S.W. The N-terminal to C-terminal motif in protein folding and function. Proc. Natl. Acad. Sci. USA 2005, 102, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Novototskaya-Vlasova, K.; Petrovskaya, L.; Rivkina, E.; Dolgikh, D.; Kirpichnikov, M. Characterization of a cold-active lipase from Psychrobacter cryohalolentisK5T and its deletion mutants. Biochemistry (Moscow) 2013, 78, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Zha, D.; Zhang, H.; Zhang, H.; Xu, L.; Yan, Y. N-terminal transmembrane domain of lipase LipA from Pseudomonas protegens Pf-5: A must for its efficient folding into an active conformation. J. Biochim. 2014, 105, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Fernando, P.; Abdulle, R.; Mohindra, A.; Guillemette, J.G.; Heikkila, J.J. Mutation or deletion of the C-terminal tail affects the function and structure of Xenopus laevis small heat shock protein, hsp30. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 133, 95–103. [Google Scholar] [CrossRef]
- Latip, W.; Rahman, R.N.Z.A.; Leow, A.T.C.; Shariff, F.M.; Ali, M.S.M. Expression and characterization of thermotolerant lipase with broad pH profiles isolated from an Antarctic Pseudomonas sp. strain AMS3. PeerJ 2016, 4, e2420. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; McArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemistry quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Hou, Y.; Hong, Y.; Han, X.; Yi, J.; Qu, J.; Lu, Y. Molecular cloning, expression and enzymatic characterization of glutathione S-transferase from Antarctic sea-ice bacteria Pseudoalteromonas sp. ANT506. Microbiol. Res. 2014, 169, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Amico, S.D.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R. Soc. B Biol. Sci. 2002, 357, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Kamarudin, N.H.A.; Rahman, R.N.Z.R.A.; Ali, M.S.M.; Thean, C.L.; Saleh, A.B.; Basri, M. Unscrambling the effect of C-terminal tail deletion on the stability of a cold-adapted, organic solvent stable lipase from Staphylococcus epidermidis AT2. Mol. Biotechnol. 2014, 56, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; Van der Spoel, D. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Inf. Comput. Sci. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Nagasundaram, N.; Zhu, H.; Liu, J.; Karthick, V.; George, P.D.C.; Chakraborty, C.; Chen, L. Analysing the effect of mutation on protein function and discovering potential inhibitors of CDK4: Molecular modelling and dynamics studies. PLoS ONE 2015, 10, e0133969. [Google Scholar]
- Chen, J.; Shen, B. Computational analysis of amino acid mutation: A proteome wide perspective. Curr. Proteom. 2009, 6, 228–234. [Google Scholar] [CrossRef]
- Kamal, M.Z.; Mohamad, T.A.S.; Krisnamoorithy, G.; Rao, N.M. Role of active site rigidity in activity: MD simulation and fluorescence study on lipase mutant. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Lawson, V.A.; Priola, S.A.; Meade-White, K.; Lawson, M.; Chesebro, B. Flexible N-terminal region of prion protein influences conformation of protease-resistant prion protein isoforms associated with cross-species scrapie infection in vivo and in vitro. J. Biol. Chem. 2004, 279, 13689–13695. [Google Scholar] [CrossRef] [PubMed]
- Frankenfield, K.N.; Powers, E.T.; Kelly, J.W. Influence of the N-terminal domain on the aggregation properties of the prion protein. Protein Sci. 2005, 14, 2154–2166. [Google Scholar] [CrossRef] [PubMed]
- Craveur, P.; Joseph, A.P.; Esque, J.; Narwani, T.J.; Noël, F.; Shinada, N.; de Brevern, A.G. Protein flexibility in the light of structural alphabets. Front. Mol. Biosci. 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Falconi, M.; Biocca, S.; Novelli, G.; Desideri, A. Molecular dynamics simulation of human LOX-1 provides an explanation for the lack of OxLDL binding to the Trp150Ala mutant. BMC Struct. Biol. 2007, 7. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh-Rahrovi, J.; Shayesteh, A.; Ebrahim-Habibi, A. Structural stability of myoglobin and glycomyoglobin: A comparative molecular dynamics simulation study. J. Biol. Phys. 2015, 41, 349–366. [Google Scholar] [CrossRef] [PubMed]
- Karjiban, R.A.; Rahman, M.B.A.; Basri, M.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Chor, A.L.T. On the importance of the small domain in the thermostability of thermoalkalophilic lipases from L1 and T1: Insights from molecular dynamic simulation. Protein Pept. Lett. 2009, 17, 699–707. [Google Scholar] [CrossRef]
- Amini-Bayat, Z.; Hosseinkhani, S.; Jafari, R.; Khajeh, K. Relationship between stability and flexibility in the most flexible region of Photinus pyralis luciferase. Biochim. Biophys. Acta 2012, 1824, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Choo, D.-W.; Kurihara, T.; Suzuki, T.; Soda, K.; Esaki, N. A Cold-adapted lipase of an Alaskan Psychrotroph, Pseudomonas sp. Strain B11-1: Gene cloning and enzyme purification and characterization. Appl. Environ. Microbiol. 1998, 64, 486–491. [Google Scholar] [PubMed]
- Hongfei, S.U.; Zhimao, M.A.I.; Zhang, S. Cloning, Expression and characterization of a lipase gene from marine bacterium Pseudoalteromonas lipolyticaSCSIO 04301. J. Ocean Univ. China 2016, 15, 1051–1058. [Google Scholar]
- Liu, Z.Q.; Zheng, X.B.; Zhang, S.P.; Zheng, Y.G. Cloning, expression and characterization of a lipase gene from the Candida antarctica ZJB09193 and its application in biosynthesis of vitamin A esters. Microbiol. Res. 2012, 167, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Jiewei, T.; Zuchao, L.; Peng, Q.; Lei, W.; Yongqiang, T. Purification and characterization of a cold-adapted lipase from Oceanobacillus strain PT-11. PLoS ONE 2014, 9, e101343. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Hiroshima, S.; Hirose, S.; Yasuda, M.; Ishimi, K.; Ishikawa, H. Cloning, expression and characterization of a lipase gene (lip3) from Pseudomonas aeruginosa LST-03. Mol. Genet. Genom. 2004, 1, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Mimitsuka, T.; Muto, T.; Matsumura, M.; Yasuda, M.; Ishimi, K.; Ishikawa, H. Cloning, expression, and characterization of a lipolytic enzyme gene (lip8) from Pseudomonas aeruginosa LST-03. J. Mol. Microbiol. Biotechnol. 2004, 7, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Ogino, H.; Nakagawa, S.; Shinya, K.; Muto, T.; Fujimura, N.; Yasuda, M.; Ishikawa, H. Purification and characterization of organic solvent-stable lipase from organic solvent-tolerant Pseudomonas aeruginosa LST-03. J. Biosci. Bioeng. 2000, 89, 451–457. [Google Scholar] [CrossRef]
- Ramania, K.; John Kennedy, L.; Ramakrishnana, M.; Sekaran, K. Purification, characterization and application of acidic lipase from Pseudomonas gessardii using beef tallow as a substrate for fats and oil hydrolysis. Process Biochem. 2010, 10, 1683–1691. [Google Scholar] [CrossRef]
- Goodarzi, N.; Karkhane, A.A.; Mirlohi, A.; Tabandeh, F.; Torktas, I.; Aminzadeh, S.; Yakhchali, B.; Shamsara, M.; Ghafouri, M.A. Protein engineering of Bacillus thermocatenulatus lipase via deletion of the α5 helix. Appl. Biochem. Biotechnol. 2014, 174, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.T.; Kim, H.K.; Kim, S.J.; Pan, J.G.; Oh, T.K.; Ryu, S.E. Novel zinc-binding center and temperature switch in the Bacillus stearothermophilus L1 lipase. J. Biol. Chem. 2002, 277, 17041–17047. [Google Scholar] [CrossRef] [PubMed]
- Maiangwa, J.; Mohamad Ali, M.S.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Normi, Y.M.; Mohd Shariff, F.; Leow, T.C. Lid opening and conformational stability of T1 lipase is mediated by increasing chain length polar solvents. PeerJ 2017, 5, e3341. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Lin, Y.T.; Liang, T.W.; Chio, S.H.; Ming, L.J.; Wu, P.C. Purification and characterization of extracellular lipases from Pseudomonas monteilii TKU009 by the use of soybeans as the substrate. J. Ind. Microbiol. Biotechnol. 2009, 36, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kikon, K.; Upadhyay, A.; Kanwar, S.S.; Gupta, R. Production, purification, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein Expr. Purif. 2005, 41, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jang, S.H.; Lee, C. An organic solvent-tolerant alkaline lipase from cold-adapted Pseudomonas mandelii: Cloning, expression, and characterization. Biosci. Biotechnol. Biochem. 2013, 77, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritschi, E.F.; Maniatis, T. Molecular Cloning: A Laboratorymanual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Laemmli, U.K. Cleavage of structure protein during assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Rhee, J.S. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J. Am. Oil Chem. Soc. 1986, 63, 89–92. [Google Scholar] [CrossRef]












| MD Simulation (Time) | Distance (Å) | |||
|---|---|---|---|---|
| Temperature | ||||
| 10 °C | 20 °C | 30 °C | 40 °C | |
| 10 ns | 33.55 | 17.17 | 14.84 | 18.91 |
| 15 ns | 9.59 | 22.80 | 11.73 | 17.48 |
| 20 ns | 12.84 | 14.13 | 4.53 | 4.91 |
| Step | Protein Concentration (mg/mL) | Total Protein (mg) | Lipase Activity (U/mL) | Total Activity (U) | Specific Activity (U/mg) | Fold | Recovery (%) |
|---|---|---|---|---|---|---|---|
| Crude | 1.20 | 60 | 130.7 | 6535 | 108.9 | 1 | 100 |
| Affinity | 0.52 | 10.4 | 287.5 | 5750 | 552.8 | 5.07 | 87 |
| Gel filtration | 0.28 | 5.6 | 213.71 | 4274.2 | 763.25 | 7 | 65.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latip, W.; Raja Abd Rahman, R.N.Z.; Leow, A.T.C.; Mohd Shariff, F.; Kamarudin, N.H.A.; Mohamad Ali, M.S. The Effect of N-Terminal Domain Removal towards the Biochemical and Structural Features of a Thermotolerant Lipase from an Antarctic Pseudomonas sp. Strain AMS3. Int. J. Mol. Sci. 2018, 19, 560. https://doi.org/10.3390/ijms19020560
Latip W, Raja Abd Rahman RNZ, Leow ATC, Mohd Shariff F, Kamarudin NHA, Mohamad Ali MS. The Effect of N-Terminal Domain Removal towards the Biochemical and Structural Features of a Thermotolerant Lipase from an Antarctic Pseudomonas sp. Strain AMS3. International Journal of Molecular Sciences. 2018; 19(2):560. https://doi.org/10.3390/ijms19020560
Chicago/Turabian StyleLatip, Wahhida, Raja Noor Zaliha Raja Abd Rahman, Adam Thean Chor Leow, Fairolniza Mohd Shariff, Nor Hafizah Ahmad Kamarudin, and Mohd Shukuri Mohamad Ali. 2018. "The Effect of N-Terminal Domain Removal towards the Biochemical and Structural Features of a Thermotolerant Lipase from an Antarctic Pseudomonas sp. Strain AMS3" International Journal of Molecular Sciences 19, no. 2: 560. https://doi.org/10.3390/ijms19020560
APA StyleLatip, W., Raja Abd Rahman, R. N. Z., Leow, A. T. C., Mohd Shariff, F., Kamarudin, N. H. A., & Mohamad Ali, M. S. (2018). The Effect of N-Terminal Domain Removal towards the Biochemical and Structural Features of a Thermotolerant Lipase from an Antarctic Pseudomonas sp. Strain AMS3. International Journal of Molecular Sciences, 19(2), 560. https://doi.org/10.3390/ijms19020560