Abstract
Tumor necrosis-factor related apoptosis-inducing ligand, also known as TRAIL or APO2L (Apo-2 ligand), is a cytokine of the TNF superfamily acknowledged for its ability to trigger selective apoptosis in tumor cells while being relatively safe towards normal cells. Its binding to its cognate agonist receptors, namely death receptor 4 (DR4) and/or DR5, can induce the formation of a membrane-bound macromolecular complex, coined DISC (death-signaling inducing complex), necessary and sufficient to engage the apoptotic machinery. At the very proximal level, TRAIL DISC formation and activation of apoptosis is regulated both by antagonist receptors and by glycosylation. Remarkably, though, despite the fact that all membrane-bound TRAIL receptors harbor putative glycosylation sites, only pro-apoptotic signaling through DR4 and DR5 has, so far, been found to be regulated by N- and O-glycosylation, respectively. Because putative N-glycosylation sequons and O-glycosylation sites are also found and conserved in all these receptors throughout all animal species (in which these receptors have been identified), glycosylation is likely to play a more prominent role than anticipated in regulating receptor/receptor interactions or trafficking, ultimately defining cell fate through TRAIL stimulation. This review aims to present and discuss these emerging concepts, the comprehension of which is likely to lead to innovative anticancer therapies.
Keywords:
TRAIL; death-receptor; ligand; glycosylation; apoptosis; galectin; aggregation; trafficking; evolution; carbohydrate binding proteins; DR4; DR5; DcR1; DcR2; TRAIL-R1; TRAIL-R2; TRAIL-R3; TRAIL-R4; TNFRSF10A; TNFRSF10B; TNFRSF10C; TNFRSF10D 1. Introduction
All ligands and receptors of the TNF superfamily, with the exception of TRAIL (TNF-Related apoptosis-inducing ligand or APO2L), harbor putative N- and/or O-linked glycosylation sites [1]. Yet, only a limited number of studies have investigated whether these post-translational modifications play a role in regulating their signal transduction capabilities. Protein glycosylation is a complex process involving hundreds of distinct genes. It is estimated that more than 50% of the human proteome is glycosylated [2]. It is probably the most common and ubiquitous post-translational protein modification, resulting in the covalent linkage of complex oligosaccharide chains to transmembrane or secreted proteins. The most abundantly occurring forms of carbohydrate modifications are linked to asparagine (Asn) [3] and serine (Ser) or threonine (Thr) amino acids [4].
These carbohydrate chains are not solely involved in protein folding control—i.e., ensuring proper synthesis of polypeptides prior to their addressing at the cell surface or secretion—but are also directly involved in the fine-tuning of membrane-bound receptor signaling capabilities. They could affect TRAIL receptors directly by changing the folding or flexibility of the cysteine-rich domain (CRD), similar to the recent findings on the ectodomain of the LDL-receptor-related protein 6 (LRP6), the N-glycosylation of which was found to be critical to its folding and aggregation potential [5]. Alternatively, depending on their location and quality, carbohydrate moieties could also serve as binding domains for lectins, thus allowing changes in transmembrane receptor trafficking or cell surface arrangement. Since most membrane-bound proteins harbor these post-translational modifications, carbohydrate chains may also allow unexpected glycoprotein/glycoprotein interactions, resulting in the positive or negative regulation of a given pathway and, in particular, signal transduction induced by TRAIL receptors.
2. Membrane Proximal TRAIL DISC Formation and Signaling Regulation
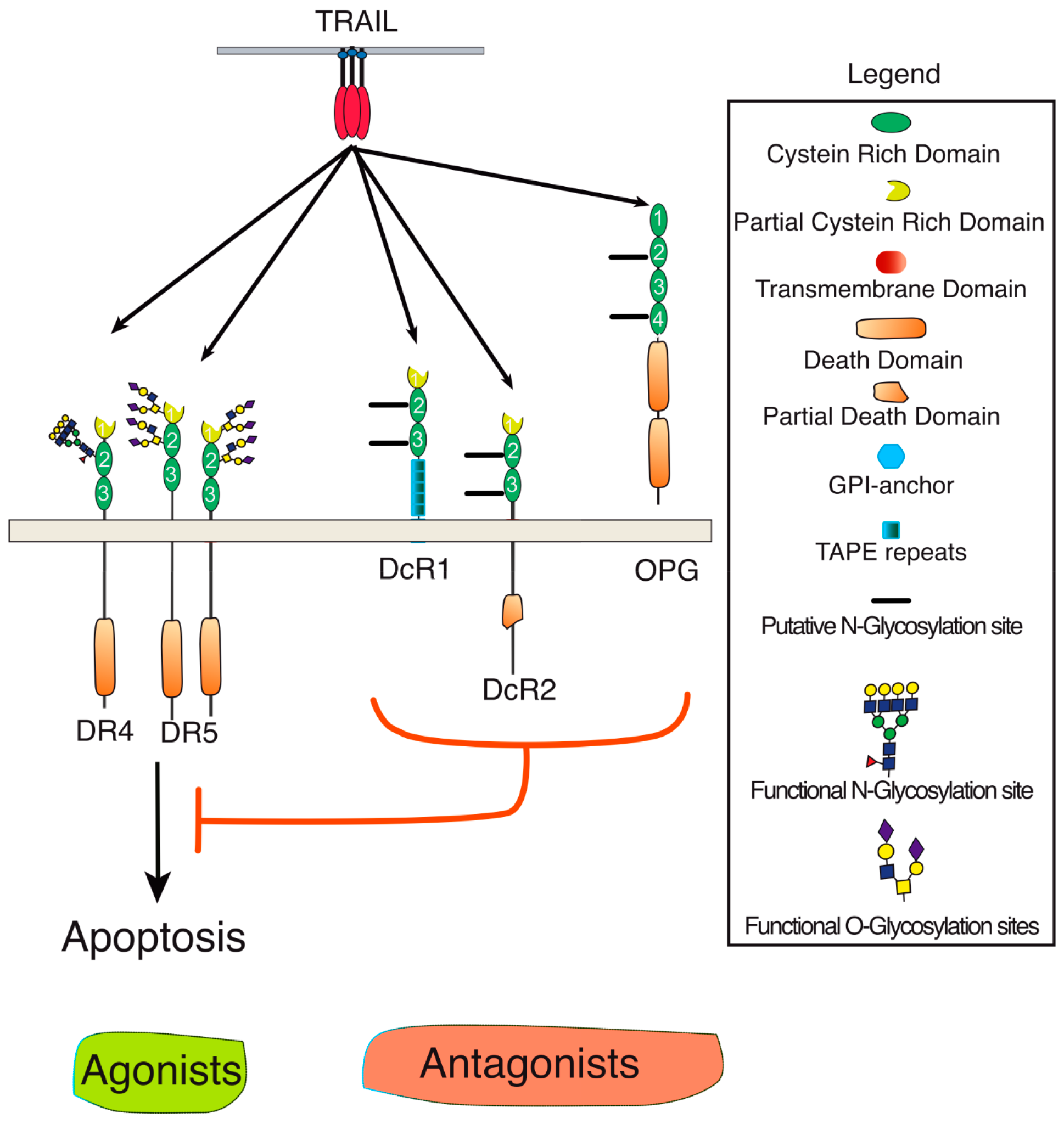
TRAIL belongs to the TNF ligand superfamily. This family is composed of 19 ligands, each capable of binding to at least one of the 29 receptors described so far [1]. TRAIL is unique for its ability to bind with high affinity to four distinct receptors, namely DR4 (TRAIL-R1), DR5 (TRAIL-R2), DcR1 (TRAIL-R3), and DcR2 (TRAIL-R4). It can also bind with the soluble receptor osteoprotegerin (OPG), albeit with much lower affinity (Figure 1) [6,7]. Because signal transduction induced by TRAIL has been associated with the induction of apoptosis in tumor cells early on, the targeting of this cytokine or its receptors has prompted major interest in oncology [8,9]. TRAIL-induced apoptosis is solely induced through DR4 and DR5, owing to the presence of the death domain (DD) within their intracellular domain. DD is a homotypic protein interaction domain [10]. The DD is necessary and sufficient for the recruitment of the adaptor protein FADD which, in turn, enables the association of the initiator caspases [11], including caspase-8 (Figure 2). Recruitment of these initiator caspases to DR4 and/or DR5 leads to their activation within the so-called DISC (death-inducing signaling complex). Once activated, initiator caspases are released into the cytosol, allowing for the cleavage and activation of effector caspases, such as caspase-3, which act directly on specific cellular substrates to dismantle the cells by apoptosis [12]. DcR1 and DcR2 are unable to transduce apoptosis upon TRAIL binding due to the lack of a functional death domain (Figure 1). Like OPG (osteoprotegerin) (which is a secreted soluble receptor) and despite the fact that OPG harbors two DD, these receptors have long been considered as decoy receptors [13,14]. While most of these antagonist receptors behave as decoy receptors when co-expressed with agonist receptors on the cell surface or within the tumor microenvironment [15], DcR2 has, in addition, been found to be able to interact directly with both DR4 and DR5, restraining caspase-8 recruitment and activation [16,17]. DcR2 has also been found to be capable of transducing signaling pathways such as NF-κB or AKT (Protein kinase B) [18,19,20].
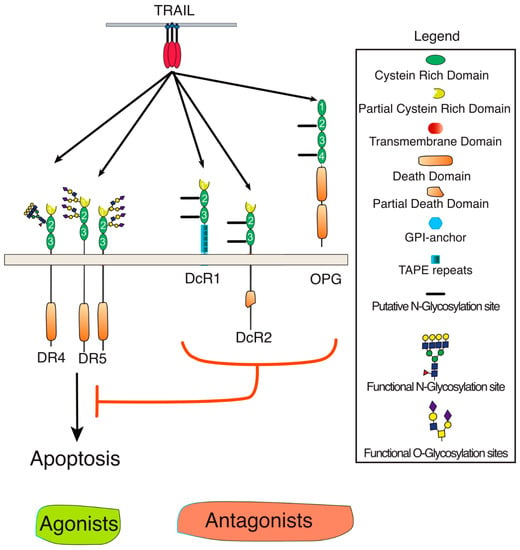
Figure 1.
Schematic representation of TRAIL and its receptors. TRAIL and its agonist (DR4 and DR5) or antagonist (DcR1, Dcr2, or OPG) receptors are membrane-bound glycoproteins of the tumor necrosis factor (TNF) superfamily. DR stands for death receptor and DcR for decoy receptor. Specific domains or putative and described O- and N-glycosylation sites are depicted in the legend. Red T bar means inhibition.
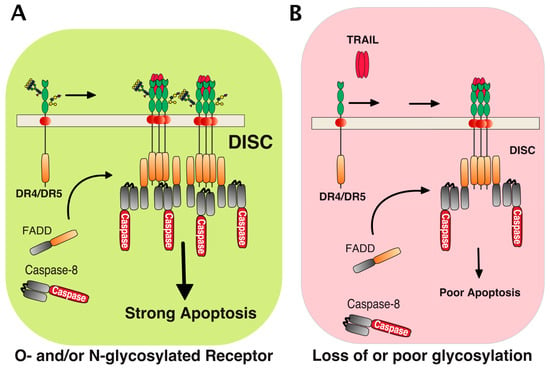
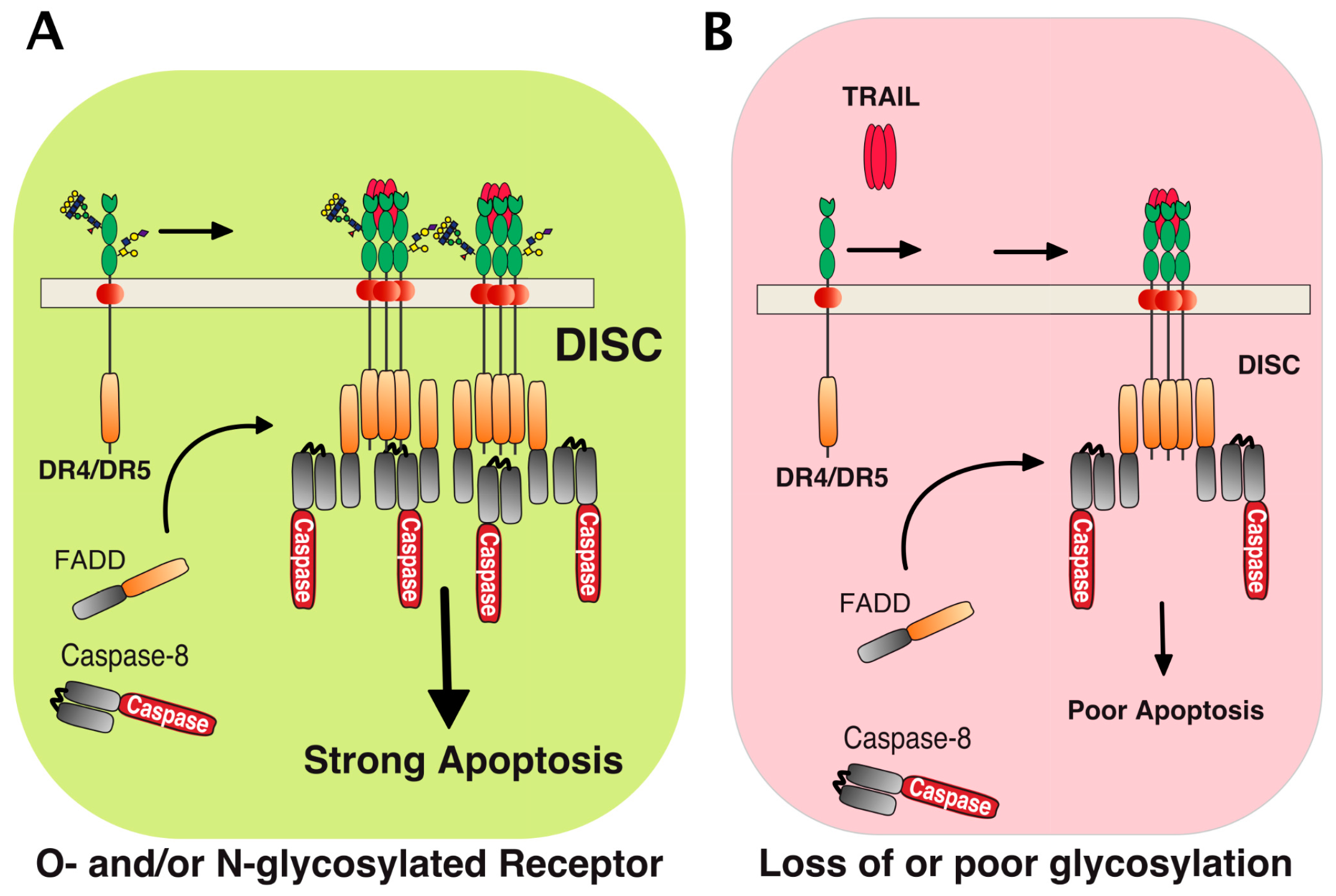
Figure 2.
Schematic representation of TRAIL-induced death-inducing signaling complex DISC formation. TRAIL induced apoptosis in cancer cells is tightly associated with glycosylation of its agonist receptors, DR4 and DR5. (A) Stimulation of glycosylated DR4 or DR5 by TRAIL induces recruitment of the adaptor protein FADD and caspase-8, hence forming the so-called TRAIL DISC (or death-inducing signaling complex), where processing of the caspase-8 occurs. This allows the triggering of apoptosis. Carbohydrate transferases, including N-acetylgalactosaminyl-, fucosyl-, or sialyl-transferases, as well as galectins could act directly at the receptor level to regulate TRAIL DISC formation and activation (see text for details). (B) In cells displaying poor polypeptide N-acetylgalactosaminyltransferase activity [42] or expressing a non-glycosylable receptor [43], binding of TRAIL to the receptors is not altered, but DISC formation and processing of caspase-8 are restrained. This impairs TRAIL’s ability to trigger apoptosis.
As inferred from studies on Fas/CD95, another member of the TNF family which also harbors a DD, caspase-8 activation within the TRAIL DISC is thought to occur by proximity [21,22]. Quantitative mass spectrometric studies suggest that recruitment and activation of caspase-8 within the DISC occurs via the formation of caspase-8 chains [23,24]. Irrespective of the stoichiometry, the requirement for a high oligomerization order for proper caspase-8 activation implies that a specific arrangement of the receptor is required to allow efficient apoptosis triggering. Consistent with this model, forced aggregation of DD-containing receptors of the TNF superfamily is often sufficient to increase caspase-8 activation [25,26], whereas, on the other hand, co-recruitment of DcR2 within the TRAIL DISC decreases caspase-8 recruitment and arrangement. This, thus, prevents its activation and apoptosis induced by TRAIL [17,27,28].
Within the TRAIL DISC, other proteins are also co-recruited with the caspase-8 [12,29], including, but not restricted to c-FLIP. This is probably the most important intracellular inhibitor caspase involved in the initiation of the extrinsic pathway [27,30,31,32]. However, unlike glycosylation or TRAIL antagonist receptors, downstream DISC components are not specific to TRAIL signaling. Most of them are also recruited to and regulate signal transduction induced by other receptors of the TNF superfamily [33,34,35,36,37,38,39] and beyond. An example of other such receptors are the toll-like receptors, including TLR3 [40,41].
3. Agonist TRAIL Receptors are Glycosylated
Proper arrangement of the TRAIL DISC scaffold has been found to be controlled by glycosylation. In 2007, the first demonstration showing that O-linked glycosylation contributes to DR5 pro-apoptotic potential was published (Figure 2 and [42]). Using a whole genome-profiling approach in a large collection of tumor cell lines, the authors found that cell sensitivity to TRAIL-induced cell death was tightly associated with elevated levels of polypeptide N-acetylgalactosaminyltransferases, such as GALNT14. Consistent with this observation, it was found that inhibition of this O-glycosyltransferase using siRNAs impairs TRAIL-induced cell death (Table 1). DR5 was demonstrated in this study to be O-glycosylated on two stretches of serines and threonines within or surrounding the CRDs 2 and 3 (Figure 3). Mutagenesis of these serines and threonines to alanine prevented O-glycosylation of DR5 and limited its ability to transduce apoptosis [42]. Importantly, it was also found that TRAIL binding affinity to the receptor was neither increased nor decreased whether DR5 was O-glycosylated or not. Mechanistically, albeit it remains unclear how glycosylation precisely regulates apoptosis induced by TRAIL, the authors could show that the sugar moieties could directly affect caspase-8 recruitment and activation at the level of the TRAIL DISC, indicating that these carbohydrate chains are likely to play a direct role in the fine structuration of the DISC and/or the devenir of the membrane-bound complex. It should be emphasized here that, in contrast to DR5, physiological and biological evidence for DR4 O-glycosylation remains poor in this study, raising the question of whether O-glycosylation also accounts for efficient signal transduction of apoptosis through DR4.

Table 1.
Enzymes and inhibitors involved or regulating protein glycosylation found to alter TRAIL-induced cell death (see text for details). ⬆ stands for an increase whereas ⬇ indicates a decrease in protein expression or complex formation.
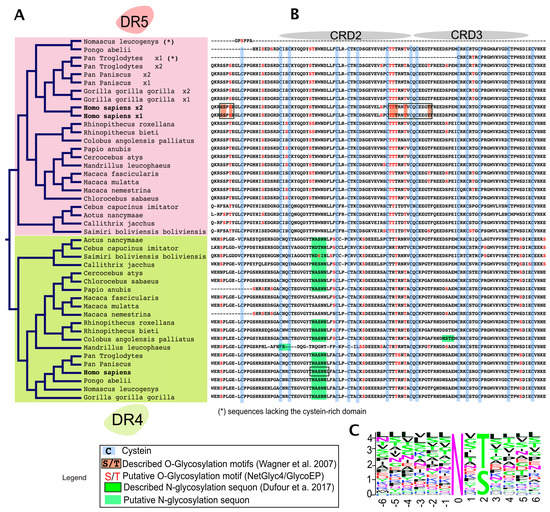
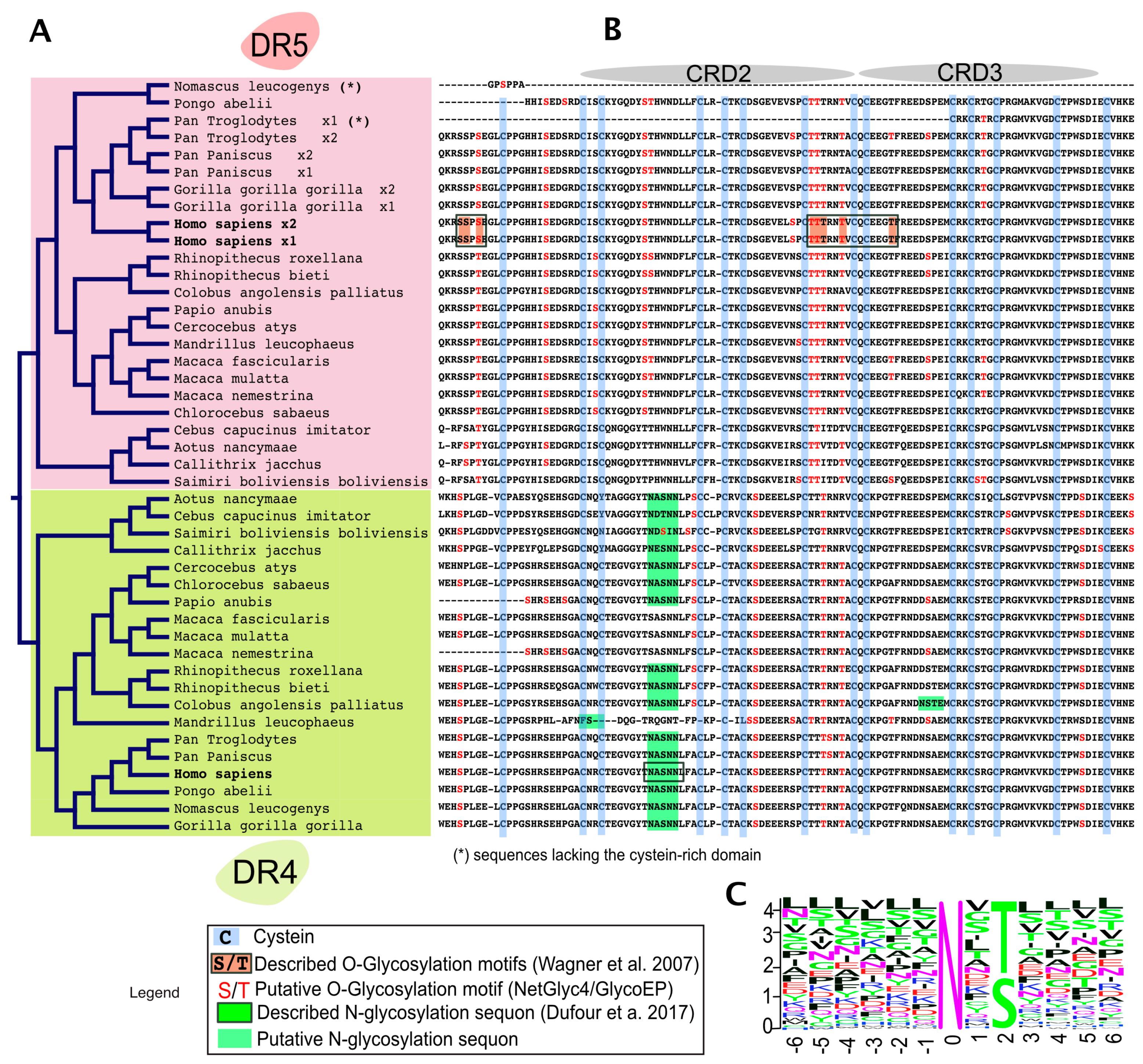
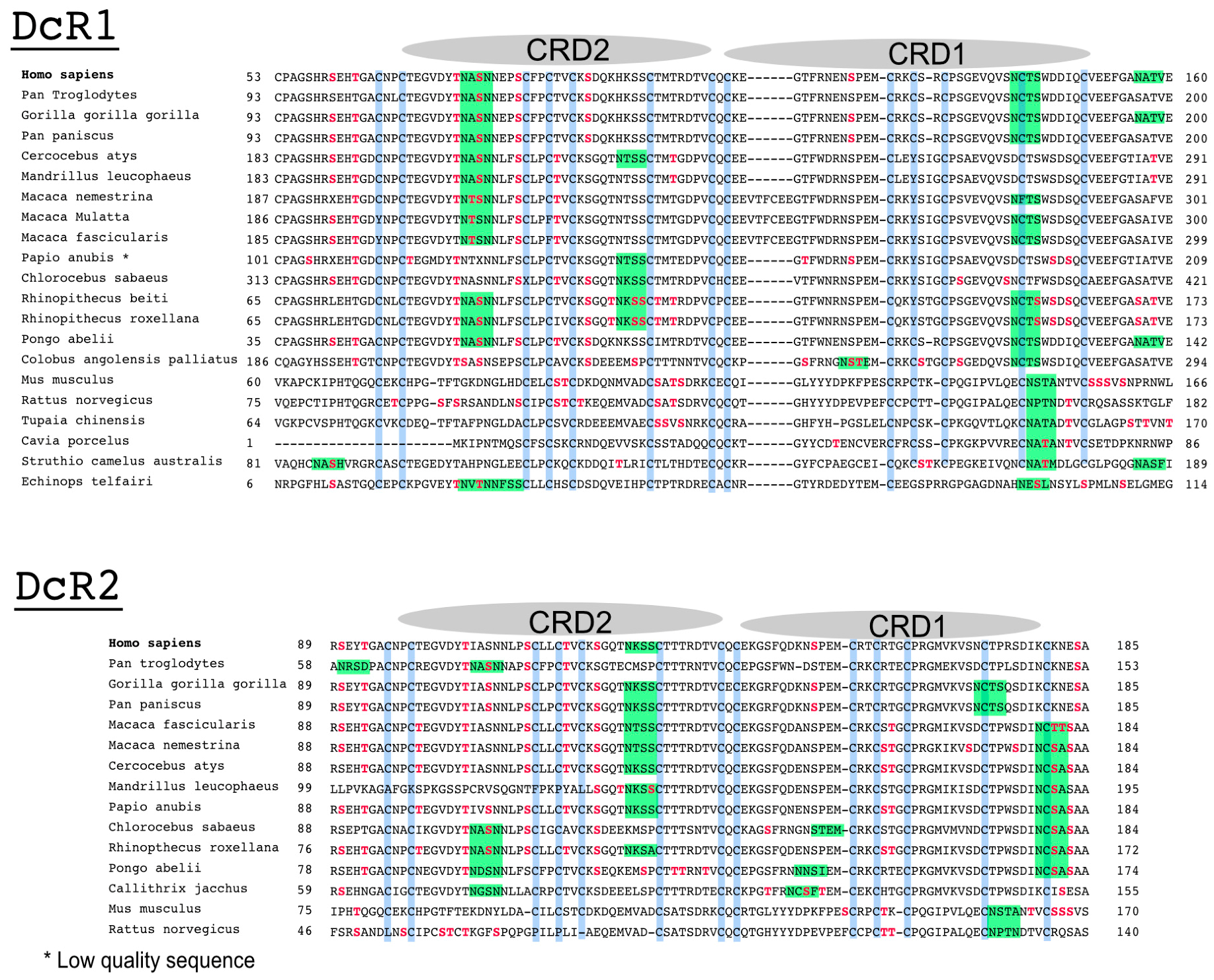
Figure 3.
Alignment comparison of the cysteine-rich domains of DR4 and DR5 in primates. (A) Dendrogram of DR4 and DR5 full primate sequences; (B) a corresponding Clustal Omega multiple sequence alignment showing the region encompassing the two-cysteine rich domains of DR4 and DR5 [66]. The legend illustrates how cysteine residues and putative or described O- or N-glycans are depicted in the alignment; (C) sequence logo displaying residues preferentially placed at occupied N-glycan sequons, adapted from Weng et al. [67]. Neighboring residues located downstream (positions +6) and upstream (positions −6) from the asparagine residue (position 0) are also shown. The height of each letter represents the residue prevalence or frequency at the putative position.
Sequence alignment of the extracellular domains, encompassing CRD2 and CRD3, of DR4 and DR5 in primates, indicates that both receptors share amino acid sequence homology with respect to serines and threonines as well as predicted putative O-glycosylation sites (Figure 3). Strikingly, however, while DR5 primate sequences lack putative N-glycosylation sites, DR4 sequences (with the exception of Macacas) contain at least one sequon, suggesting that N-glycosylation may occur and regulate signal transduction through this receptor.
Consistent with a regulatory function associated with the N-glycosylation of these receptors, DR4 was found to be N-glycosylated in human tumor cell lines [44] and this translational modification was demonstrated to be required for apoptosis induced by this receptor ([43] and Figure 2). This carbohydrate modification also occurs within the CRD2 on an asparagine located in position 156 of human DR4 (Figure 3). Like DR5, TRAIL binding affinity for DR4 was found to remain unchanged, irrespective of whether the receptor is glycosylated or not [43]. Mechanistically, it was found that receptor aggregation, initiator caspase recruitment and activation within the DISC, and apoptosis were increased when DR4 was N-glycosylated. These events were reduced or impaired in human cancer cells expressing a non-glycosylable form of DR4 [43]. These findings are in sharp contrast to previous findings demonstrating that the inhibition of N-glycosylation [45,46,47,48,49], endoplasmic reticulum (ER) and Golgi stressors [50,51,52,53], or glucose deprivation [54,55,56,57] increase tumor cell sensitivity to TRAIL-induced cell death (Table 1). Keeping in mind that these compounds are either pan-inhibitors of N-glycosylation and of the unfolded protein response (UPR) pathway or competitive inhibitors of glucose metabolism—such as the tunicamycin, thapsigargin or 2-deoxyglucose (2DG)—conclusions drawn from these experiments may not apply to a given transmembrane protein such as DR4. Indeed, these inhibitors are not specific to TRAIL agonist receptors and, albeit some of them might coincidentally induce cell death via DR4 and/or DR5 (Table 1), they often induce drastic changes of glycosylation in all proteins expressed by the targeted cells or induce the collapse of the Golgi apparatus and/or the ER. Mechanistically, these compounds have been found to induce the upregulation of DR5, as well as to decrease the expression levels of important inhibitors such as c-FLIP [52,54,56], IAPs [48], or Bcl-2 inhibitors [51,55]. Some of them also induce, alone, a ligand-independent cell death program that involves DR4 and DR5 [58,59,60,61]. By perturbing or inducing the collapse of the ER or the Golgi apparatus, these drugs also alter the glycosylation of transmembrane receptors. In particular, they alter O- or N-linked glycosylation of receptors such as DR4 and DR5, but not only these (Figure 4). It thus appears relatively clear that only gene editing studies followed by rescue experiments with point mutations of the putative glycosylated sites, such as that published by Dufour et al. [43], can address the importance of these carbohydrate chains in regulating TRAIL-induced cell death.
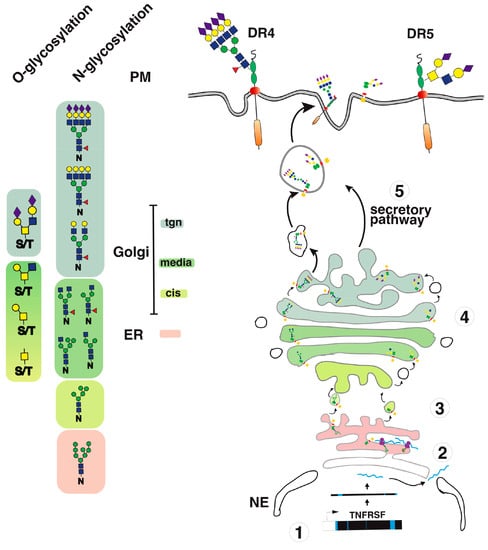
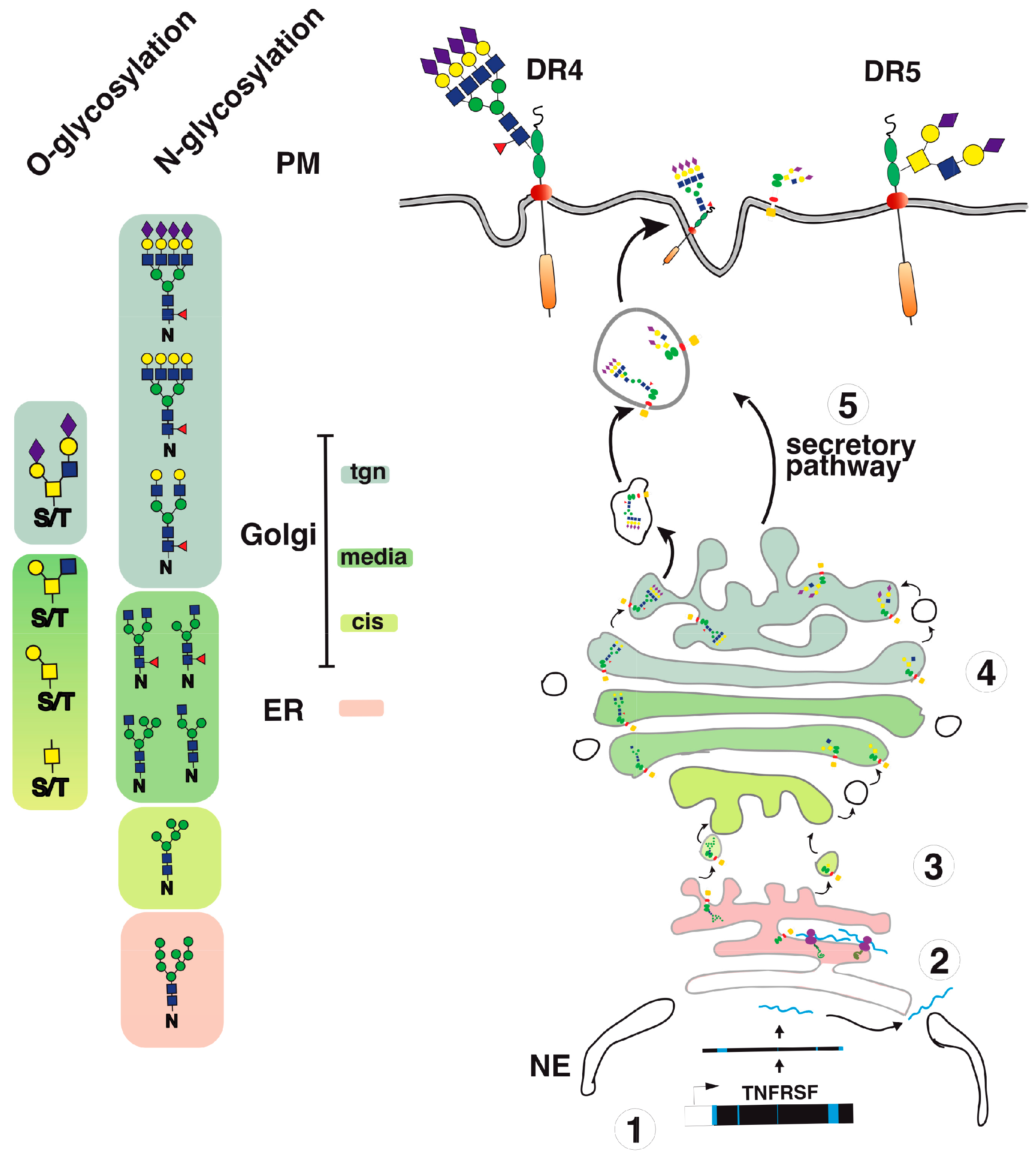
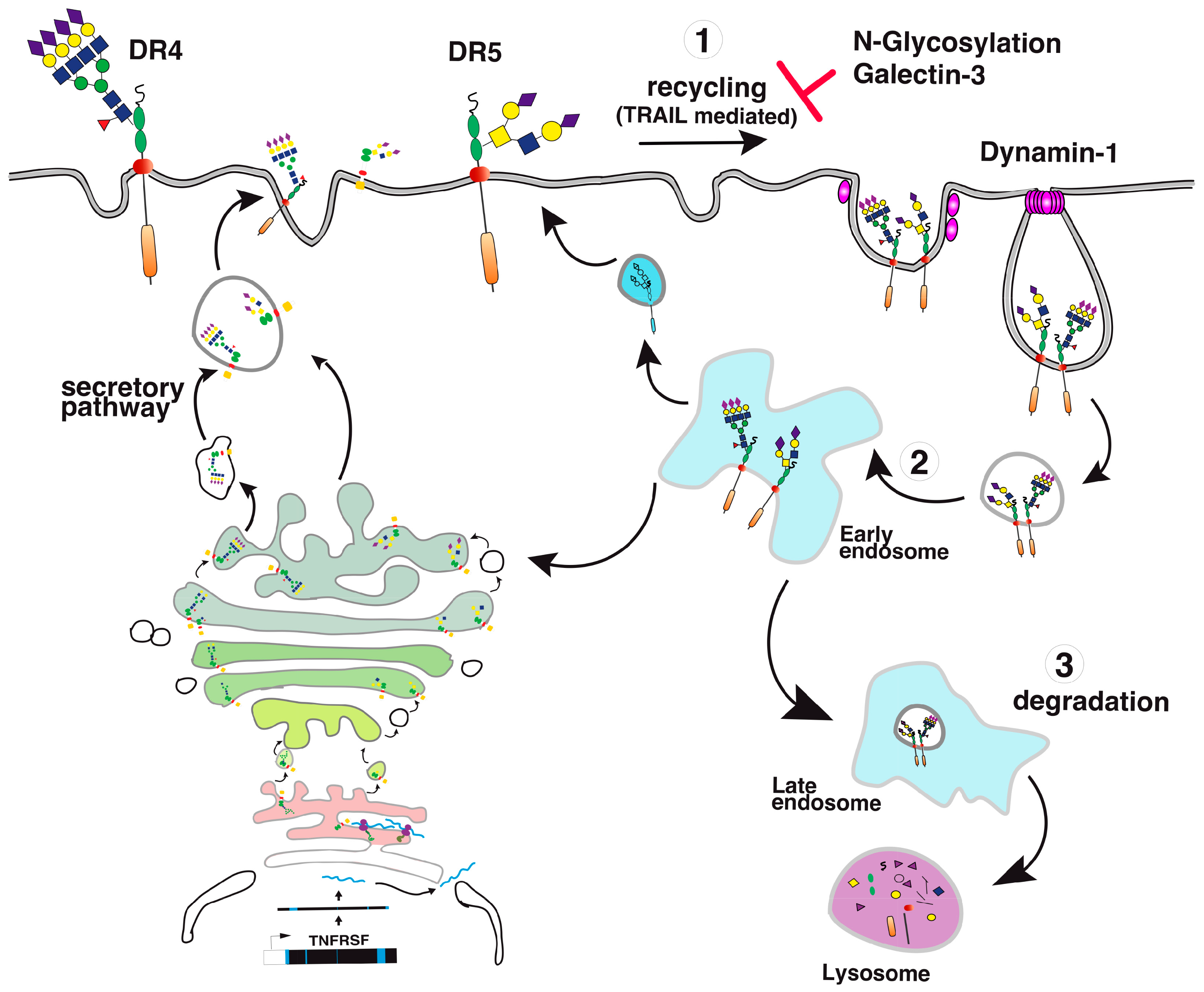
Figure 4.
TRAIL receptor glycosylation and trafficking. A simplified illustration of DR4 and DR5 glycosylation and trafficking to the cell surface. (1) Once transcribed; (2) upon translation; (3–4) TRAIL receptors undergo glycosylation from the ER to the Golgi apparatus. (5) This post-translational modification occurs stepwise in the Golgi apparatus, allowing glycan maturation and complexification prior to cell surface transport via vesicular trafficking.
4. TRAIL Receptor Glycosylation during Evolution
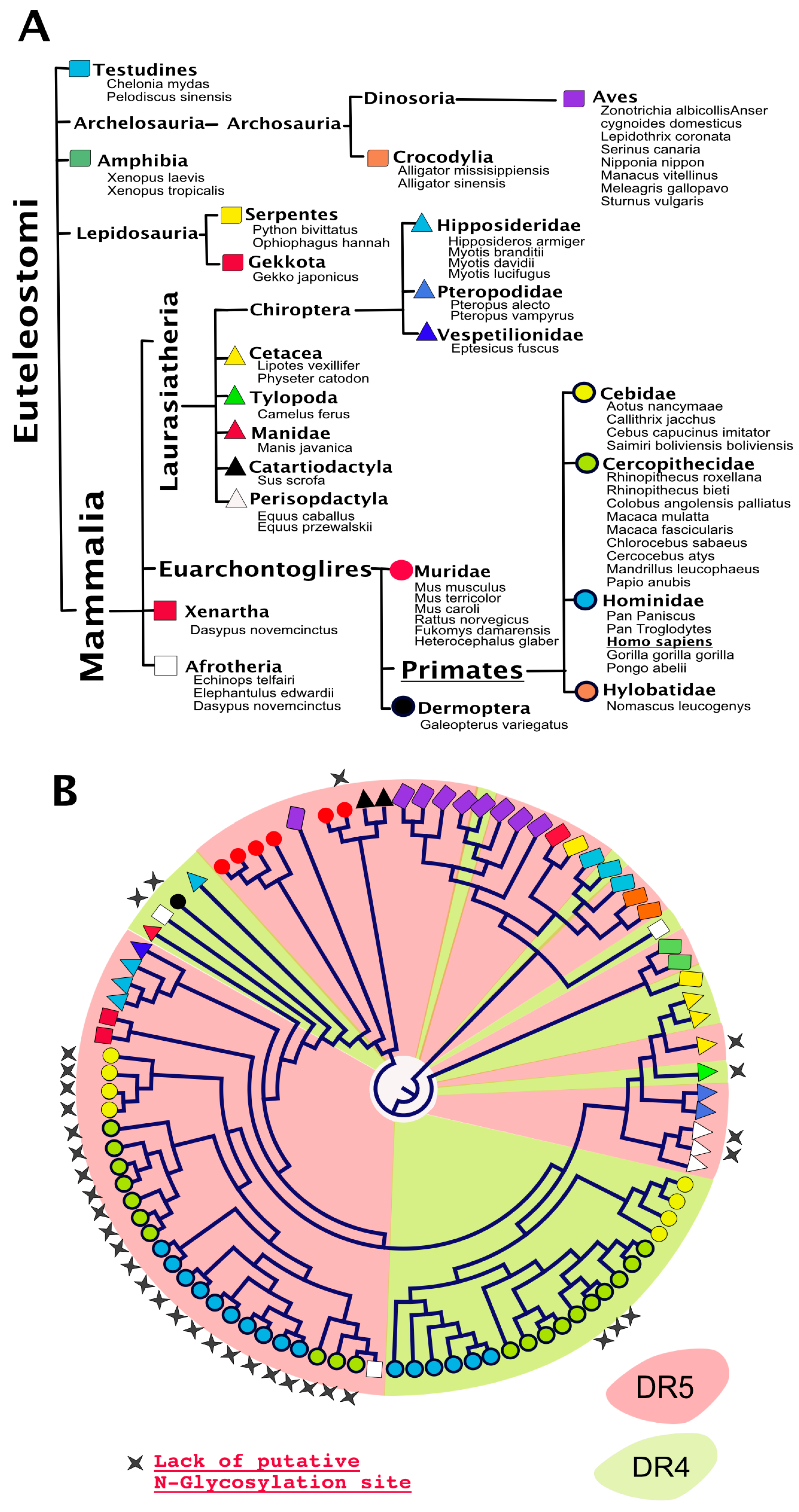
Analysis of putative N-glycosylation sites in orthologue DR4 and/or DR5 sequences reveal that, with a few exceptions, almost all TRAIL agonist receptors harbor putative N-glycosylation sites (Figure 5). Moreover, it is also worth noting that the presence of conserved putative N-glycosylation sites extends beyond agonist receptors—since such sequons are also found in TRAIL antagonist receptors—within the extracellular region encompassing CRD1 and CRD2 (Figure 6). Like many other genes found in higher organisms, DR4 and DR5 arise from a gene duplication during evolution. DR4 and DR5 are the two flavors of an original gene which remains present as a single version in lower organisms such as rodents (see also Figure 5). Interestingly enough, with the exception of DR5 primate sequences and a few others, most TRAIL agonist receptor sequences harbor at least one N-glycosylation site, including the unique TRAIL agonist receptor expressed in lower organisms which is often defined as the orthologue of DR5. It would thus be tempting to speculate that N-glycosylation represents the most ancient form of carbohydrate modification linked to the original TRAIL agonist receptor and beyond the TNFR superfamily. Because this feature has been preserved during evolution, it was not surprising to find that N-glycosylation plays also a major regulatory function with regards to TRAIL receptor/receptor interactions and signal transduction in lower organisms. Accordingly, the unique mouse TRAIL agonist receptor has also been demonstrated to be N-glycosylated on three asparagines located in positions 99, 122, and 150. This corresponds to CRD2 and CRD3 [43]. Similar to DR4, N-glycosylation of mTRAIL-R was found to increase apoptosis induced by TRAIL, without inhibiting or enhancing TRAIL binding to its receptor [43]. Rather, like DR4 and DR5, the glycosylation of mTRAIL-R was associated with better receptor aggregation at the cell surface.
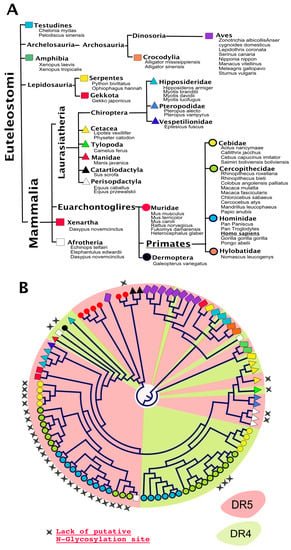
Figure 5.
Phylogeny of DR4 and/or DR5 in “bony vertebrates”. (A) DR4 and DR5 are, so far, exclusively reported in the Euteleostomi clade. Orders and families in which DR4 and DR5 are expressed are depicted here using distinctly colored symbols; (B) phylogenetic distribution of DR4 (green) and DR5 (red). Stars above the circular cladogram point to sequences devoid of putative N-glycosylation sites.
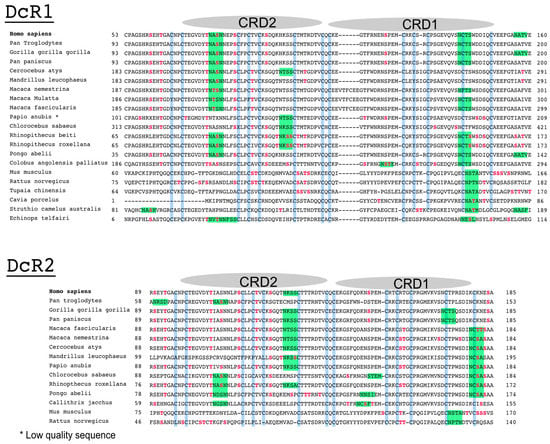
Figure 6.
Alignment comparison of the cysteine-rich domains of DcR1 and DcR2. Corresponding Clustal Omega multiple sequence alignment showing the region encompassing the two cysteine-rich domains of DcR1 and DcR2 in different species [66].
5. TRAIL Receptor Clustering
How glycosylation favors receptor clustering or helps proper formation of the TRAIL DISC scaffold remains unclear (Figure 7A). Several models of receptor cluster/aggregation formation have been proposed so far, based on structure/function studies of TNF/TNF receptors [68,69,70], as well as crystallographic studies of TRAIL or agonist antibodies binding to TRAIL receptors [71]. While ligands of the TNF superfamily are known to preassemble spontaneously as trimers [72], TNF receptors, albeit long thought to be monomeric, are able to form dimers on the cell surface [68,73]. Spontaneous assembly of TNF receptor homo- or hetero-dimers was found to be mediated by the pre-ligand assembly domain or PLAD [16,39,70], a stretch of amino acids residing within the partial CRD1 (Figure 1). The PLAD is particularly interesting with respect to TRAIL receptors, since this dimeric binding-domain has been demonstrated to allow heteromeric interactions between DR5 and DcR2 in the absence of TRAIL [74]. It should be noted, however, that this interaction is not only maintained but also increased in the presence of TRAIL, as demonstrated experimentally by native TRAIL DISC immunoprecipitation [17]. This implies that models describing receptor arrangement through dimeric interactions are likely to apply to heteromers too.
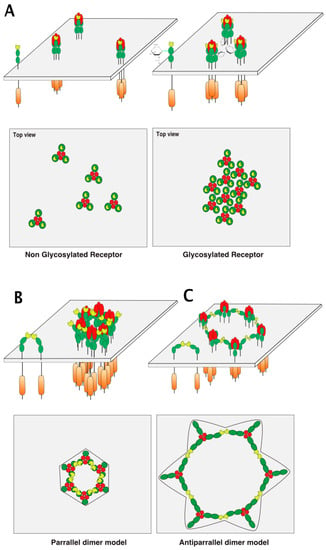
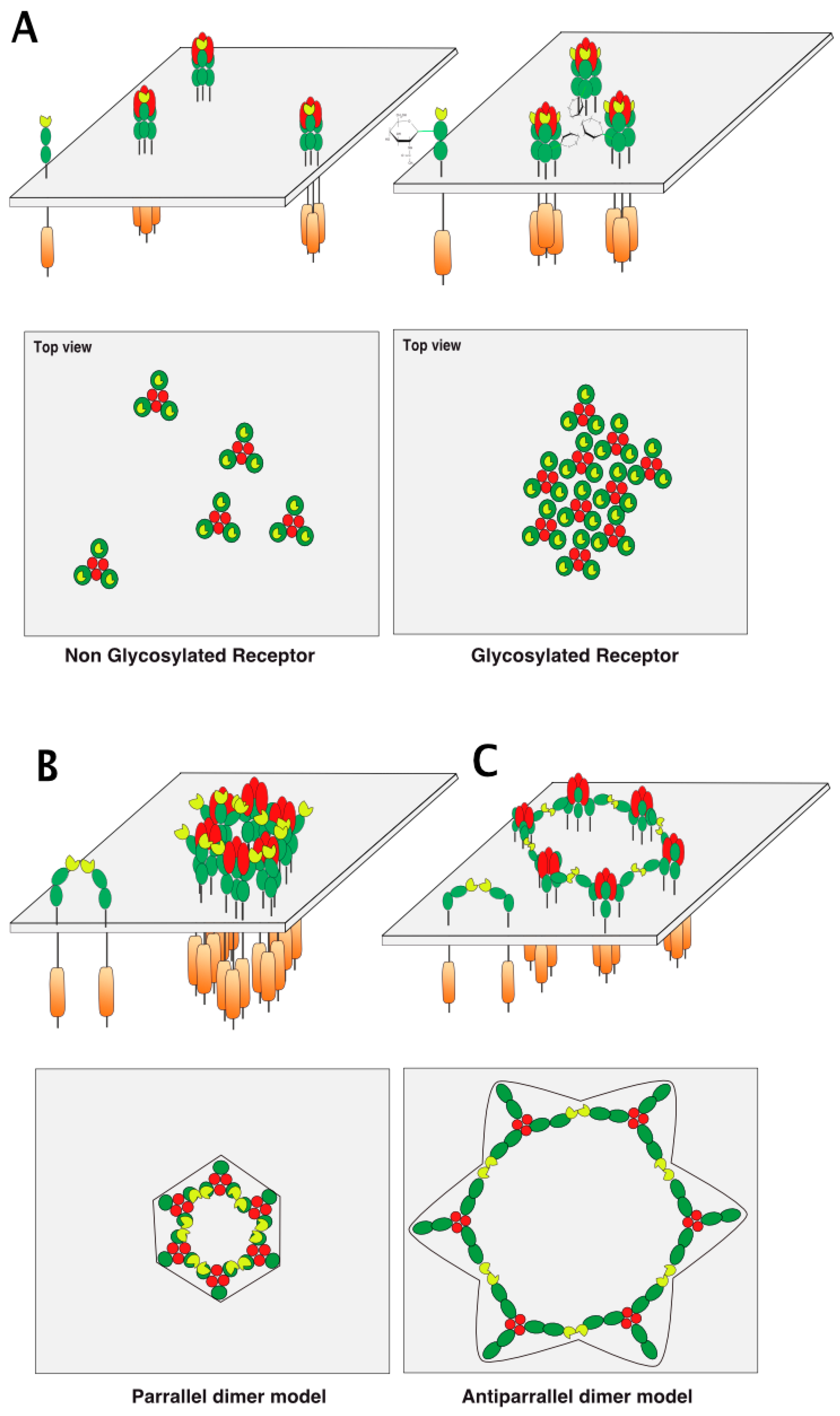
Figure 7.
TRAIL receptor aggregation models. (A) Illustration of the basic ligand/receptor organization consisting of a trimeric ligand binding three distinct receptors. The carton below is a hypothetical representation of the clustering of DR4 or mTRAIL-R according to respective glycosylation status viewed from the top of the cell membrane; (B) PLAD-based parallel and (C) antiparallel dimer models adapted from Vanamee et al. [71].
Hexameric arrangement models, based on parallel or antiparallel dimers of TRAIL receptors have been proposed, explaining how receptor clustering leads to efficient apoptotic signal transduction (Figure 7B,C). These models are inferred from crystallographic analysis of the ternary complex formed by the anti-DR5 (AMG655) Fab fragment, TRAIL, and DR5 [75] and mutational analysis of DR5 [76]. It should be kept in mind that activation of caspase-8 occurs through proximity [22], irrespective of whether the caspase-8 is able to form chains within the DISC [23,24]. Therefore, arrangement of TRAIL receptors and, thus, caspase-8 vicinity within the antiparallel model is likely to be less favorable for efficient caspase-8 activation than the parallel model, which supports closer caspase-8/caspase-8 interactions (Figure 7).
Whether glycosylation directly affects these arrangements remains to be determined. However, it may be possible to envision the possibility that carbohydrate chains allow the binding of lectins to regulate receptor lattice formation.
6. TRAIL Signal Transduction Regulation by Carbohydrate-Binding or Modifying Proteins
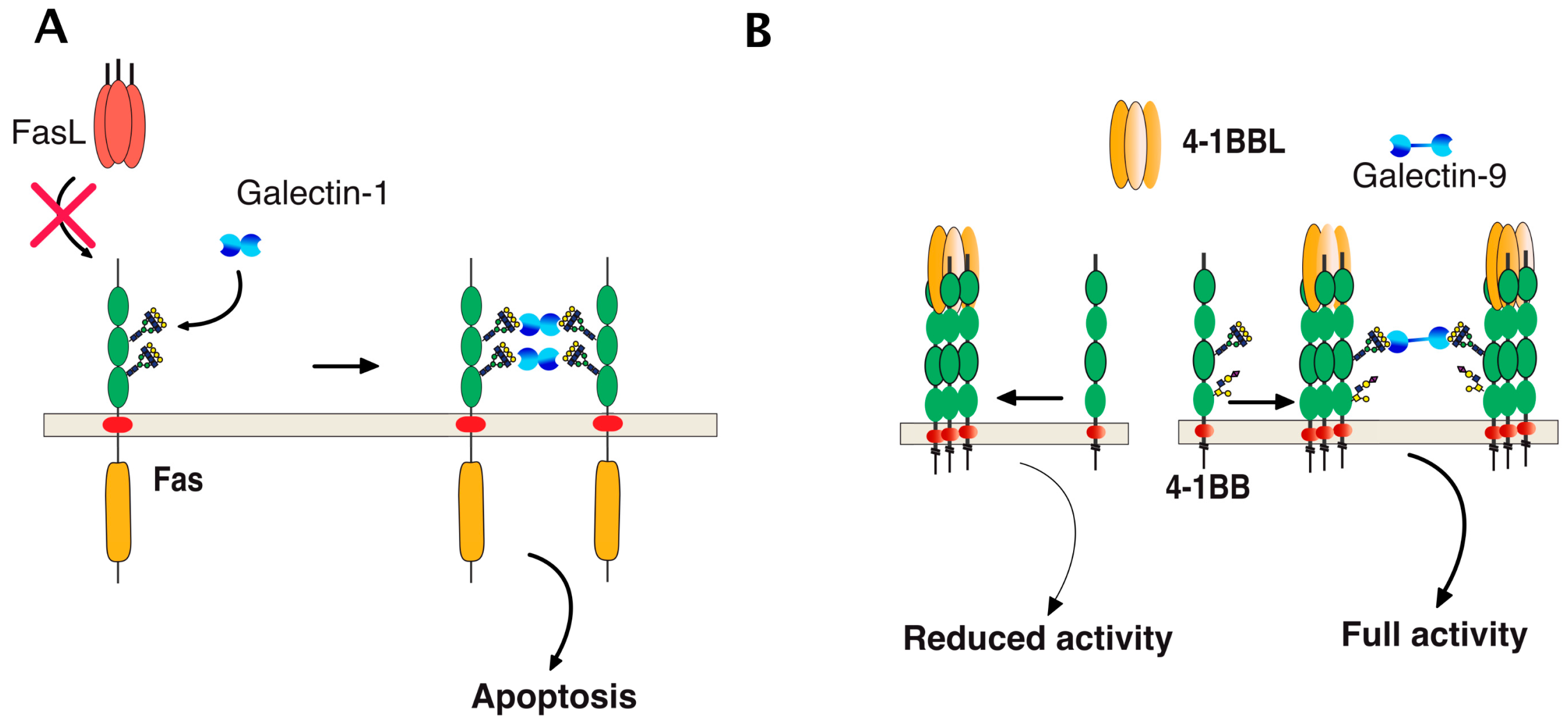
Direct binding of galectins to glycosylated TNF receptors has seldom been studied. However, increasing evidence suggests that these interactions are likely to be more frequent and important than expected. It has been found, for example, that galectin-1 could induce apoptosis in two leukemic T cell lines, namely Jurkat and CEM, owing to its ability to bind directly to Fas [77]. Since Fas is N-glycosylated [78,79], the binding of galectin-1 to Fas in these cells is thought to be sufficient to induce Fas clustering, caspase-8 recruitment, and activation of the initiator caspase in the absence of its cognate ligand (Figure 8). More recently, 4-1BB, another receptor of the TNF superfamily which is also N-glycosylated [80], has been demonstrated to interact with and to require galectin-9 (Figure 8) for efficient signal transduction [81,82]. Regulation of TRAIL-induced signaling by galectins has also been described. However, direct interactions of β-galactoside-binding proteins with TRAIL receptors have not been described to date, with the exception of a publication in 2012 describing the inhibition of TRAIL receptors endocytosis by galectin-3 [83]. In this work, Mazurek et al. could demonstrate that a colon cancer cell line, stably generated for TRAIL-resistance after repetitive exposition to TRAIL, gained insensitivity to TRAIL-induced apoptosis by elevating cell surface expression of galectin-3 (Figure 9). Elevation of galectin-3 expression was associated with reduced TRAIL receptor endocytosis and a resistance to TRAIL. Inhibition of galectin-3 using shRNAs or selective inhibitors restored TRAIL sensitivity and receptor internalization [83]. Moreover, inhibition of DR4 and DR5 endocytosis after TRAIL stimulation was directly associated with galectin-3 cell surface expression and physical interaction between TRAIL receptors and galectin-3. This was demonstrated by the immunoprecipitation of the receptors and immunoblotting or by immunofluorescence [83]. Galectins are devoid of peptide signal and transmembrane domain, but they can be found both in the cytosol or on the cell surface after secretion. Despite it remaining unclear how the latter are being secreted, their retention on the cell surface has recently been found to require N-glycosylated plasma membrane receptors or lipids [84], which would support the finding that galectin-3 could act directly at the cell surface to regulate apoptosis induced by TRAIL. The findings of Mazurek et al. do not rule out the possibility that the accumulation of galectin-3, which is associated with a reduction of TRAIL receptor endocytosis after TRAIL stimulation in their clone, may be driven by an alteration of the clathrin-dynamin-1 endocytic pathway [85] (Figure 9). It is worth mentioning here that a number of studies have been produced describing the pro-apoptotic properties of galectins [86,87,88,89] or their ability to interfere with cell death signal transduction, including apoptosis induced by ligands of the TNF superfamily ([83,90,91,92,93,94,95] and Table 2). One of them has provided evidence, for instance, that galectin-3 can bind to the intracellular domain of Fas [96] and account for the so-called type I or type II cell subtypes which define cells according to their requirement to induce apoptosis through an amplification loop induced by mitochondria [97]. In this study, it was found that galectin-3 would transform type II cells to type I and, therefore, allow complete apoptosis in a mitochondria-independent manner [96]. Studies focused on TRAIL have mostly reported indirect modulation of the extrinsic pathway by galectins. Likewise, in the reconstitution of galectin-3 in the TRAIL-resistant breast cancer cell line BT459, expressing low levels of the lectin was found to restore apoptosis induced by TRAIL through a mechanism associated with a decrease in AKT phosphorylation [90]. It was next demonstrated that a restoration of TRAIL-induced cell death by galectin-3 in BT459 cells required its phosphorylation on serine-6 [98] and that TRAIL sensitivity may be influenced by a sequence polymorphism commonly found in galectin-3 and associated with breast cancer incidence [94]. However, in contrast, in the human bladder carcinoma cell line J82 or in papillary thyroid cancer cells, overexpression of galectin-3 was found to impair cell death induced by TRAIL through an increase of phosphorylation of Akt on serine 473 [91,93]. Lastly, it has also been described that galectin-1 could impair TRAIL-induced apoptosis in human hepatocellular carcinoma cells, a finding associated with a change in survivin and Bcl-2 expression [95]. Unlike the studies demonstrating functional regulation of 4-1BB signal transduction by galectin-9 [81,82], none of the studies mentioned above has addressed whether the galectins, due to their potential ability to bind to glycosylated TRAIL receptors, are likely to specifically regulate apoptosis by interfering or enhancing receptor aggregation.
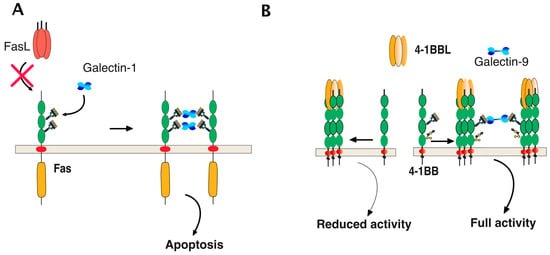
Figure 8.
Schematic representation of galectin-mediated Fas and 4-1BB clusterin, two receptors of the TNF receptor superfamily. (A) Galectin-1 induced apoptosis can occur through its binding with Fas in a ligand-independent manner due to the fact that Fas is N-glycosylated [77]. The red cross illustrates the lack of requirement of FasL for this interaction to occur; (B) binding of 4-1BBL to its cognate receptor 4-1BB, a glycoprotein harboring N-glycosylation sites promoting CD4 and CD8 T-cell activation. Full activation of 4-1BB has recently been found to require direct binding of galectin-9 to 4-1BB [81,82].
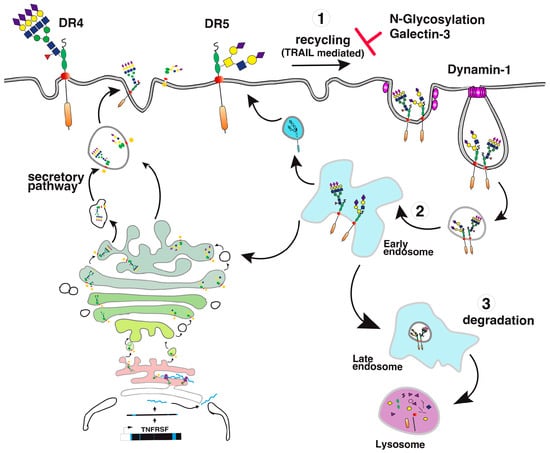
Figure 9.
TRAIL receptor internalization and recycling after TRAIL stimulation. A simplified illustration of DR4 and DR5 internalization. (1) After TRAIL binding, the receptor aggregates and undergoes fast internalization in a clathrin/dynamin-1 dependent manner. Receptor internalization has been found to be negatively regulated by galectin-3 [83] and restrained by glycosylation [43]. (2) In early endosomes, the receptors can then be recycled to the cell surface (3) or routed to late endosomes for degradation.

Table 2.
Carbohydrate-binding proteins found to alter TRAIL-induced cell death (see text for details). ⬆ stands for an increase whereas ⬇ indicates a decrease in protein expression.
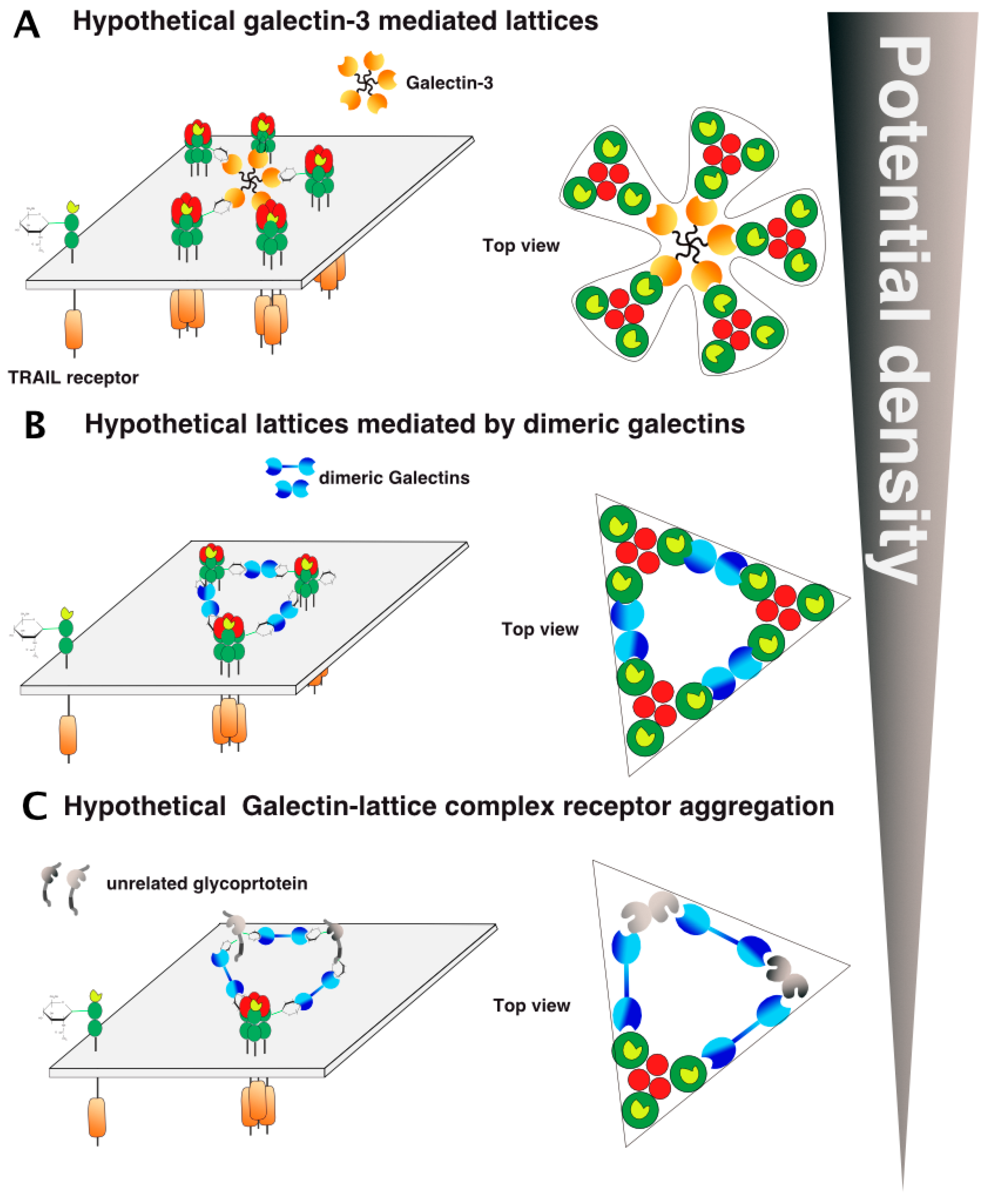
However, keeping in mind that the arrangement and clustering of TRAIL receptors at the cell surface after TRAIL stimulation represents a crucial step to induce efficient caspase-8 activation, proteins displaying affinity with carbohydrates, such as galectins, are likely to play a crucial regulatory role in the process. Similar to the PLAD dimeric models (Figure 7B,C), galectins, owing to their affinity for β-galactosides, are likely to interact directly with DR4 or DR5 and to contribute to their spatial organization within the plasma membrane. As illustrated in Figure 10, at least two hypothetical models that may also enrich or exclude the PLAD dimeric models could explain how galectins may enhance or inhibit TRAIL signaling. This can be done merely by reducing the distance between TRAIL/receptor-based units or, in contrast, by increasing the distance between trimerized receptors (Figure 10). As illustrated in this figure, depending on their valency, galectins are likely to differentially shape TRAIL receptor arrangement on the cell surface, either increasing or inhibiting signal transduction. Sugar moieties may be involved or required for heteromeric TRAIL receptor formation. Such a scenario could apply to TRAIL inhibitory receptors, as these also harbor putative N-glycosylation sites (Figure 6).
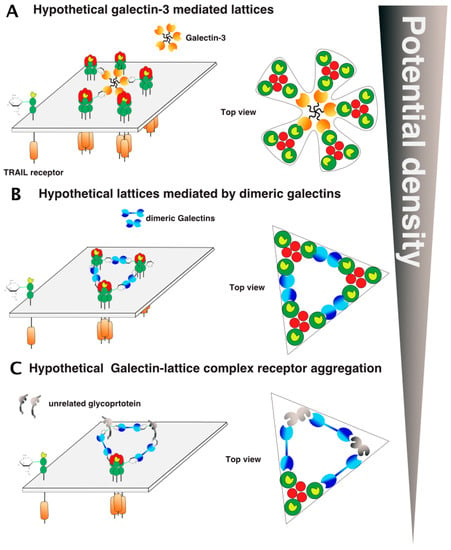
Figure 10.
Galectin/TRAIL receptor hypothetical aggregation models. Several models involving galectins could explain how glycosylation regulates TRAIL DISC formation and activation, including carbohydrate-mediated interactions with galectins such as lattice formation. (A) An illustration of the hypothetic galectin-glycan-receptor lattice organization is shown with the pentameric galectin-3 and (B) dimeric galectins. (C) Inclusion of non-TRAIL receptor-related glycoproteins to TRAIL receptor clusters, gathered by galectins. Potential density here stands for cell membrane density of the receptors.
Alternatively, since all TRAIL receptors are glycosylated and are thus potential binding-partners for galectins, other arrangements may be proposed, including unexpected interactions with non-TNF receptor-related glycoproteins (Figure 10). In the figure, these interactions have been illustrated as inhibitory, but the latter could, contrarily, increase TRAIL receptor aggregation and density on the cell surface or stabilize the TRAIL DISC, thus allowing proper caspase-8 activation and apoptosis triggering.
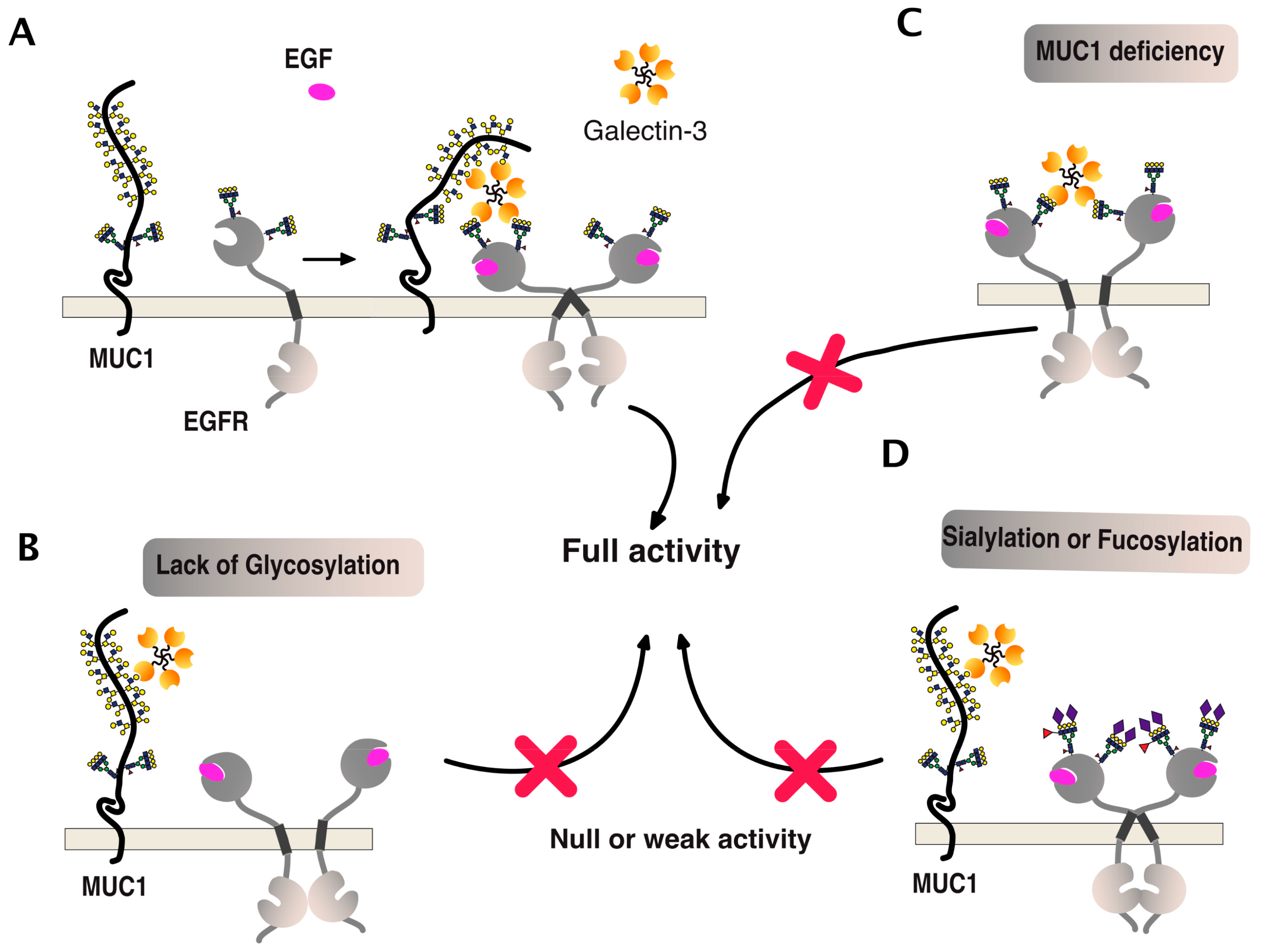
One of the best examples of such a positive association can be illustrated by the epithelial growth factor receptor (EGFR) signaling (Figure 11). This receptor is both O- and N-glycosylated and, like DR5, its signal transduction activity has been described to be tightly associated with polypeptide N-acetylgalactosaminyltransferases [99] and galectins [100,101]. As illustrated in Figure 11, EGFR carbohydrate sugar chains support complex ternary interactions between the receptor itself, galectin-3, and MUC1. A deficiency of any of these partners or changes in glycosylation, such as sialylation of fucosylation, can disrupt the fine organization of the scaffold, thus leading to reduced EGFR dimerization and signal transduction [100,102,103,104,105,106,107,108].
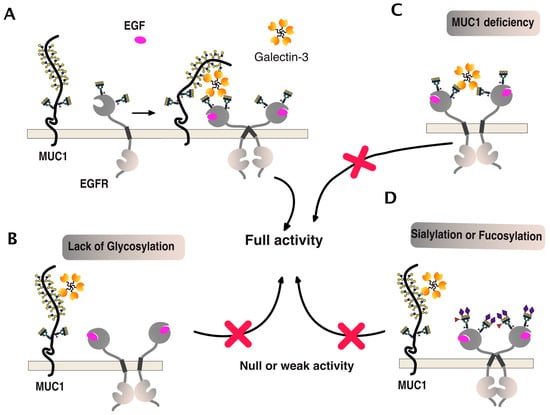
Figure 11.
EGFR multimerization and signaling, illustrating how carbohydrate-binding proteins or carbohydrate-transferases can directly affect transmembrane receptor signal transduction. (A) To be optimal, activation of EGFR signaling was found to involve both a galectin, here being galectin-3, and the glycoprotein MUC1 [100,106]. (B–D) Illustrations depict how the qualitative carbohydrate decoration of EGFR glycosylation or MUC1 deficiency can impair EGFR signaling. Red crosses indicate that activation of this signaling pathway is compromised, as compared to the situation presented in panel A.
On this topic, it is worth noting that glycan-modifying enzymes can also alter tumor necrosis factor receptor superfamily (TNFRSF) members’ signal transduction capabilities, including cell death induced by DD-containing receptors [109]. Likewise, it was found, early on, that Fas could be sialylated in B and T cell lymphoma cell lines. Mere removal of sialic acids in these cell lines using a neuraminidase was found to be sufficient to restore sensitivity to Fas-induced cell death [110,111]. Because the carbohydrate binding affinity of galectins is usually compromised by sialylation [112], these findings would support the hypothesis that galectins may positively contribute to Fas ligand-induced cell death. Consistent with this possibility, it has been demonstrated that the glycosyltransferase, ST6Gal-I (which adds sialic acid in α-2,6 to N-glycans), induces Fas sialylation and inhibits apoptosis-induced by this receptor [113]. Intriguingly, however, although the authors of this study suggested that sialylation would not affect TRAIL signaling, the results presented in this manuscript suggest that sialylation may also prevent TRAIL-induced cell death, at least to some extent. It shall be kept in mind, though, that ST6Gal-I would only be effective on DR4 which is N-glycosylated [43] but not on DR5 which is O-glycosylated [42]. This differential glycosylation status could, therefore, explain why the effects of sialylation were less pronounced towards TRAIL-induced apoptosis than Fas ligand in this study. This is because DR5, which is unlikely to be affected by ST6Gal-I, would remain fully capable of transducing apoptosis upon TRAIL stimulation. In agreement with this hypothesis, TNFR1 (which is also subject to N-glycosylation [114]) has also been found to be modified by ST6Gal-I and, similar to Fas, its signal transduction efficacy was also found to be impaired by sialylation [115].
Conversely, fucosylation, another oligosaccharide modification, may contribute to apoptosis induced by TRAIL [42,70,71]. A deficiency in GDP-mannose-4,6-dehydratase (GMDS), a GDP-mannose converting enzymes essential for de novo fucosylation in colorectal cancer cells, was found to confer resistance to TRAIL [70]. Of interest, the authors of this study demonstrated that fucosylation of DR4, but not DR5, restored TRAIL sensitivity in these cells [71].
7. Conclusions
DD-containing receptors of the TNF super family have been known, since the beginning, for their propensity to self-aggregate and to trigger apoptosis, even in the absence of their cognate ligand [10,116]. TRAIL agonist receptors also display this tendency [117,118]. Investigators have been searching a long time for an explanation for the lack of self-receptor aggregation at the cell surface, explaining why cells harboring the latter fail to undergo spontaneous apoptosis in the absence of the ligand. In the late 1990s, a protein coined silencer of death domain (SODD) or silencer of death domain was proposed to bind the DD-containing receptor TNFR1, thus avoiding recruitment and activation of the caspase-8 [116]. It turned-out, however, that SODD knock-out mice were viable and born at the expected Mendelian ratio, and that cells deficient for SODD would not undergo spontaneous apoptosis nor display differential sensitivity to TNF-induced apoptosis [119]. Given that a growing body of evidence indicate that glycosylation is likely to contribute to the regulation of TNF receptor family signaling and aggregation, it could be interesting to determine whether and how these post-translational modifications control receptor self- or ligand-induced aggregation. Keeping in mind that sialylation and fucosylation, depending on the lactosamine modification [120], can differentially impair galectin binding to carbohydrate chains, the results gathered so far (albeit apparently discrepant) may be unified into a more general model explaining how glycosylation plays a prominent role in the fine-tuning of these potentially harmful receptors. Future studies will be required to determine whether this interplay offers the cell the ability to control receptor aggregation, cell surface arrangement, trafficking, and apoptosis. Increasing our understanding of these complex interactions could additionally open unexpected therapeutic options valuable in oncology and in autoimmune diseases.
Acknowledgments
Olivier Micheau is supported by grants from the ANR (Agence Nationale de la Recherche) program “Investissements d’Avenir” Labex LipSTIC (ANR-11-LABX-0021-01), ANR grants (07-PCV-0031 and SphingoDR), INCA (Institut National du Cancer), the Conseil Regional de Bourgogne, the European commission FEDER (Fonds Européen de Développement Régional, BG0004892) and RISE (DISCOVER, 777995), and the foundation ARC (Association pour la Recherche sur le cancer). Olivier Micheau is grateful to Rafael Saraiva for constructive discussions.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AKT | protein kinase B |
| Asn | asparagine |
| c-FLIP | cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein |
| CRD | cysteine-rich domain |
| DcR | decoy receptor |
| DcR1 | decoy receptor 1, TRAIL-R3, TNFRSF10C |
| DcR2 | decoy receptor 2, TRAIL-R4, TNFRSF10D |
| DD | death domain |
| DISC | death-inducing signaling complex |
| DR | death receptor |
| DR4 | death receptor 4, TRAIL-R1, TNFRSF10A |
| DR5 | death receptor 5, TRAIL-R2, TNFRSF10B |
| EGFR | epithelial growth factor receptor |
| ER | endoplasmic reticulum |
| Fas | fibroblast associated surface antigen, also known as CD95 |
| FADD | Fas associated death domain |
| Fas-L | Fas ligand |
| NE | nuclear envelope |
| NF-κB | nuclear factor-kappa B |
| OPG | osteoprotegerin |
| PLAD | pre-ligand assembly domain |
| PM | plasma membrane |
| Ser | serine |
| SODD | silencer of death domain |
| shRNA | short hairpin RNA |
| siRNA | small interfering RNA |
| Thr | threonine |
| TNF | tumor necrosis factor |
| TNFR1 | tumor necrosis factor receptor 1 |
| TRAIL | TNF related apoptosis inducing ligand, also known as APO2L |
References
- Micheau, O. Posttranslational Modifications and Death Receptor Signalling. In TRAIL, Fas Ligand, TNF and TLR3 in Cancer; Micheau, O., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 247–290. [Google Scholar]
- Varki, A.; Sharon, N. Historical Background and Overview. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: Nassau, NY, USA, 2009. [Google Scholar]
- Stanley, P.; Schachter, H.; Taniguchi, N. N-Glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: Nassau, NY, USA, 2009. [Google Scholar]
- Brockhausen, I.; Schachter, H.; Stanley, P. O-GalNAc Glycans. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E., Eds.; Cold Spring Harbor: Nassau, NY, USA, 2009. [Google Scholar]
- Matoba, K.; Mihara, E.; Tamura-Kawakami, K.; Miyazaki, N.; Maeda, S.; Hirai, H.; Thompson, S.; Iwasaki, K.; Takagi, J. Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1. Cell Rep. 2017, 18, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Emery, J.G.; McDonnell, P.; Burke, M.B.; Deen, K.C.; Lyn, S.; Silverman, C.; Dul, E.; Appelbaum, E.R.; Eichman, C.; DiPrinzio, R.; et al. Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J. Biol. Chem. 1998, 273, 14363–14367. [Google Scholar] [CrossRef] [PubMed]
- Truneh, A.; Sharma, S.; Silverman, C.; Khandekar, S.; Reddy, M.P.; Deen, K.C.; McLaughlin, M.M.; Srinivasula, S.M.; Livi, G.P.; Marshall, L.A.; et al. Temperature-sensitive differential affinity of TRAIL for its receptors. DR5 is the highest affinity receptor. J. Biol. Chem. 2000, 275, 23319–23325. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O.; Shirley, S.; Dufour, F. Death receptors as targets in cancer. Br. J. Pharmacol. 2013, 169, 1723–1744. [Google Scholar] [CrossRef] [PubMed]
- Dubuisson, A.; Micheau, O. Antibodies and Derivatives Targeting DR4 and DR5 for Cancer Therapy. Antibodies 2017, 6, 16. [Google Scholar] [CrossRef]
- Boldin, M.P.; Mett, I.L.; Varfolomeev, E.E.; Chumakov, I.; Shemer-Avni, Y.; Camonis, J.H.; Wallach, D. Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J. Biol. Chem. 1995, 270, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, J.L.; Holler, N.; Reynard, S.; Vinciguerra, P.; Schneider, P.; Juo, P.; Blenis, J.; Tschopp, J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2000, 2, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O.; Shirley, S.; Morizot, A. TRAIL Receptor-Induced Cell Death Regulation: An Update to Our Deadly Discussion. In Topics in Anti-Cancer Research; Rahman, A., Zaman, K., Eds.; Bentham Science Publishers: Busum, Germany, 2014; Volume 3, pp. 3–36. [Google Scholar]
- Pan, G.; Ni, J.; Yu, G.; Wei, Y.F.; Dixit, V.M. TRUNDD, a new member of the TRAIL receptor family that antagonizes TRAIL signalling. FEBS Lett. 1998, 424, 41–45. [Google Scholar] [CrossRef]
- Pan, G.; Ni, J.; Wei, Y.F.; Yu, G.; Gentz, R.; Dixit, V.M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997, 277, 815–818. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, L.; van der Sloot, A.M.; Reis, C.R.; Deegan, S.; Ryan, A.E.; Dhami, S.P.; Murillo, L.S.; Cool, R.H.; de Sampaio, P.C.; Thompson, K.; et al. Decoy receptors block TRAIL sensitivity at a supracellular level: The role of stromal cells in controlling tumour TRAIL sensitivity. Oncogene 2016, 35, 1261. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Hasenauer, J.; Pollak, N.; Scheurich, P. Dominant negative effects of tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) receptor 4 on TRAIL receptor 1 signaling by formation of heteromeric complexes. J. Biol. Chem. 2014, 289, 16576–16587. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.; Lalaoui, N.; Morizot, A.; Schneider, P.; Solary, E.; Micheau, O. Differential inhibition of TRAIL-mediated DR5-DISC formation by decoy receptors 1 and 2. Mol. Cell. Biol. 2006, 26, 7046–7055. [Google Scholar] [CrossRef] [PubMed]
- Anees, M.; Horak, P.; Schiefer, A.I.; Vanhara, P.; El-Gazzar, A.; Perco, P.; Kiesewetter, B.; Mullauer, L.; Streubel, B.; Raderer, M.; et al. The potential evasion of immune surveillance in mucosa associated lymphoid tissue lymphoma by DcR2-mediated up-regulation of nuclear factor-kappaB. Leuk. Lymphoma 2015, 56, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Lalaoui, N.; Morle, A.; Merino, D.; Jacquemin, G.; Iessi, E.; Morizot, A.; Shirley, S.; Robert, B.; Solary, E.; Garrido, C.; et al. TRAIL-R4 promotes tumor growth and resistance to apoptosis in cervical carcinoma HeLa cells through AKT. PLoS ONE 2011, 6, e19679. [Google Scholar] [CrossRef] [PubMed]
- Degli-Esposti, M.A.; Dougall, W.C.; Smolak, P.J.; Waugh, J.Y.; Smith, C.A.; Goodwin, R.G. The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 1997, 7, 813–820. [Google Scholar] [CrossRef]
- Muzio, M.; Stockwell, B.R.; Stennicke, H.R.; Salvesen, G.S.; Dixit, V.M. An induced proximity model for caspase-8 activation. J. Biol. Chem. 1998, 273, 2926–2930. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Dixit, V.M. Caspase activation: The induced-proximity model. Proc. Natl. Acad. Sci. USA 1999, 96, 10964–10967. [Google Scholar] [CrossRef] [PubMed]
- Schleich, K.; Krammer, P.H.; Lavrik, I.N. The chains of death: A new view on caspase-8 activation at the DISC. Cell Cycle 2013, 12, 193–194. [Google Scholar] [CrossRef] [PubMed]
- Dickens, L.S.; Boyd, R.S.; Jukes-Jones, R.; Hughes, M.A.; Robinson, G.L.; Fairall, L.; Schwabe, J.W.; Cain, K.; Macfarlane, M. A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death. Mol. Cell 2012, 47, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Berg, D.; Lehne, M.; Muller, N.; Siegmund, D.; Munkel, S.; Sebald, W.; Pfizenmaier, K.; Wajant, H. Enforced covalent trimerization increases the activity of the TNF ligand family members TRAIL and CD95L. Cell Death Differ. 2007, 14, 2021–2034. [Google Scholar] [CrossRef] [PubMed]
- Holler, N.; Tardivel, A.; Kovacsovics-Bankowski, M.; Hertig, S.; Gaide, O.; Martinon, F.; Tinel, A.; Deperthes, D.; Calderara, S.; Schulthess, T.; et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 2003, 23, 1428–1440. [Google Scholar] [CrossRef] [PubMed]
- Shirley, S.; Morizot, A.; Micheau, O. Regulating TRAIL Receptor-Induced Cell Death at the Membrane: A Deadly Discussion. Recent Pat. Anti-Cancer Drug Discov. 2011, 6, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Morizot, A.; Merino, D.; Lalaoui, N.; Jacquemin, G.; Granci, V.; Iessi, E.; Lanneau, D.; Bouyer, F.; Solary, E.; Chauffert, B.; et al. Chemotherapy overcomes TRAIL-R4-mediated TRAIL resistance at the DISC level. Cell Death Differ. 2011, 18, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Lafont, E.; Hartwig, T.; Walczak, H. Paving TRAIL’s Path with Ubiquitin. Trends Biochem. Sci. 2018, 43, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.D.; Franco, A.; Myers, K.; Gray, C.; Nguyen, T.; Hersey, P. Relation of TNF-related apoptosis-inducing ligand (TRAIL) receptor and FLICE-inhibitory protein expression to TRAIL-induced apoptosis of melanoma. Cancer Res. 1999, 59, 2747–2753. [Google Scholar] [PubMed]
- Olsson, A.; Diaz, T.; Aguilar-Santelises, M.; Osterborg, A.; Celsing, F.; Jondal, M.; Osorio, L.M. Sensitization to TRAIL-induced apoptosis and modulation of FLICE-inhibitory protein in B chronic lymphocytic leukemia by actinomycin D. Leukemia 2001, 15, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Irmler, M.; Thome, M.; Hahne, M.; Schneider, P.; Hofmann, K.; Steiner, V.; Bodmer, J.L.; Schroter, M.; Burns, K.; Mattmann, C.; et al. Inhibition of death receptor signals by cellular FLIP. Nature 1997, 388, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Morle, A.; Garrido, C.; Micheau, O. Hyperthermia restores apoptosis induced by death receptors through aggregation-induced c-FLIP cytosolic depletion. Cell Death Dis. 2015, 6, e1633. [Google Scholar] [CrossRef] [PubMed]
- Kreuz, S.; Siegmund, D.; Scheurich, P.; Wajant, H. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol. Cell. Biol. 2001, 21, 3964–3973. [Google Scholar] [CrossRef] [PubMed]
- Haas, T.L.; Emmerich, C.H.; Gerlach, B.; Schmukle, A.C.; Cordier, S.M.; Rieser, E.; Feltham, R.; Vince, J.; Warnken, U.; Wenger, T.; et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 2009, 36, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O. FLIP. In Cancer Therapeutic Targets; Marshall, J.L., Ed.; Springer: New York, NY, USA, 2017; pp. 881–891. [Google Scholar]
- Lavrik, I.N. Systems biology of death receptor networks: Live and let die. Cell Death Dis. 2014, 5, e1259. [Google Scholar] [CrossRef] [PubMed]
- Walczak, H. Death receptor-ligand systems in cancer, cell death, and inflammation. Cold Spring Harb. Perspect. Biol. 2013, 5, a008698. [Google Scholar] [CrossRef] [PubMed]
- Sessler, T.; Healy, S.; Samali, A.; Szegezdi, E. Structural determinants of DISC function: New insights into death receptor-mediated apoptosis signalling. Pharmacol. Ther. 2013, 140, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Estornes, Y.; Toscano, F.; Virard, F.; Jacquemin, G.; Pierrot, A.; Vanbervliet, B.; Bonnin, M.; Lalaoui, N.; Mercier-Gouy, P.; Pacheco, Y.; et al. dsRNA induces apoptosis through an atypical death complex associating TLR3 to caspase-8. Cell Death Differ. 2012, 19, 1482–1494. [Google Scholar] [CrossRef] [PubMed]
- Estornes, Y.; Micheau, O.; Renno, T.; Lebecque, S. Dual role of TLR3 in Inflammation and Cancer. Cell Apoptosis Oncogene Cancer 2013. [Google Scholar] [CrossRef]
- Wagner, K.W.; Punnoose, E.A.; Januario, T.; Lawrence, D.A.; Pitti, R.M.; Lancaster, K.; Lee, D.; von Goetz, M.; Yee, S.F.; Totpal, K.; et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 2007, 13, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Rattier, T.; Shirley, S.; Picarda, G.; Constantinescu, A.A.; Morle, A.; Zakaria, A.B.; Marcion, G.; Causse, S.; Szegezdi, E.; et al. N-glycosylation of mouse TRAIL-R and human TRAIL-R1 enhances TRAIL-induced death. Cell Death Differ. 2017, 24, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Shiraishi, T.; Horinaka, M.; Wakada, M.; Sakai, T. Glycosylation modulates TRAIL-R1/death receptor 4 protein: Different regulations of two pro-apoptotic receptors for TRAIL by tunicamycin. Oncol. Rep. 2007, 18, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Yoshida, T.; Nakata, S.; Horinaka, M.; Wakada, M.; Mizutani, Y.; Miki, T.; Sakai, T. Tunicamycin enhances tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis in human prostate cancer cells. Cancer Res. 2005, 65, 6364–6370. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.C.; Chen, L.H.; Gillespie, S.; Kiejda, K.A.; Mhaidat, N.; Wang, Y.F.; Thorne, R.; Zhang, X.D.; Hersey, P. Tunicamycin sensitizes human melanoma cells to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by up-regulation of TRAIL-R2 via the unfolded protein response. Cancer Res. 2007, 67, 5880–5888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Du, Z.X.; Liu, B.Q.; Gao, Y.Y.; Meng, X.; Guan, Y.; Deng, W.W.; Wang, H.Q. Tunicamycin enhances TRAIL-induced apoptosis by inhibition of cyclin D1 and the subsequent downregulation of survivin. Exp. Mol. Med. 2009, 41, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Lim, E.J.; Heo, J.; Kwon, T.K.; Kim, Y.H. Tunicamycin sensitizes human prostate cells to TRAIL-induced apoptosis by upregulation of TRAIL receptors and downregulation of cIAP2. Int. J. Oncol. 2012, 40, 1941–1948. [Google Scholar] [PubMed]
- Guo, X.; Meng, Y.; Sheng, X.; Guan, Y.; Zhang, F.; Han, Z.; Kang, Y.; Tai, G.; Zhou, Y.; Cheng, H. Tunicamycin enhances human colon cancer cells to TRAIL-induced apoptosis by JNK-CHOP-mediated DR5 upregulation and the inhibition of the EGFR pathway. Anticancer Drugs 2017, 28, 66–74. [Google Scholar] [CrossRef] [PubMed]
- van Roosmalen, I.A.M.; Reis, C.R.; Setroikromo, R.; Yuvaraj, S.; Joseph, J.V.; Tepper, P.G.; Kruyt, F.A.E.; Quax, W.J. The ER stress inducer DMC enhances TRAIL-induced apoptosis in glioblastoma. Springerplus 2014, 3, 495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Jiang, C.C.; Kiejda, K.A.; Wang, Y.F.; Thorne, R.F.; Zhang, X.D.; Hersey, P. Thapsigargin sensitizes human melanoma cells to TRAIL-induced apoptosis by up-regulation of TRAIL-R2 through the unfolded protein response. Carcinogenesis 2007, 28, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Martin-Perez, R.; Niwa, M.; Lopez-Rivas, A. ER stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. Apoptosis 2012, 17, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Siegelin, M.D. Utilization of the cellular stress response to sensitize cancer cells to TRAIL-mediated apoptosis. Expert Opin. Ther. Targets 2012, 16, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Pinedo, C.; Ruiz-Ruiz, C.; Ruiz de Almodovar, C.; Palacios, C.; Lopez-Rivas, A. Inhibition of glucose metabolism sensitizes tumor cells to death receptor-triggered apoptosis through enhancement of death-inducing signaling complex formation and apical procaspase-8 processing. J. Biol. Chem. 2003, 278, 12759–12768. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, M.; Robinson, G.L.; Cain, K. Glucose—A sweet way to die: Metabolic switching modulates tumor cell death. Cell Cycle 2012, 11, 3919–3925. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nam, S.Y.; Amoscato, A.A.; Lee, Y.J. Low glucose-enhanced TRAIL cytotoxicity is mediated through the ceramide-Akt-FLIP pathway. Oncogene 2002, 21, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.Z.; Xin, H.; Nickoloff, B.J. 2-deoxyglucose sensitizes melanoma cells to TRAIL-induced apoptosis which is reduced by mannose. Biochem. Biophys. Res. Commun. 2010, 401, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Lawrence, D.A.; Marsters, S.; Acosta-Alvear, D.; Kimmig, P.; Mendez, A.S.; Paton, A.W.; Paton, J.C.; Walter, P.; Ashkenazi, A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014, 345, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Dufour, F.; Rattier, T.; Constantinescu, A.A.; Zischler, L.; Morle, A.; Ben Mabrouk, H.; Humblin, E.; Jacquemin, G.; Szegezdi, E.; Delacote, F.; et al. TRAIL receptor gene editing unveils TRAIL-R1 as a master player of apoptosis induced by TRAIL and ER stress. Oncotarget 2017, 8, 9974–9985. [Google Scholar] [CrossRef] [PubMed]
- Iurlaro, R.; Puschel, F.; Leon-Annicchiarico, C.L.; O’Connor, H.; Martin, S.J.; Palou-Gramon, D.; Lucendo, E.; Munoz-Pinedo, C. Glucose Deprivation Induces ATF4-Mediated Apoptosis through TRAIL Death Receptors. Mol. Cell. Biol. 2017, 37, e00479-16. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Pinedo, C.; Lopez-Rivas, A. A role for caspase-8 and TRAIL-R2/DR5 in ER-stress-induced apoptosis. Cell Death Differ. 2018, 25, 226. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Li, Y.; Pu, L.; Xia, F.; Jiang, C.; Liu, H.; Jiang, Z. [Glycosylation inhibitor 2-deoxy-d-glucose sensitizes oral cancer cells to TRAIL-induced apoptosis]. J. South. Med. Univ. 2013, 33, 524–527. [Google Scholar]
- Liu, H.; Jiang, C.C.; Lavis, C.J.; Croft, A.; Dong, L.; Tseng, H.Y.; Yang, F.; Tay, K.H.; Hersey, P.; Zhang, X.D. 2-Deoxy-d-glucose enhances TRAIL-induced apoptosis in human melanoma cells through XBP-1-mediated up-regulation of TRAIL-R2. Mol. Cancer 2009, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Noda, K.; Furukawa, Y.; Ohshima, K.; Uchiyama, A.; Nakagawa, T.; Taniguchi, N.; Daigo, Y.; Nakamura, Y.; Hayashi, N.; et al. Deficiency of GMDS leads to escape from NK cell-mediated tumor surveillance through modulation of TRAIL signaling. Gastroenterology 2009, 137, 188–198. [Google Scholar] [CrossRef] [PubMed]
- Moriwaki, K.; Shinzaki, S.; Miyoshi, E. GDP-mannose-4,6-dehydratase (GMDS) deficiency renders colon cancer cells resistant to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor- and CD95-mediated apoptosis by inhibiting complex II formation. J. Biol. Chem. 2011, 286, 43123–43133. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.; Sui, Z.; Jiang, H.; Shan, Y.; Chen, L.; Zhang, S.; Zhang, L.; Zhang, Y. Releasing N-glycan from peptide N-terminus by N-terminal succinylation assisted enzymatic deglycosylation. Sci. Rep. 2015, 5, 9770. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K. The pre-ligand binding assembly domain: A potential target of inhibition of tumour necrosis factor receptor function. Ann. Rheum. Dis. 2000, 59 (Suppl. S1), i50–i53. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K. Three is better than one: Pre-ligand receptor assembly in the regulation of TNF receptor signaling. Cytokine 2007, 37, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Edmond, V.; Ghali, B.; Penna, A.; Taupin, J.L.; Daburon, S.; Moreau, J.F.; Legembre, P. Precise Mapping of the CD95 Pre-Ligand Assembly Domain. PLoS ONE 2012, 7, e46236. [Google Scholar] [CrossRef] [PubMed]
- Vanamee, E.S.; Faustman, D.L. Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal. 2018, 11, eaao4910. [Google Scholar] [CrossRef] [PubMed]
- Eck, M.J.; Beutler, B.; Kuo, G.; Merryweather, J.P.; Sprang, S.R. Crystallization of trimeric recombinant human tumor necrosis factor (cachectin). J. Biol. Chem. 1988, 263, 12816–12819. [Google Scholar] [PubMed]
- Chan, F.K.; Chun, H.J.; Zheng, L.; Siegel, R.M.; Bui, K.L.; Lenardo, M.J. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 2000, 288, 2351–2354. [Google Scholar] [CrossRef] [PubMed]
- Clancy, L.; Mruk, K.; Archer, K.; Woelfel, M.; Mongkolsapaya, J.; Screaton, G.; Lenardo, M.J.; Chan, F.K. Preligand assembly domain-mediated ligand- independent association between TRAIL receptor 4 (TR4) and TR2 regulates TRAIL-induced apoptosis. Proc. Natl. Acad. Sci. USA 2005, 102, 18099–18104. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.D.; Kordich, J.J.; Huang, T.H.; Piasecki, J.; Bush, T.L.; Sullivan, T.; Foltz, I.N.; Chang, W.; Douangpanya, H.; Dang, T.; et al. Apo2L/TRAIL and the Death Receptor 5 Agonist Antibody AMG 655 Cooperate to Promote Receptor Clustering and Antitumor Activity. Cancer Cell 2014, 26, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Valley, C.C.; Lewis, A.K.; Mudaliar, D.J.; Perlmutter, J.D.; Braun, A.R.; Karim, C.B.; Thomas, D.D.; Brody, J.R.; Sachs, J.N. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces death receptor 5 networks that are highly organized. J. Biol. Chem. 2012, 287, 21265–21278. [Google Scholar] [CrossRef] [PubMed]
- Brandt, B.; Buchse, T.; Abou-Eladab, E.F.; Tiedge, M.; Krause, E.; Jeschke, U.; Walzel, H. Galectin-1 induced activation of the apoptotic death-receptor pathway in human Jurkat T lymphocytes. Histochem. Cell Biol. 2008, 129, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Dorrie, J.; Sapala, K.; Zunino, S.J. Interferon-gamma increases the expression of glycosylated CD95 in B-leukemic cells: An inducible model to study the role of glycosylation in CD95-signalling and trafficking. Cytokine 2002, 18, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Shatnyeva, O.M.; Kubarenko, A.V.; Weber, C.E.; Pappa, A.; Schwartz-Albiez, R.; Weber, A.N.; Krammer, P.H.; Lavrik, I.N. Modulation of the CD95-induced apoptosis: The role of CD95 N-glycosylation. PLoS ONE 2011, 6, e19927. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Kim, S.; Hurtado, J.; Lee, Z.H.; Kim, K.K.; Pollok, K.E.; Kwon, B.S. Characterization of human homologue of 4-1BB and its ligand. Immunol. Lett. 1995, 45, 67–73. [Google Scholar] [CrossRef]
- Madireddi, S.; Eun, S.Y.; Lee, S.W.; Nemcovicova, I.; Mehta, A.K.; Zajonc, D.M.; Nishi, N.; Niki, T.; Hirashima, M.; Croft, M. Galectin-9 controls the therapeutic activity of 4-1BB-targeting antibodies. J. Exp. Med. 2014, 211, 1433–1448. [Google Scholar] [CrossRef] [PubMed]
- Bitra, A.; Doukov, T.; Wang, J.; Picarda, G.; Benedict, C.A.; Croft, M.; Zajonc, D.M. Crystal structure of murine 4-1BB and its interaction with 4-1BBL support a role for galectin-9 in 4-1BB signaling. J. Biol. Chem. 2018, 293, 1317–1329. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, N.; Byrd, J.C.; Sun, Y.; Hafley, M.; Ramirez, K.; Burks, J.; Bresalier, R.S. Cell-surface galectin-3 confers resistance to TRAIL by impeding trafficking of death receptors in metastatic colon adenocarcinoma cells. Cell Death Differ. 2012, 19, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.E.; Menzies, S.A.; Popa, S.J.; Savinykh, N.; Petrunkina Harrison, A.; Lehner, P.J.; Moreau, K. A genome-wide CRISPR screen reconciles the role of N-linked glycosylation in galectin-3 transport to the cell surface. J. Cell Sci. 2017, 130, 3234–3247. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.R.; Chen, P.H.; Bendris, N.; Schmid, S.L. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc. Natl. Acad. Sci. USA 2017, 114, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Hadari, Y.R.; Arbel-Goren, R.; Levy, Y.; Amsterdam, A.; Alon, R.; Zakut, R.; Zick, Y. Galectin-8 binding to integrins inhibits cell adhesion and induces apoptosis. J. Cell Sci. 2000, 113, 2385–2397. [Google Scholar] [PubMed]
- Perillo, N.L.; Pace, K.E.; Seilhamer, J.J.; Baum, L.G. Apoptosis of T cells mediated by galectin-1. Nature 1995, 378, 736–739. [Google Scholar] [CrossRef] [PubMed]
- Perillo, N.L.; Uittenbogaart, C.H.; Nguyen, J.T.; Baum, L.G. Galectin-1, an endogenous lectin produced by thymic epithelial cells, induces apoptosis of human thymocytes. J. Exp. Med. 1997, 185, 1851–1858. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Y.; Hsu, D.K.; Liu, F.T. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc. Natl. Acad. Sci. USA 1996, 93, 6737–6742. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Song, Y.K.; Song, J.J.; Siervo-Sassi, R.R.; Kim, H.R.; Li, L.; Spitz, D.R.; Lokshin, A.; Kim, J.H. Reconstitution of galectin-3 alters glutathione content and potentiates TRAIL-induced cytotoxicity by dephosphorylation of Akt. Exp. Cell Res. 2003, 288, 21–34. [Google Scholar] [CrossRef]
- Oka, N.; Nakahara, S.; Takenaka, Y.; Fukumori, T.; Hogan, V.; Kanayama, H.O.; Yanagawa, T.; Raz, A. Galectin-3 inhibits tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by activating Akt in human bladder carcinoma cells. Cancer Res. 2005, 65, 7546–7553. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, N.; Sun, Y.J.; Liu, K.F.; Gilcrease, M.Z.; Schober, W.; Nangia-Makker, P.; Raz, A.; Bresalier, R.S. Phosphorylated galectin-3 mediates tumor necrosis factor-related apoptosis-inducing ligand signaling by regulating phosphatase and tensin homologue deleted on chromosome 10 in human breast carcinoma cells. J. Biol. Chem. 2007, 282, 21337–21348. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.I.; Whang, E.E.; Abramson, M.A.; Donner, D.B.; Bertagnolli, M.M.; Moore, F.D., Jr.; Ruan, D.T. Galectin-3 regulates apoptosis and doxorubicin chemoresistance in papillary thyroid cancer cells. Biochem. Biophys. Res. Commun. 2009, 379, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, N.; Byrd, J.C.; Sun, Y.; Ueno, S.; Bresalier, R.S. A galectin-3 sequence polymorphism confers TRAIL sensitivity to human breast cancer cells. Cancer 2011, 117, 4375–4380. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, R.R.; Yu, Z.J.; Liang, H.; Shen, S.; Kan, Q. Galectin-1 Modulates the Survival and Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand (TRAIL) Sensitivity in Human Hepatocellular Carcinoma Cells. Cancer Biother. Radiopharm. 2015, 30, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Fukumori, T.; Takenaka, Y.; Oka, N.; Yoshii, T.; Hogan, V.; Inohara, H.; Kanayama, H.O.; Kim, H.R.; Raz, A. Endogenous galectin-3 determines the routing of CD95 apoptotic signaling pathways. Cancer Res. 2004, 64, 3376–3379. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, C.; Schmitz, I.; Zha, J.; Korsmeyer, S.J.; Krammer, P.H.; Peter, M.E. Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J. Biol. Chem. 1999, 274, 22532–22538. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, N.; Jie Sun, Y.; Feng Liu, K.; Gilcrease, M.Z.; Schober, W.; Nangia-Makker, P.; Raz, A.; Bresalier, R.S. Phosphorylated galectin-3 mediates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) signaling by regulating PTEN in human breast carcinoma cells. J. Biol. Chem. 2007, 282, 21337–21348. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C.; Chen, S.T.; Huang, M.C.; Huang, J.; Hsu, C.L.; Juan, H.F.; Lin, H.H.; Chen, C.H. GALNT6 expression enhances aggressive phenotypes of ovarian cancer cells by regulating EGFR activity. Oncotarget 2017, 8, 42588–42601. [Google Scholar] [CrossRef] [PubMed]
- Merlin, J.; Stechly, L.; de Beauce, S.; Monte, D.; Leteurtre, E.; van Seuningen, I.; Huet, G.; Pigny, P. Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene 2011, 30, 2514–2525. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Hsu, D.K.; Chen, H.Y.; Yang, R.Y.; Carraway, K.L., 3rd; Isseroff, R.R.; Liu, F.T. Galectin-3 regulates intracellular trafficking of EGFR through Alix and promotes keratinocyte migration. J. Investig. Dermatol. 2012, 132, 2828–2837. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, J.; Ihara, H.; Miyoshi, E.; Honke, K.; Taniguchi, N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J. Biol. Chem. 2006, 281, 2572–2577. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Yokote, H.; Arao, T.; Maegawa, M.; Tanaka, K.; Fujita, Y.; Shimizu, C.; Hanafusa, T.; Fujiwara, Y.; Nishio, K. N-Glycan fucosylation of epidermal growth factor receptor modulates receptor activity and sensitivity to epidermal growth factor receptor tyrosine kinase inhibitor. Cancer Sci. 2008, 99, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Yen, H.Y.; Chen, C.Y.; Chen, C.H.; Cheng, P.F.; Juan, Y.H.; Chen, C.H.; Khoo, K.H.; Yu, C.J.; Yang, P.C.; et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11332–11337. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, J.T.; de Matos, A.J.; Santos, A.L.; Pinto, R.; Gomes, J.; Hespanhol, V.; Chammas, R.; Manninen, A.; Bernardes, E.S.; Albuquerque Reis, C.; et al. Sialylation regulates galectin-3/ligand interplay during mammary tumour progression--a case of targeted uncloaking. Int. J. Dev. Biol. 2011, 55, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Piyush, T.; Chacko, A.R.; Sindrewicz, P.; Hilkens, J.; Rhodes, J.M.; Yu, L.G. Interaction of galectin-3 with MUC1 on cell surface promotes EGFR dimerization and activation in human epithelial cancer cells. Cell Death Differ. 2017, 24, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, A.; Forcella, M.; Riva, A.; Difrancesco, C.; Molinari, F.; Martin, V.; Papini, N.; Bernasconi, B.; Nonnis, S.; Tedeschi, G.; et al. NEU3 activity enhances EGFR activation without affecting EGFR expression and acts on its sialylation levels. Glycobiology 2015, 25, 855–868. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yen, H.Y.; Liu, Y.C.; Chen, N.Y.; Tsai, C.F.; Wang, Y.T.; Chen, Y.J.; Hsu, T.L.; Yang, P.C.; Wong, C.H. Effect of sialylation on EGFR phosphorylation and resistance to tyrosine kinase inhibition. Proc. Natl. Acad. Sci. USA 2015, 112, 6955–6960. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, R.G.; Rabinovich, G.A. Glycobiology of cell death: When glycans and lectins govern cell fate. Cell Death Differ. 2013, 20, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.E.; Hellbardt, S.; Schwartz-Albiez, R.; Westendorp, M.O.; Walczak, H.; Moldenhauer, G.; Grell, M.; Krammer, P.H. Cell surface sialylation plays a role in modulating sensitivity towards APO-1-mediated apoptotic cell death. Cell Death Differ. 1995, 2, 163–171. [Google Scholar] [PubMed]
- Keppler, O.T.; Peter, M.E.; Hinderlich, S.; Moldenhauer, G.; Stehling, P.; Schmitz, I.; Schwartz-Albiez, R.; Reutter, W.; Pawlita, M. Differential sialylation of cell surface glycoconjugates in a human B lymphoma cell line regulates susceptibility for CD95 (APO-1/Fas)-mediated apoptosis and for infection by a lymphotropic virus. Glycobiology 1999, 9, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Galvan, M.; He, J.; Baum, L.G. The ST6Gal I sialyltransferase selectively modifies N-glycans on CD45 to negatively regulate galectin-1-induced CD45 clustering, phosphatase modulation, and T cell death. J. Biol. Chem. 2003, 278, 7469–7475. [Google Scholar] [CrossRef] [PubMed]
- Swindall, A.F.; Bellis, S.L. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J. Biol. Chem. 2011, 286, 22982–22990. [Google Scholar] [CrossRef] [PubMed]
- Corti, A.; Merli, S.; Bagnasco, L.; D’Ambrosio, F.; Marino, M.; Cassani, G. Identification of two forms (31-33 and 48 kD) of the urinary soluble p55 tumor necrosis factor receptor that are differentially N- and O-glycosylated. J. Interferon Cytokine Res. 1995, 15, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Swindall, A.F.; Kesterson, R.A.; Schoeb, T.R.; Bullard, D.C.; Bellis, S.L. ST6Gal-I regulates macrophage apoptosis via alpha2-6 sialylation of the TNFR1 death receptor. J. Biol. Chem. 2011, 286, 39654–39662. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Woronicz, J.D.; Liu, W.; Goeddel, D.V. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science 1999, 283, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Kitson, J.; Raven, T.; Jiang, Y.P.; Goeddel, D.V.; Giles, K.M.; Pun, K.T.; Grinham, C.J.; Brown, R.; Farrow, S.N. A death-domain-containing receptor that mediates apoptosis. Nature 1996, 384, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Walczak, H.; Degli-Esposti, M.A.; Johnson, R.S.; Smolak, P.J.; Waugh, J.Y.; Boiani, N.; Timour, M.S.; Gerhart, M.J.; Schooley, K.A.; Smith, C.A.; et al. TRAIL-R2: A novel apoptosis-mediating receptor for TRAIL. Embo J. 1997, 16, 5386–5397. [Google Scholar] [CrossRef] [PubMed]
- Takada, H.; Chen, N.J.; Mirtsos, C.; Suzuki, S.; Suzuki, N.; Wakeham, A.; Mak, T.W.; Yeh, W.C. Role of SODD in regulation of tumor necrosis factor responses. Mol. Cell. Biol. 2003, 23, 4026–4033. [Google Scholar] [CrossRef] [PubMed]
- Kamili, N.A.; Arthur, C.M.; Gerner-Smidt, C.; Tafesse, E.; Blenda, A.; Dias-Baruffi, M.; Stowell, S.R. Key regulators of galectin-glycan interactions. Proteomics 2016, 16, 3111–3125. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).