Platforms for Single-Cell Collection and Analysis
Abstract
:1. Introduction
2. Identification of Cells of Interest
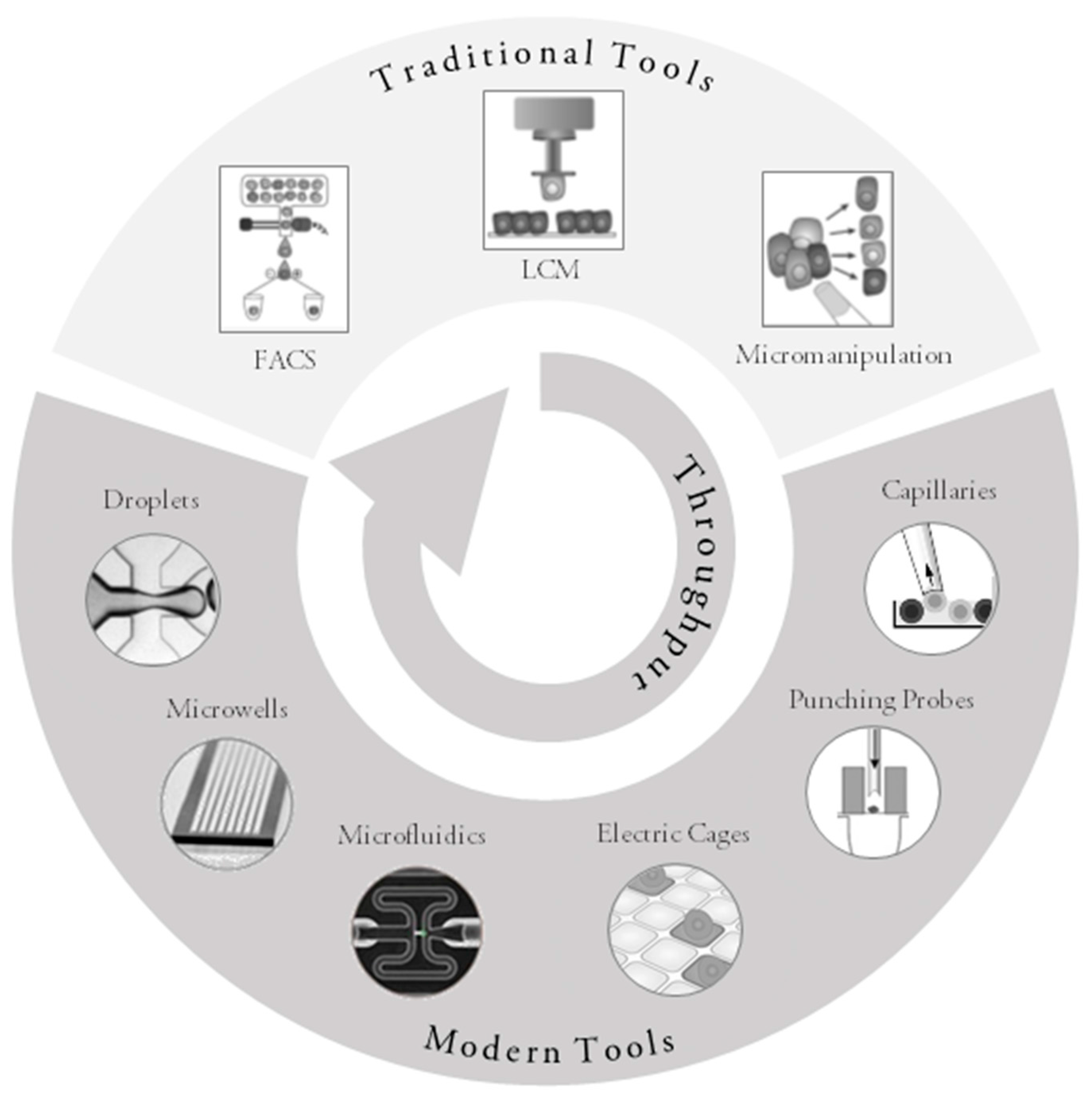
3. Traditional Approaches for Single-Cell Collection
4. Modern Approaches to Single-Cell Collection
4.1. High-Throughput Devices
4.1.1. Chromium System (10x Genomics)
4.1.2. Nadia and RNA-Seq System (Dolomite Bio)
4.1.3. InDrop System (1CellBio)
4.1.4. Single-Cell Sequencing Solution (Illumina, Bio-Rad)
4.1.5. Tapestri Platform (MissionBio)
4.1.6. Rhapsody Single-Cell Analysis System/Resolve (BD)
4.2. Mid-Throughput Devices
4.2.1. ICELL8 Single-Cell System (Takara)
4.2.2. C1 System and Polaris (Fluidigm)
4.3. Low-Throughput Devices
4.3.1. Puncher Platform (Vycap)
4.3.2. CellRaft AIR System (CellMicrosystems)
4.3.3. DEPArray NxT and DEPArray System (Menarini Silicon Biosystems)
4.3.4. AVISO CellCelector (ALS)
5. Enrichment Technologies
6. Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| FACS | Fluorescence-activated cell sorting |
| CTC | Circulating tumor cells |
| qPCR | Quantitative polymerase chain reaction |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| RNA/DNA-Seq | RNA/DNA sequencing |
| IHC | Immunohistochemistry |
| MS | Mass spectroscopy |
| ddPCR | Digital droplet PCR |
| LCM | Laser capture microdissection |
| ICP | Inductively coupled plasma |
| WGA | Whole genome amplification |
| CE-IVD | European Conformity for In Vitro Diagnostic Medical Devices |
| FDA | Food and Drug Administration |
| WTA BD | Whole transcriptome amplification Becton, Dickinson and Company |
| RT | Reverse transcription |
| Drop-seq | Sequencing in droplets |
| inDrop-seq | Indexing in DROPlets and sequencing |
| SCRB-seq | Single-Cell RNA Barcoding and Sequencing |
| UMI | Unique molecular identifier |
| DroNc-Seq | Droplet single-nucleus RNA sequencing |
| PACS UV | PCR-activated cell sorting Ultraviolet radiation |
| SNV | Single nucleotide variant |
| MSND | Multi-sample nano-dispenser |
| IFC | Integrated fluidic circuits |
| DEP ALS | Dielectrophoresis ALS Automated Lab Solutions |
| MACS | Magnetic-activated cell sorting |
| EpCAM | Epithelial cell adhesion molecule |
References
- Almendro, V.; Marusyk, A.; Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 2013, 8, 277–302. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Saadatpour, A.; Lai, S.; Guo, G.; Yuan, G.C. Single-cell analysis in cancer genomics. Trends Genet. 2015, 31, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Kalisky, T.; Blainey, P.; Quake, S.R. Genomic analysis at the single-cell level. Annu. Rev. Genet. 2011, 45, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Lovett, M. The applications of single-cell genomics. Hum. Mol. Genet. 2013, 22, R22–R26. [Google Scholar] [CrossRef] [PubMed]
- Stahlberg, A.; Kubista, M.; Aman, P. Single-cell gene-expression profiling and its potential diagnostic applications. Expert Rev. Mol. Diagn. 2011, 11, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Stahlberg, A.; Rusnakova, V.; Kubista, M. The added value of single-cell gene expression profiling. Brief. Funct. Genom. 2013, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.C.; Lao, K.Q.; Surani, M.A. Development and applications of single-cell transcriptome analysis. Nat. Methods 2011, 8, S6–S11. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.J.; Bodovitz, S. Single cell analysis: The new frontier in ‘omics’. Trends Biotechnol. 2010, 28, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Mishra, L.; Li, S.L. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabieres, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Yang, X.R.; Zhou, J.; Qiu, S.J.; Fan, J.; Xu, Y. Circulating tumor cells: Advances in detection methods, biological issues, and clinical relevance. J. Cancer Res. Clin. 2011, 137, 1151–1173. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Stoecklein, N.H.; Lin, P.P.; Gires, O. Circulating and disseminated tumor cells: Diagnostic tools and therapeutic targets in motion. Oncotarget 2017, 8, 1884–1912. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ramnath, N.; Nagrath, S. Current status of CTCs as liquid biopsy in lung cancer and future directions. Front. Oncol. 2015, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Kuske, A.; Rock, K.; Mauermann, O.; Muller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef] [PubMed]
- Navin, N.E. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015, 25, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Proserpio, V.; Lonnberg, T. Single-cell technologies are revolutionizing the approach to rare cells. Immunol. Cell Biol. 2016, 94, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Hodzic, E. Single-cell analysis: Advances and future perspectives. Bosn. J. Basic Med. Sci. 2016, 16, 313–314. [Google Scholar] [CrossRef] [PubMed]
- Kleparnik, K.; Foret, F. Recent advances in the development of single cell analysis—A review. Anal. Chim. Acta 2013, 800, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.C.; Cai, L.; Elowitz, M.; Enver, T.; Fan, G.; Guo, G.; Irizarry, R.; Kharchenko, P.; Kim, J.; Orkin, S.; et al. Challenges and emerging directions in single-cell analysis. Genome Biol. 2017, 18, 84. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Zhang, W.; Xin, H.; Deng, G. Single cell isolation and analysis. Front. Cell Dev. Biol. 2016, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Hodne, K.; Weltzien, F.A. Single-cell isolation and gene analysis: Pitfalls and possibilities. Int. J. Mol. Sci. 2015, 16, 26832–26849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Singh, A.K. Single-cell protein analysis. Curr. Opin. Biotechnol. 2012, 23, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Ponting, C.P.; Voet, T. Single-cell multiomics: Multiple measurements from single cells. Trends Genet. 2017, 33, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Gallant, C.J.; Marinescu, V.D.; Niklasson, M.; Segerman, A.; Flamourakis, G.; Fredriksson, S.; Assarsson, E.; Lundberg, M.; Nelander, S.; et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 2016, 14, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Stahlberg, A.; Thomsen, C.; Ruff, D.; Aman, P. Quantitative PCR analysis of DNA, RNAs, and proteins in the same single cell. Clin. Chem. 2012, 58, 1682–1691. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Shi, Q.; Wei, W. Single cell proteomics in biomedicine: High-dimensional data acquisition, visualization, and analysis. Proteomics 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Trapnell, C. Single-cell transcriptome sequencing: Recent advances and remaining challenges. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Kanter, I.; Kalisky, T. Single cell transcriptomics: Methods and applications. Front. Oncol. 2015, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Kim, J.K.; Svensson, V.; Marioni, J.C.; Teichmann, S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell 2015, 58, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for single-cell isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Malhotra, L.; Dickerson, R.; Chaffee, S.; Sen, C.K.; Roy, S. Laser capture microdissection: Big data from small samples. Histol. Histopathol. 2015, 30, 1255–1269. [Google Scholar] [PubMed]
- Lee, L.M.; Liu, A.P. The application of micropipette aspiration in molecular mechanics of single cells. J. Nanotechnol. Eng. Med. 2014, 5, 0408011–0408016. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, P.K.; Roederer, M. Cytometry: Today’s technology and tomorrow’s horizons. Methods 2012, 57, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.J.; Li, Q.; Lim, C.T. Manipulation and isolation of single cells and nuclei. Methods Cell Biol. 2010, 98, 79–96. [Google Scholar] [PubMed]
- Lindstrom, S.; Andersson-Svahn, H. Overview of single-cell analyses: Microdevices and applications. Lab Chip 2010, 10, 3363–3372. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Heiby, M.; Pierobon, M.; Liotta, L.A. Laser capture microdissection technology. Expert Rev. Mol. Diagn. 2007, 7, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Andersson, H.; van den Berg, A. Microtechnologies and nanotechnologies for single-cell analysis. Curr. Opin. Biotechnol. 2004, 15, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Walch, A.; Specht, K.; Smida, J.; Aubele, M.; Zitzelsberger, H.; Hofler, H.; Werner, M. Tissue microdissection techniques in quantitative genome and gene expression analyses. Histochem. Cell Biol. 2001, 115, 269–276. [Google Scholar] [PubMed]
- Orfao, A.; RuizArguelles, A. General concepts about cell sorting techniques. Clin. Biochem. 1996, 29, 5–9. [Google Scholar] [CrossRef]
- Wiedenmann, J.; Oswald, F.; Nienhaus, G.U. Fluorescent proteins for live cell imaging: Opportunities, limitations, and challenges. IUBMB Life 2009, 61, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef] [PubMed]
- Day, R.N.; Schaufele, F. Fluorescent protein tools for studying protein dynamics in living cells: A review. J. Biomed. Opt. 2008, 13, 031202. [Google Scholar] [CrossRef] [PubMed]
- Abraham, P.; Maliekal, T.T. Single cell biology beyond the era of antibodies: Relevance, challenges, and promises in biomedical research. Cell. Mol. Life Sci. 2017, 74, 1177–1189. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, A.; Pluckthun, A. Standardize antibodies used in research. Nature 2015, 518, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Reproducibility crisis: Blame it on the antibodies. Nature 2015, 521, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Gopalan, V.; Islam, F.; Lam, A.K. Surface markers for the identification of cancer stem cells. Methods Mol. Biol. 2018, 1692, 17–29. [Google Scholar] [PubMed]
- Ansari, M.A. Temporal profile of m1 and m2 responses in the hippocampus following early 24 h of neurotrauma. J. Neurol. Sci. 2015, 357, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Rusnakova, V.; Honsa, P.; Dzamba, D.; Stahlberg, A.; Kubista, M.; Anderova, M. Heterogeneity of astrocytes: From development to injury—Single cell gene expression. PLoS ONE 2013, 8, e69734. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.J.; Abidi, S.N.F.; Tian, Y.; Skinner, A.; Smith-Bolton, R.K. A rapid, gentle and scalable method for dissociation and fluorescent sorting of imaginal disc cells for mRNA sequencing. Fly 2016, 10, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.L.; De Jesus, J.; Pennell, M.; Troiani, M.; Haun, J.B. Microfluidic device for mechanical dissociation of cancer cell aggregates into single cells. Lab Chip 2015, 15, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Phelan, K.; May, K.M. Basic techniques in mammalian cell tissue culture. Curr. Protoc. Toxicol. 2016, 70, A3B1–A3B22. [Google Scholar]
- Zeng, J.; Mohammadreza, A.; Gao, W.; Merza, S.; Smith, D.; Kelbauskas, L.; Meldrum, D.R. A minimally invasive method for retrieving single adherent cells of different types from cultures. Sci. Rep. 2014, 4, 5424. [Google Scholar] [CrossRef] [PubMed]
- Bebarova, M. Advances in patch clamp technique: Towards higher quality and quantity. Gen. Physiol. Biophys. 2012, 31, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Rubaiy, H.N. A short guide to electrophysiology and ion channels. J. Pharm. Pharm. Sci. 2017, 20, 48–67. [Google Scholar] [CrossRef] [PubMed]
- Podgorny, O.V. Live cell isolation by laser microdissection with gravity transfer. J. Biomed. Opt. 2013, 18, 055002. [Google Scholar] [CrossRef] [PubMed]
- Decarlo, K.; Emley, A.; Dadzie, O.E.; Mahalingam, M. Laser capture microdissection: Methods and applications. Methods Mol. Biol. 2011, 755, 1–15. [Google Scholar] [PubMed]
- Aggerholm-Pedersen, N.; Safwat, A.; Baerentzen, S.; Nordsmark, M.; Nielsen, O.S.; Alsner, J.; Sorensen, B.S. The importance of reference gene analysis of formalin-fixed, paraffin-embedded samples from sarcoma patients—An often underestimated problem. Transl. Oncol. 2014, 7, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Kashofer, K.; Viertler, C.; Pichler, M.; Zatloukal, K. Quality control of RNA preservation and extraction from paraffin-embedded tissue: Implications for RT-PCR and microarray analysis. PLoS ONE 2013, 8, e70714. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A. Formalin fixation in the ‘-omics’ era: A primer for the surgeon-scientist. ANZ J. Surg. 2012, 82, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Zhou, H.; Li, W.; Hu, L.; Zhang, Y. Evaluation of RNA quality in fixed and unembedded mouse embryos by different methods. Exp. Mol. Pathol. 2013, 95, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Dotti, I.; Bonin, S.; Basili, G.; Nardon, E.; Balani, A.; Siracusano, S.; Zanconati, F.; Palmisano, S.; De Manzini, N.; Stanta, G. Effects of formalin, methacarn, and finefix fixatives on RNA preservation. Diagn. Mol. Pathol. 2010, 19, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Mansor, M.A.; Ahmad, M.R. Single cell electrical characterization techniques. Int. J. Mol. Sci. 2015, 16, 12686–12712. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.M.; Loveall, B.; Ewer, J.; Deitcher, D.L.; Sucher, N.J. Characterization of mRNA expression in single neurons. Methods Mol. Biol. 2007, 399, 133–152. [Google Scholar] [PubMed]
- Sucher, N.J.; Deitcher, D.L.; Baro, D.J.; Warrick, R.M.H.; Guenther, E. Genes and channels: Patch/voltage-clamp analysis and single-cell RT-PCR. Cell Tissue Res. 2000, 302, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Gregori, G.; Patsekin, V.; Rajwa, B.; Jones, J.; Ragheb, K.; Holdman, C.; Robinson, J.P. Hyperspectral cytometry at the single-cell level using a 32-channel photodetector. Cytom. Part A 2012, 81, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Nolan, G.P.; Roederer, M.; Chattopadhyay, P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012, 33, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Svec, D.; Andersson, D.; Pekny, M.; Sjoback, R.; Kubista, M.; Stahlberg, A. Direct cell lysis for single-cell gene expression profiling. Front. Oncol. 2013, 3, 274. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.B.; Audet, J. Current techniques for single-cell lysis. J. R. Soc. Interface 2008, 5, S131–S138. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Qing, T.; Zheng, Y.; Jin, L.; Shi, L. Advances in single-cell RNA sequencing and its applications in cancer research. Oncotarget 2017, 8, 53763–53779. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, E.; Deng, Q. Single-cell RNA sequencing: Technical advancements and biological applications. Mol. Asp. Med. 2018, 59, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.E.; Westermann, A.J.; Gorski, S.A.; Vogel, J. Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014, 42, 8845–8860. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.R.; Wang, J.; Streets, A.M.; Huang, Y. Single-cell transcriptional analysis. Annu. Rev. Anal. Chem. 2017, 10, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Kalisky, T.; Oriel, S.; Bar-Lev, T.H.; Ben-Haim, N.; Trink, A.; Wineberg, Y.; Kanter, I.; Gilad, S.; Pyne, S. A brief review of single-cell transcriptomic technologies. Brief. Funct. Genom. 2017, 17, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lonnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Bacher, R.; Kendziorski, C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol. 2016, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Grun, D.; van Oudenaarden, A. Design and analysis of single-cell sequencing experiments. Cell 2015, 163, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Cai, W.; Sun, Z. Single-cell sequencing technologies: Current and future. J. Genet. Genom. 2014, 41, 513–528. [Google Scholar] [CrossRef] [PubMed]
- Bheda, P.; Schneider, R. Epigenetics reloaded: The single-cell revolution. Trends Cell Biol. 2014, 24, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 2017, 65, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lonnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 2014, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Kivioja, T.; Vaharautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2012, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.K.; Hu, J.; Wang, P.H.; Fodor, S.P.A. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc. Natl. Acad. Sci. USA 2011, 108, 9026–9031. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.; Sciambi, A.; Yates, J.L.; Mast, J.D.; Silver, C.; Eastburn, D.J. RNA-seq following PCR-based sorting reveals rare cell transcriptional signatures. BMC Genom. 2016, 17, 361. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.D.; Chen, Y.J.; Dunne, J.; Mir, A.; Hubschle, H.; Guillory, J.; Yuan, W.; Zhang, J.; Stinson, J.; Jaiswal, B.; et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC Genom. 2017, 18, 519. [Google Scholar] [CrossRef] [PubMed]
- Swennenhuis, J.F.; Tibbe, A.G.; Stevens, M.; Katika, M.R.; van Dalum, J.; Tong, H.D.; van Rijn, C.J.; Terstappen, L.W. Self-seeding microwell chip for the isolation and characterization of single cells. Lab Chip 2015, 15, 3039–3046. [Google Scholar] [CrossRef] [PubMed]
- Attayek, P.J.; Waugh, J.P.; Hunsucker, S.A.; Grayeski, P.J.; Sims, C.E.; Armistead, P.M.; Allbritton, N.L. Automated microraft platform to identify and collect non-adherent cells successfully gene-edited with crispr-cas9. Biosens. Bioelectron. 2017, 91, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Phillips, C.; Xu, W.; Pai, J.H.; Dhopeshwarkar, R.; Sims, C.E.; Allbritton, N. Micromolded arrays for separation of adherent cells. Lab Chip 2010, 10, 2917–2924. [Google Scholar] [CrossRef] [PubMed]
- Abonnenc, M.; Manaresi, N.; Borgatti, M.; Medoro, G.; Fabbri, E.; Romani, A.; Altomare, L.; Tartagni, M.; Rizzo, R.; Baricordi, O.; et al. Programmable interactions of functionalized single bioparticles in a dielectrophoresis-based microarray chip. Anal. Chem. 2013, 85, 8219–8224. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, M.; Altomare, L.; Abonnec, M.; Fabbri, E.; Manaresi, N.; Medoro, G.; Romani, A.; Tartagni, M.; Nastruzzi, C.; Di Croce, S.; et al. Dielectrophoresis-based ‘lab-on-a-chip’ devices for programmable binding of microspheres to target cells. Int. J. Oncol. 2005, 27, 1559–1566. [Google Scholar] [PubMed]
- Haupt, S.; Grutzner, J.; Thier, M.C.; Kallweit, T.; Rath, B.H.; Laufenberg, I.; Forgber, M.; Eberhardt, J.; Edenhofer, F.; Brustle, O. Automated selection and harvesting of pluripotent stem cell colonies. Biotechnol. Appl. Biochem. 2012, 59, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Ogunniyi, A.O.; Du, M.; Du, M.; Kretschmann, M.; Eberhardt, J.; Love, J.C. Development and optimization of a process for automated recovery of single cells identified by microengraving. Biotechnol. Prog. 2010, 26, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, M. Startups use short-read data to expand long-read sequencing market. Nat. Biotechnol. 2015, 33, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Eastburn, D.J.; Sciambi, A.; Abate, A.R. Identification and genetic analysis of cancer cells with PCR-activated cell sorting. Nucleic Acids Res. 2014, 42, e128. [Google Scholar] [CrossRef] [PubMed]
- Habib, N.; Avraham-Davidi, I.; Basu, A.; Burks, T.; Shekhar, K.; Hofree, M.; Choudhury, S.R.; Aguet, F.; Gelfand, E.; Ardlie, K.; et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 2017, 14, 955–958. [Google Scholar] [CrossRef] [PubMed]
- BD. BD Expands Genomics Portfolio with New Single Cell Platform for RNA Expression Analysis; Becton Dickinson: Franklin Lakes, NJ, USA, 2017. [Google Scholar]
- De Wit, S.; van Dalum, G.; Lenferink, A.T.; Tibbe, A.G.; Hiltermann, T.J.; Groen, H.J.; van Rijn, C.J.; Terstappen, L.W. The detection of epcam(+) and epcam(−) circulating tumor cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Rho, H.S.; Stevens, M.; Tibbe, A.G.; Gardeniers, H.; Terstappen, L.W. Microfluidic device for DNA amplification of single cancer cells isolated from whole blood by self-seeding microwells. Lab Chip 2015, 15, 4331–4337. [Google Scholar] [CrossRef] [PubMed]
- Attayek, P.J.; Hunsucker, S.A.; Sims, C.E.; Allbritton, N.L.; Armistead, P.M. Identification and isolation of antigen-specific cytotoxic t lymphocytes with an automated microraft sorting system. Integr. Biol. 2016, 8, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E.L.; Rader, J.; Ruden, J.; Rappaport, E.F.; Hunter, K.N.; Hallberg, P.L.; Krytska, K.; O’Dwyer, P.J.; Mosse, Y.P. Dielectrophoretic capture and genetic analysis of single neuroblastoma tumor cells. Front. Oncol. 2014, 4, 201. [Google Scholar] [CrossRef] [PubMed]
- Paterlini-Brechot, P.; Benali, N.L. Circulating tumor cells (CTC) detection: Clinical impact and future directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Barres, B. Reactive astrocytes: Production, function, and therapeutic potential. Immunity 2017, 46, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The m1 and m2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Harouaka, R.A.; Nisic, M.; Zheng, S.Y. Circulating tumor cell enrichment based on physical properties. J. Lab. Autom. 2013, 18, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Low, W.S.; Wan Abas, W.A. Benchtop technologies for circulating tumor cells separation based on biophysical properties. Biomed. Res. Int. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.H.; Hong, S. Microfluidic devices to enrich and isolate circulating tumor cells. Lab Chip 2015, 15, 4500–4511. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Balic, M.; Datar, R.; Cote, R. Size-based enrichment technologies for CTC detection and characterization. Recent Results Cancer Res. 2012, 195, 87–95. [Google Scholar] [PubMed]
- Yeo, T.; Tan, S.J.; Lim, C.L.; Lau, D.P.X.; Chua, Y.W.; Krisna, S.S.; Iyer, G.; Tan, G.S.; Lim, T.K.H.; Tan, D.S.W.; et al. Microfluidic enrichment for the single cell analysis of circulating tumor cells. Sci. Rep. 2016, 6, 22076. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Romani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Tian, T.; Shi, Y.Z.; Liu, W.L.; Zou, Y.; Khajvand, T.; Wang, S.L.; Zhu, Z.; Yang, C.Y. Enrichment and single-cell analysis of circulating tumor cells. Chem. Sci. 2017, 8, 1736–1751. [Google Scholar] [CrossRef] [PubMed]
- Miltenyi, S.; Muller, W.; Weichel, W.; Radbruch, A. High-gradient magnetic cell-separation with macs. Cytometry 1990, 11, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Frey, O.; Petrow, P.K.; Gajda, M.; Siegmund, K.; Huehn, J.; Scheffold, A.; Hamann, A.; Radbruch, A.; Brauer, R. The role of regulatory t cells in antigen-induced arthritis: Aggravation of arthritis after depletion and amelioration after transfer of CD4+CD25+ t cells. Arthritis Res. Ther. 2005, 7, R291–R301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holt, L.M.; Olsen, M.L. Novel applications of magnetic cell sorting to analyze cell-type specific gene and protein expression in the central nervous system. PLoS ONE 2016, 11, e0150290. [Google Scholar] [CrossRef] [PubMed]
- Prieto, P.A.; Durflinger, K.H.; Wunderlich, J.R.; Rosenberg, S.A.; Dudley, M.E. Enrichment of cd8+ cells from melanoma tumor-infiltrating lymphocyte cultures reveals tumor reactivity for use in adoptive cell therapy. J. Immunother. 2010, 33, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, R.; Alunni-Fabbroni, M. The single-cell lab or how to perform single-cell molecular analysis. Whole Genome Amplif. Methods Protoc. 2015, 1347, 43–55. [Google Scholar]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating tumor cells: A review of non-epcam-based approaches for cell enrichment and isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Kenny, L.; Perry, C.; Thiery, J.P.; Jovanovic, L.; Vela, I.; Nelson, C.; Punyadeera, C. Impact of label-free technologies in head and neck cancer circulating tumour cells. Oncotarget 2016, 7, 71223–71234. [Google Scholar] [CrossRef] [PubMed]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Luthra, R.; Goswami, R.S.; Singh, R.R.; Roy-Chowdhuri, S. Analysis of pre-analytic factors affecting the success of clinical next-generation sequencing of solid organ malignancies. Cancers 2015, 7, 1699–1715. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M.; Sciacovelli, L.; Aita, A.; Padoan, A.; Chiozza, M.L. Quality indicators to detect pre-analytical errors in laboratory testing. Clin. Chim. Acta 2014, 432, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Daugharthy, E.R.; Scheiman, J.; Kalhor, R.; Ferrante, T.C.; Terry, R.; Turczyk, B.M.; Yang, J.L.; Lee, H.S.; Aach, J.; et al. Fluorescent in situ sequencing (fisseq) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 2015, 10, 442–458. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.L.; Salmen, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [PubMed]
| Properties | Micromanipulation | Fluorescence-Activated Cell Sorting | Laser Capture Microdissection |
|---|---|---|---|
| Typical Type of Sample | Viable cells | Viable cells | Non-viable |
| Throughput | Low | High | Low |
| Starting Amount of Cells | Low | High | Low |
| Capability to Capture Rare Cells | Low | High | Low |
| Analysis | Slow | Fast | Slow |
| Dissociation | Required | Required | Optional |
| Visual Inspection (Imaging) | Yes | No (Usually) | Yes |
| Information about Morphology | Depends on dissociation | No | Yes |
| Additional Analysis of Sample | No | No | Yes |
| Contamination Hazard | Yes | No | Yes |
| Multi-Parameter Analysis | Yes | Yes | No |
| Laboratory Skills | High | Normal | High |
| Others | Risk perturbing expression profiles (long collection time, dissociation) | Risk perturbing expression profiles (dissociation, fast flow of medium) | May compromise RNA quality |
| Instrument | Chromium System (10x Genomics) | Nadia (Dolomite Bio) | InDrop System (1CellBio) | Illumina Bio-Rad ddSEQ Single-Cell Isolator | Tapestri Platform (MissionBio) | BD Rhapsody Single-Cell Analysis System (BD) | ICELL8 Single-Cell System (Takara) | C1 System and Polaris (Fluidigm) | Puncher Platform (Vycap) | CellRaft AIR System (CellMicrosystems) | DEPArray NxT (Menarini Silicon Biosystems) | AVISO CellCelector (ALS) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Launched in | 10/2016 | 11/2017 | 6/2016 | 1/2017 | 10/2017 | 09/2017 | 10/2015 | 2012 (2015) | 8/2015 | 2017 | 4/2016 | 2006 |
| Principles (Reference) | Droplet-based [89] | Droplet-based (Drop-Seq [84]) | Droplet-base (InDrop-Seq [85]) | Droplet-based | Droplet-based, two-step partitioning [90] | Array of 200,000 microwells, barcoded beads | 5184-well chip, pre-printed barcodes, nano-dispensor [91] | Integrated fluidic circuits for up to 800 cells | Array of 6400 microwells with a pore, filtering, punching needle [92] | Array of 44,000 paramagnetic microwells, punching probe, magnetic collection [93,94] | Microfluidic cartridge with 30,000 dielectrophoretic (DEP) cages [95,96] | Capillary-based [97,98] |
| Main Application | RNA-Seq, DNA-Seq, Immune Repertoire Profiling | RNA-Seq, DroNc-Seq, PACS, open for other | RNA-Seq | RNA-Seq | Targeted DNA-Seq | Targeted RNA-Seq | RNA-Seq | RNA-Seq, DNA-Seq, miRNA-Seq, epigenomics, RT-qPCR | Single-cell collection, rare cell analysis (CTC) | Single-cell collection, tracking cell phenotypes, clonal populations | Single-cell collection, cell–cell interaction | Single-cell collection, transfer of cell colonies |
| Throughput (# of cells analyzed) | High (>10,000) | High (>10,000) | High (>10,000) | High (>10,000) | High (>10,000) | High (>10,000) | Medium (>1000) | Low-medium (48-800) | Low (<100) | Low (<100) | Low (<100) | Low (<100) |
| Visual Control | No | No | No | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Cell Selection | No | No | No | No | No | No | Yes | Yes (C1 size based) | Yes | Yes | Yes | Yes |
| Starting Amount of Cells | High | High | High | High-medium | High | High-medium | Medium | Medium-low | Low | Medium-low | Medium-low | Medium-low |
| Flexibility (Own Protocols) | No | Yes (Nadia Innovate) | Unknown | No | Customize panels | Customize panels | Yes | Yes | Yes | Yes | Yes | Yes |
| Laboratory Skills | Easy | Advanced | Advanced | Easy | Easy | Easy | Easy | Easy | Easy | Easy | Easy | Advanced |
| End-to-End Solution | Yes | No | No | Yes | Yes | Yes | No | Yes | No | No | No | No |
| Extra | Intensive support, 10x Community | Sample chilling, cell and beads stirrer, controllable parameters | Early access program—intensive user support | Product from industry leaders, expertize, scalable (kits for different starting number of cells) | Detect mutation co-occurrence, Characterize rare subclones down to 1% | Automated cell counting, archiving, subsampling, promised upgrade to simultaneous protein-detection | Cell selection combined with high throughput | Automatic workflow, staining, library prep, cell stimulation | Established WGA/WTA protocols using Repli-G kit of Qiagen and the AMPLI-1 kit of Silicon Biosystems | CellRaft System for Inverted Microscopes, QIAscout (Qiagen) | Established WGA/WTA protocols using own kits | Customizable, common labware, various harvest modules |
| Platforms | Advantage | Limitation |
|---|---|---|
| Chromium System (10x Genomics) | High cell capture efficiency, easy to operate, end-to-end solution, multiple applications, well established platform, intensive support | High initial cell concentration required, no users modification possible |
| Nadia (Dolomite Bio) | Open platform, possibility to develop own protocols, multiple applications (PACS, DroNc-Seq) | High initial cell concentration required, lower cell capturing efficiency, no analysis software provided, skills to operate required |
| InDrop System (1CellBio) | High cell capture efficiency, open platform, possibility to develop own protocols | High initial cell concentration required, no analysis software support, skills to operate required |
| Illumina Bio-Rad ddSEQ Single-Cell Isolator | Product from industry leaders, easy to operate, end-to-end solution, kits for different starting number of cells | High initial cell concentration required, no users modification possible, single application (RNA-Seq) |
| Tapestri Platform (MissionBio) | Only platform dedicated to DNA-Seq, easy to operate, customized panels available | Single application possible (DNA-Seq) |
| BD Rhapsody Single-Cell Analysis System (BD) | Possibility to optimize costs (subsampling, archiving, targeted assays), easy to operate, end-to-end solution, protein detection promised | Single application possible (targeted RNA-Seq) |
| ICELL8 Single-Cell System (Takara) | Combined high throughput with active cell selection, easy to operate | Bioinformatics analysis not provided, single application (RNA-Seq) |
| C1 System and Polaris (Fluidigm) | Variable throughput (48–800 cells), multiple applications, customizable protocols, cell stimulation, well established platform, intensive support | Size-based cell selection (C1) |
| Puncher Platform (Vycap) | Filtering for rare cell capturing, active cell selection, visual control, high transferring efficiency, easy to operate, established WGA/WTA protocols | Low throughput, bioinformatics analysis not provided |
| CellRaft AIR System (CellMicrosystems) | Multiple applications (cultivation and tracking cell phenotypes, substance testing), active cell selection, visual control, high transfer efficiency, cost-effective manual version available | Low throughput, bioinformatics analysis not provided, adhesive properties of cells expected (although not mandatory) |
| DEPArray NxT (Menarini Silicon Biosystems) | Active cell selection, visual control, high transfer efficiency, possibility to study cell–cell interaction, established WGA/WTA protocols | Low throughput, bioinformatics analysis not provided; compared to other low-throughput instruments, a high price of consumables (chips) |
| AVISO CellCelector (ALS) | Active cell selection, visual control, multiple applications (transfer cell colonies), low price for consumables | Low throughput, bioinformatics analysis not provided, skills to operate required, adhesive properties of cells lower transfer efficiency, risk of contamination from co-transferred medium |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valihrach, L.; Androvic, P.; Kubista, M. Platforms for Single-Cell Collection and Analysis. Int. J. Mol. Sci. 2018, 19, 807. https://doi.org/10.3390/ijms19030807
Valihrach L, Androvic P, Kubista M. Platforms for Single-Cell Collection and Analysis. International Journal of Molecular Sciences. 2018; 19(3):807. https://doi.org/10.3390/ijms19030807
Chicago/Turabian StyleValihrach, Lukas, Peter Androvic, and Mikael Kubista. 2018. "Platforms for Single-Cell Collection and Analysis" International Journal of Molecular Sciences 19, no. 3: 807. https://doi.org/10.3390/ijms19030807
APA StyleValihrach, L., Androvic, P., & Kubista, M. (2018). Platforms for Single-Cell Collection and Analysis. International Journal of Molecular Sciences, 19(3), 807. https://doi.org/10.3390/ijms19030807




