Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog
Abstract
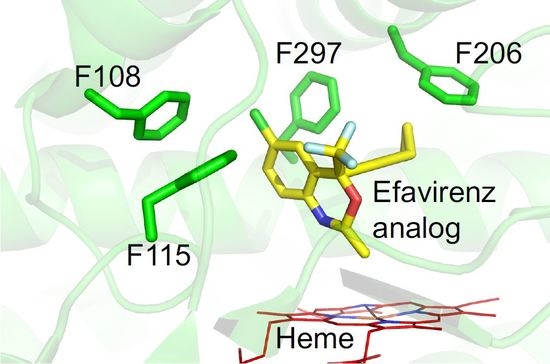
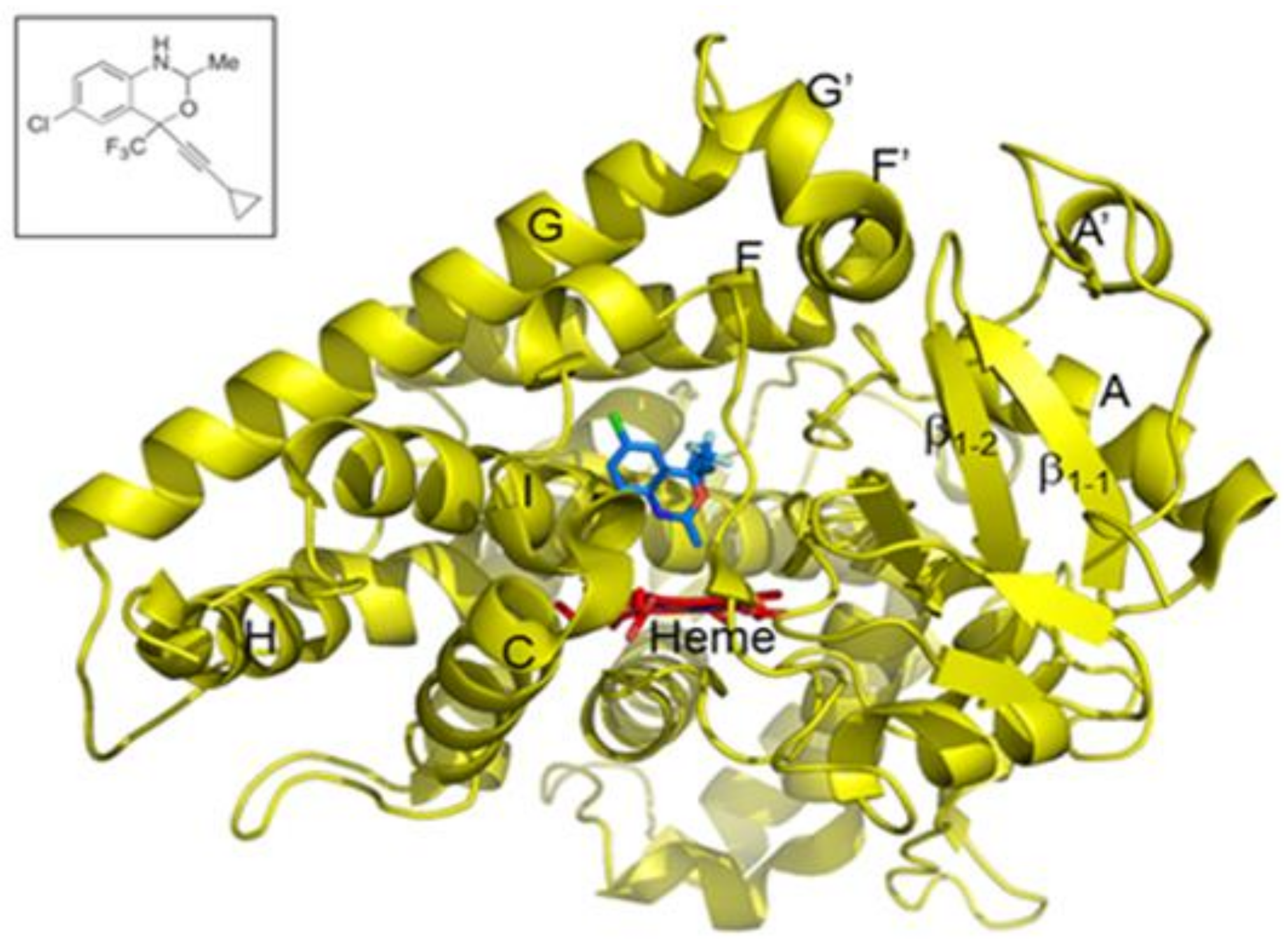
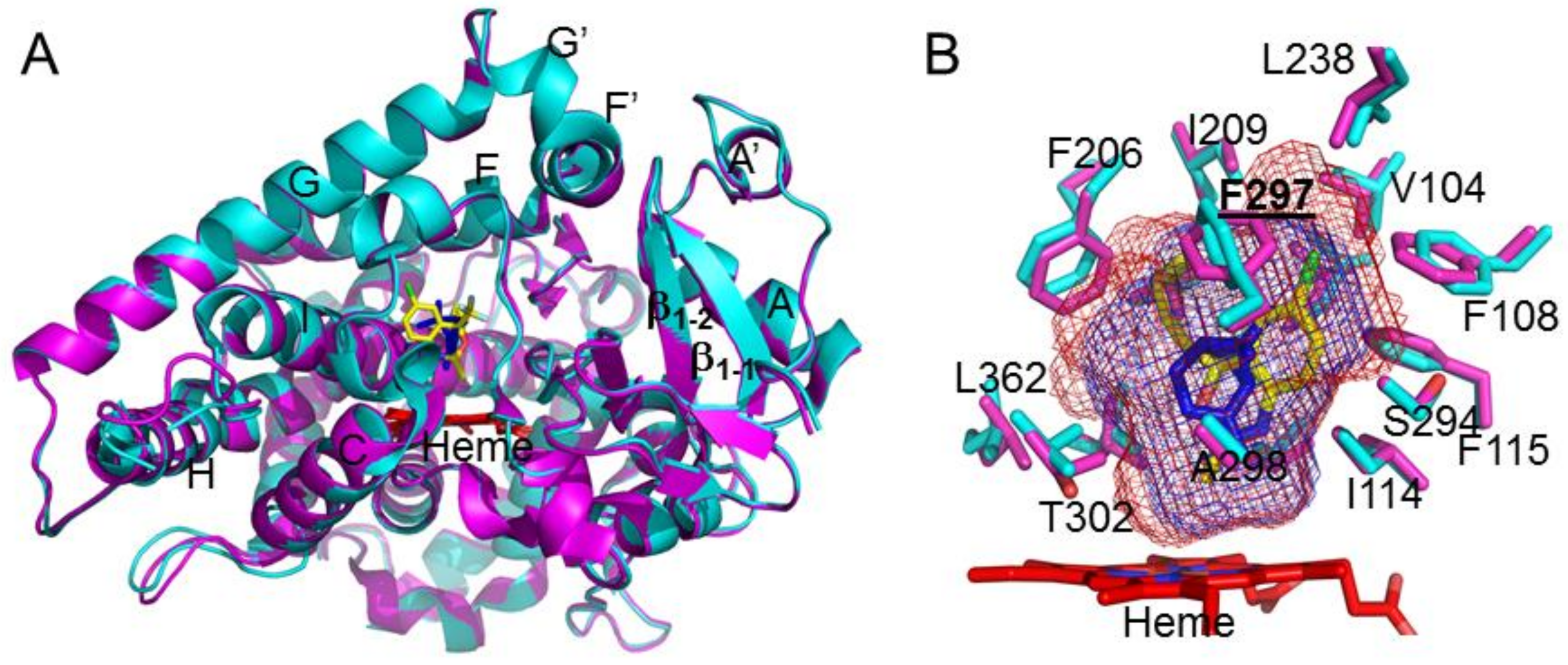
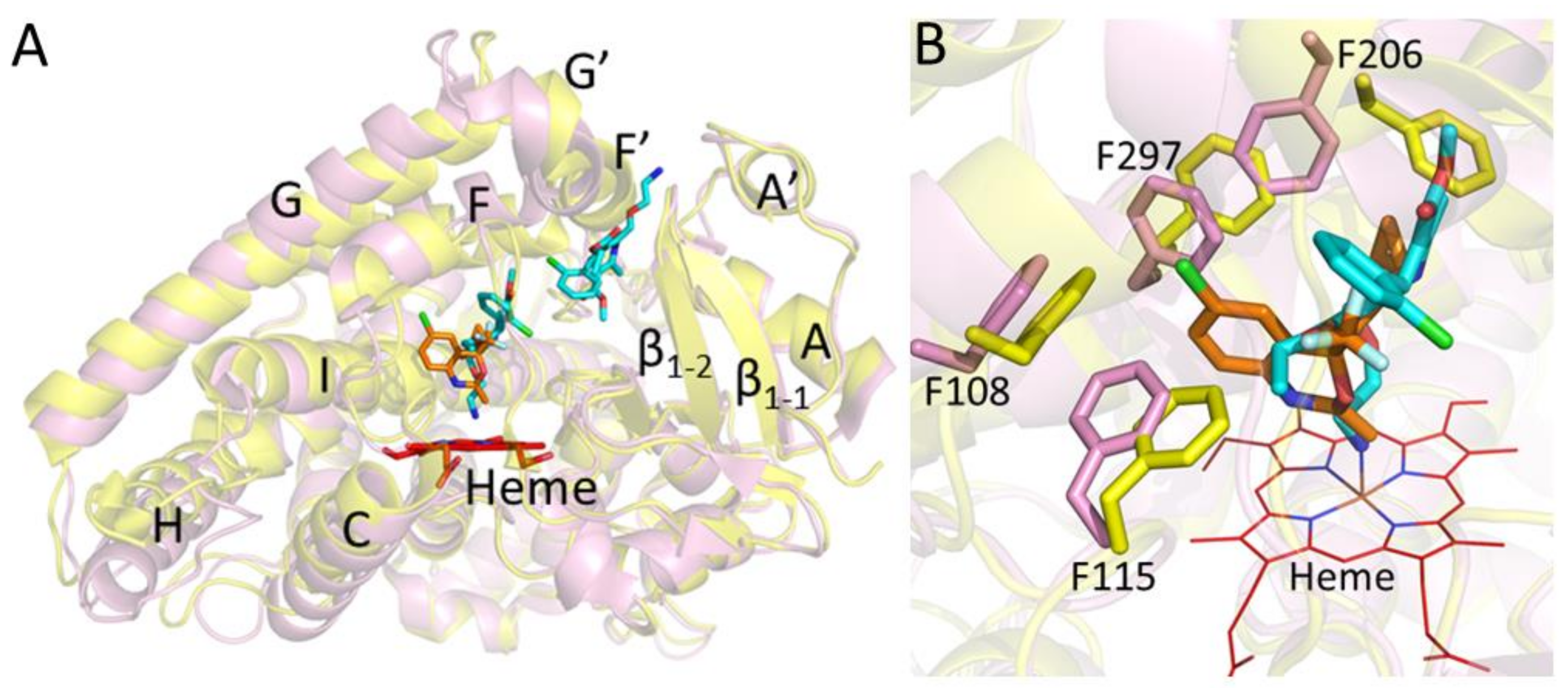
:1. Introduction
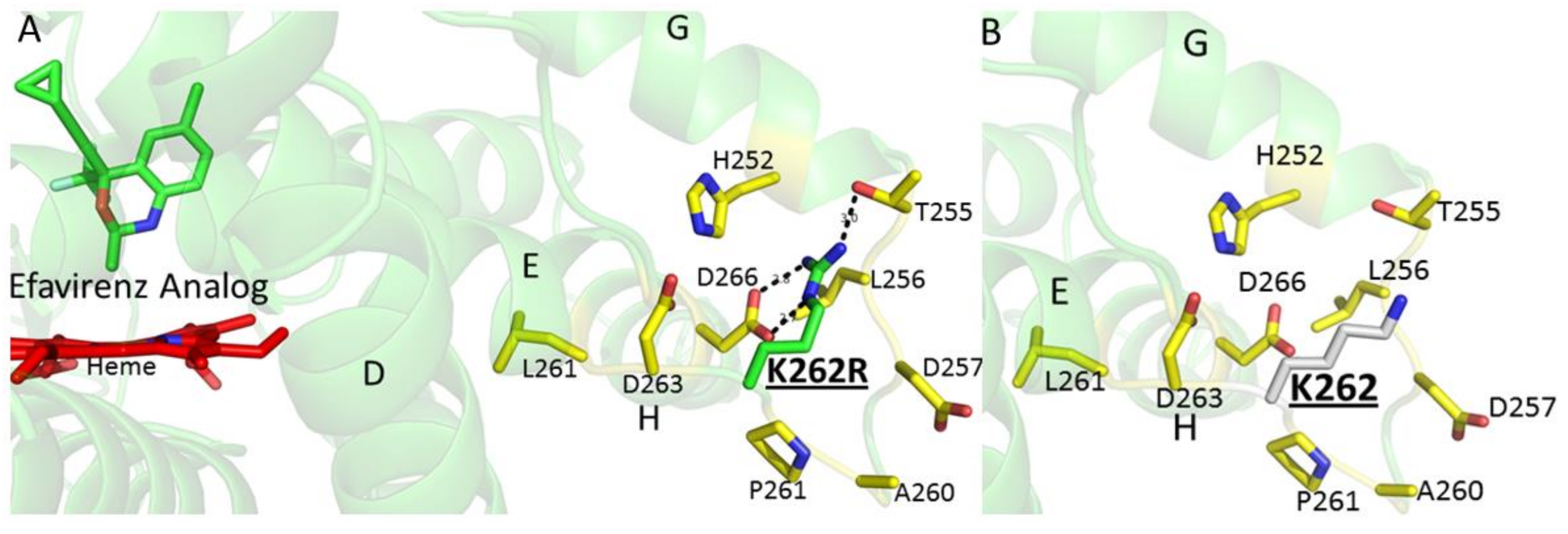
2. Results and Discussion
3. Materials and Methods
3.1. Materials
3.2. Protein Expression and Purification
3.3. Crystallization and Data Collection
3.4. Structure Determination and Refinement
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Montellano, O. Cytochrome P450: Structure, Mechanism, and Biochemistry; Kluwer Academic: Dordrecht, The Netherland, 2015. [Google Scholar]
- Guengerich, F.P.; Waterman, M.R.; Egli, M. Recent Structural Insights into Cytochrome P450 Function. Trends Pharmacol. Sci. 2016, 37, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.R.; Shah, M.B.; Jang, H.H.; Stout, C.D.; Halpert, J.R. Structural and thermodynamic basis of (+)-α-pinene binding to human cytochrome P450 2B6. J. Am. Chem. Soc. 2013, 135, 10433–10440. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.B.; Liu, J.; Huo, L.; Zhang, Q.; Dearing, M.D.; Wilderman, P.R.; Szklarz, G.D.; Stout, C.D.; Halpert, J.R. Structure-Function Analysis of Mammalian CYP2B Enzymes Using 7-Substituted Coumarin Derivatives as Probes: Utility of Crystal Structures and Molecular Modeling in Understanding Xenobiotic Metabolism. Mol. Pharmacol. 2016, 89, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Wilderman, P.R.; Jang, H.H.; Malenke, J.R.; Salib, M.; Angermeier, E.; Lamime, S.; Dearing, M.D.; Halpert, J.R. Functional characterization of cytochromes P450 2B from the desert woodrat Neotoma lepida. Toxicol. Appl. Pharmacol. 2014, 274, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chen, C.S.; Waxman, D.J.; Liu, H.; Halpert, J.R.; Kumar, S. Re-engineering cytochrome P450 2B11dH for enhanced metabolism of several substrates including the anti-cancer prodrugs cyclophosphamide and ifosfamide. Arch. Biochem. Biophys. 2007, 458, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.E.; Liu, H.; He, Y.Q.; Li, W.; Halpert, J.R. Mutagenesis and molecular dynamics suggest structural and functional roles for residues in the N-terminal portion of the cytochrome P450 2B1 I helix. Arch. Biochem. Biophys. 2004, 423, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Scott, E.E.; Liu, H.; Halpert, J.R. A rational approach to Re-engineer cytochrome P450 2B1 regioselectivity based on the crystal structure of cytochrome P450 2C5. J. Biol. Chem. 2003, 278, 17178–17184. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Klein, K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): Advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 2013, 4, 24. [Google Scholar] [CrossRef] [PubMed]
- Turpeinen, M.; Zanger, U.M. Cytochrome P450 2B6, function, genetics, and clinical relevance. Drug Metabol. Drug Interact. 2012, 27, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Ariyoshi, N.; Oguri, K.; Koga, N.; Yoshimura, H.; Funae, Y. Metabolism of highly persistent PCB congener, 2,4,5,2′,4′,5′-hexachlorobiphenyl, by human CYP2B6. Biochem. Biophys. Res. Commun. 1995, 212, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Cao, Y.; Rose, R.L.; Hodgson, E. In vitro metabolism of carbaryl by human cytochrome P450 and its inhibition by chlorpyrifos. Chem. Biol. Interact. 2002, 141, 229–241. [Google Scholar] [CrossRef]
- Feo, M.L.; Gross, M.S.; McGarrigle, B.P.; Eljarrat, E.; Barceló, D.; Aga, D.S.; Olson, J.R. Biotransformation of BDE-47 to potentially toxic metabolites is predominantly mediated by human CYP2B6. Environ. Health. Perspect. 2013, 121, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Milton, J.; Weaver, K.L.; Short, S.A.; Stuart, D.I.; Stammers, D.K. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 2000, 8, 1089–1094. [Google Scholar] [CrossRef]
- Ward, B.A.; Gorski, J.C.; Jones, D.R.; Hall, S.D.; Flockhart, D.A.; Desta, Z. The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: Implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J. Pharmacol. Exp. Ther. 2003, 306, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Mutlib, A.E.; Chen, H.; Nemeth, G.A.; Markwalder, J.A.; Seitz, S.P.; Gan, L.S.; Christ, D.D. Identification and characterization of efavirenz metabolites by liquid chromatography/mass spectrometry and high field NMR: Species differences in the metabolism of efavirenz. Drug Metab. Dispos. 1999, 27, 1319–1333. [Google Scholar] [PubMed]
- Faucette, S.R.; Zhang, T.C.; Moore, R.; Sueyoshi, T.; Omiecinski, C.J.; LeCluyse, E.L.; Negishi, M.; Wang, H. Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J. Pharmacol. Exp. Ther. 2007, 320, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Bumpus, N.N.; Kent, U.M.; Hollenberg, P.F. Metabolism of efavirenz and 8-hydroxyefavirenz by P450 2B6 leads to inactivation by two distinct mechanisms. J. Pharmacol. Exp. Ther. 2006, 318, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.M.; Bumpus, N.N. Structure-Activity Studies Reveal the Oxazinone Ring Is a Determinant of Cytochrome P450 2B6 Activity Toward Efavirenz. ACS Med. Chem. Lett. 2014, 5, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.M.; Bumpus, N.N. Single Heteroatom Substitutions in the Efavirenz Oxazinone Ring Impact Metabolism by CYP2B6. ChemMedChem 2016, 11, 2630–2637. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.B.; Liu, J.; Zhang, Q.; Stout, C.D.; Halpert, J.R. Halogen-π Interactions in the Cytochrome P450 Active Site: Structural Insights into Human CYP2B6 Substrate Selectivity. ACS Chem. Biol. 2017, 12, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.B.; Wilderman, P.R.; Pascual, J.; Zhang, Q.; Stout, C.D.; Halpert, J.R. Conformational adaptation of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 revealed upon binding multiple amlodipine molecules. Biochemistry 2012, 51, 7225–7238. [Google Scholar] [CrossRef] [PubMed]
- Gay, S.C.; Shah, M.B.; Talakad, J.C.; Maekawa, K.; Roberts, A.G.; Wilderman, P.R.; Sun, L.; Yang, J.Y.; Huelga, S.C.; Hong, W.X.; et al. Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution. Mol. Pharmacol. 2010, 77, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Klein, K.; Fischer, J.; Nüssler, A.K.; Neuhaus, P.; Hofmann, U.; Eichelbaum, M.; Schwab, M.; Zanger, U.M. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 2001, 11, 399–415. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sönnerborg, A.; Rane, A.; Josephson, F.; Lundgren, S.; Ståhle, L.; Ingelman-Sundberg, M. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet. Genom. 2006, 16, 191–198. [Google Scholar]
- Klein, K.; Lang, T.; Saussele, T.; Barbosa-Sicard, E.; Schunck, W.H.; Eichelbaum, M.; Schwab, M.; Zanger, U.M. Genetic variability of CYP2B6 in populations of African and Asian origin: Allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet. Genom. 2005, 15, 861–873. [Google Scholar] [CrossRef]
- Gatanaga, H.; Hayashida, T.; Tsuchiya, K.; Yoshino, M.; Kuwahara, T.; Tsukada, H.; Fujimoto, K.; Sato, I.; Ueda, M.; Horiba, M.; et al. Successful efavirenz dose reduction in HIV type 1-infected individuals with cytochrome P450 2B6 *6 and *26. Clin. Infect. Dis. 2007, 45, 1230–1237. [Google Scholar] [CrossRef] [PubMed]
- Radloff, R.; Gras, A.; Zanger, U.M.; Masquelier, C.; Arumugam, K.; Karasi, J.C.; Arendt, V.; Seguin-Devaux, C.; Klein, K. Novel CYP2B6 enzyme variants in a Rwandese population: Functional characterization and assessment of in silico prediction tools. Hum. Mutat. 2013, 34, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, A.; Rotger, M.; Aouri, M.; Kuster, S.P.; Telenti, A.; Décosterd, L.A.; Günthard, H.F. Efavirenz intoxication due to a new CYP2B6 constellation. Antivir. Ther. 2013, 18, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Bennett, B.C.; Hong, W.X.; Fu, Y.; Baker, K.A.; Marcoux, J.; Robinson, C.V.; Ward, A.B.; Halpert, J.R.; Stevens, R.C.; et al. Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E1203–E1211. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.E.; Spatzenegger, M.; Halpert, J.R. A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch. Biochem. Biophys. 2001, 395, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.B.; Pascual, J.; Zhang, Q.; Stout, C.D.; Halpert, J.R. Structures of cytochrome P450 2B6 bound to 4-benzylpyridine and 4-(4-nitrobenzyl)pyridine: Insight into inhibitor binding and rearrangement of active site side chains. Mol. Pharmacol. 2011, 80, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J. Biol. Chem. 1964, 239, 2370–2378. [Google Scholar] [PubMed]
- Omura, T.; Sato, R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. Ii. Solubilization, Purification, and Properties. J. Biol. Chem. 1964, 239, 2379–2385. [Google Scholar] [PubMed]
- Davydov, D.R.; Deprez, E.; Hoa, G.H.; Knyushko, T.V.; Kuznetsova, G.P.; Koen, Y.M.; Archakov, A.I. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: Evidence for conformational inhomogeneity in the oligomers. Arch. Biochem. Biophys. 1995, 320, 330–344. [Google Scholar] [CrossRef]
- Renaud, J.P.; Davydov, D.R.; Heirwegh, K.P.; Mansuy, D.; Hui Bon Hoa, G.H. Thermodynamic studies of substrate binding and spin transitions in human cytochrome P-450 3A4 expressed in yeast microsomes. Biochem. J. 1996, 319, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Battye, T.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G. iMOSFLM: A new graphical interface for diffraction-image processing with, MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, M.B.; Zhang, Q.; Halpert, J.R. Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog. Int. J. Mol. Sci. 2018, 19, 1025. https://doi.org/10.3390/ijms19041025
Shah MB, Zhang Q, Halpert JR. Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog. International Journal of Molecular Sciences. 2018; 19(4):1025. https://doi.org/10.3390/ijms19041025
Chicago/Turabian StyleShah, Manish B., Qinghai Zhang, and James R. Halpert. 2018. "Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog" International Journal of Molecular Sciences 19, no. 4: 1025. https://doi.org/10.3390/ijms19041025
APA StyleShah, M. B., Zhang, Q., & Halpert, J. R. (2018). Crystal Structure of CYP2B6 in Complex with an Efavirenz Analog. International Journal of Molecular Sciences, 19(4), 1025. https://doi.org/10.3390/ijms19041025