Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola
Abstract
:1. Introduction
2. Results
2.1. PRR Expression in Vitis amurensis ‘Shuanghong’ Infected with Incompatible and Compatible Strains of Plasmopara viticola
2.2. Characterization and Phylogenetic Analysis of VaHAESA
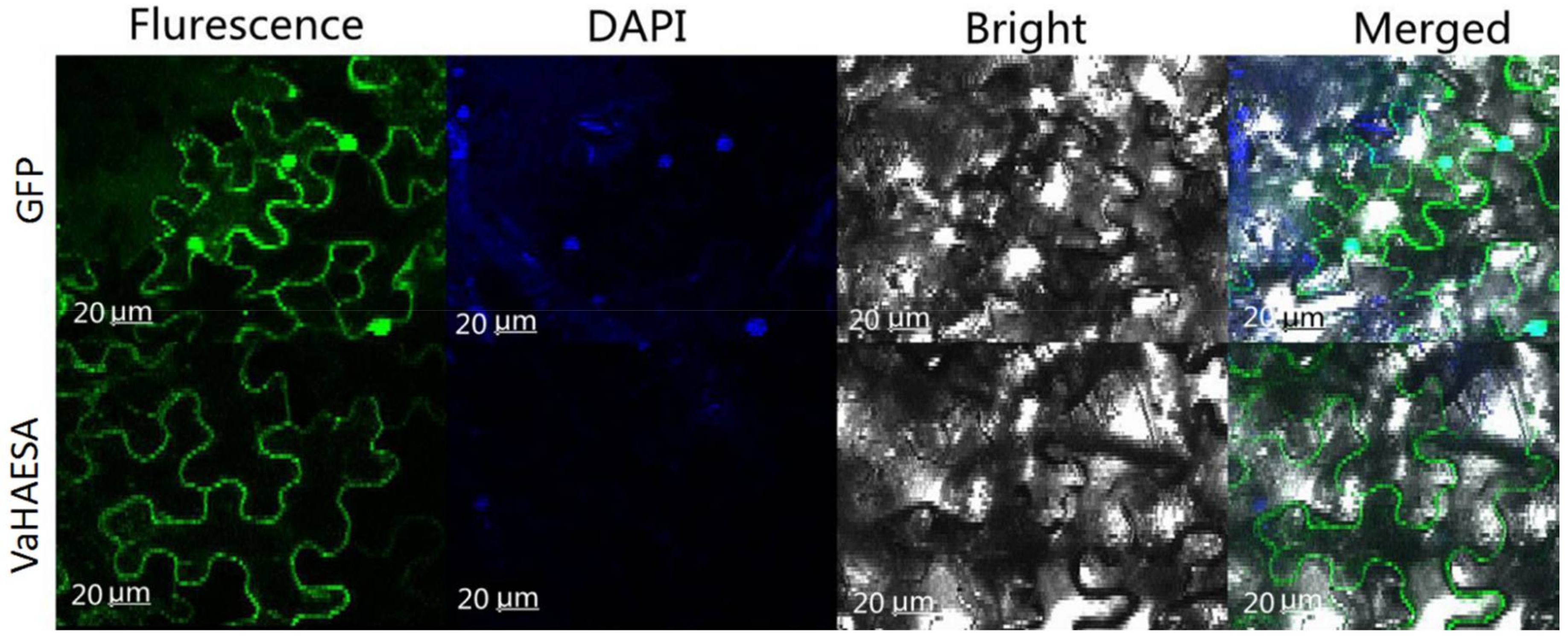
2.3. Subcellular Location of VaHAESA in Nicotiana benthamiana
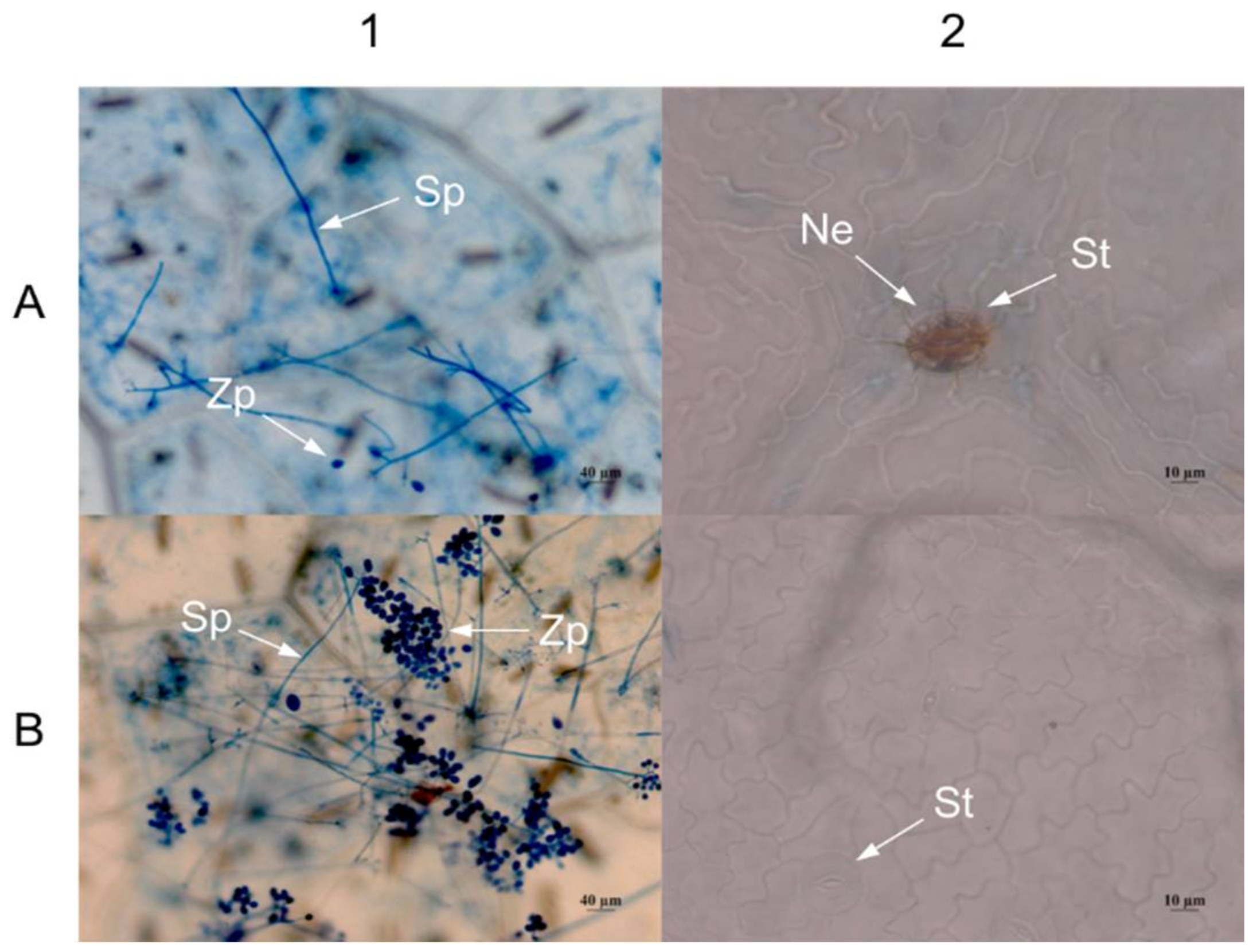
2.4. Expression of VaHAESA Promoted Resistance against Plasmopara viticola in Grapevine
2.5. Measurements of H2O2, NO, and Callose in Vitis vinifera Transiently Expressing VaHAESA
2.6. Identification and Analysis of Disease Resistance in Transgenic Arabidopsis thaliana
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Plasmopara viticola Strains, and Pathogen Infection
4.2. qRT-PCR
4.3. Cloning, Sequencing, and Phylogenetic Characterization of VaHAESA
4.4. Agrobacterium-Mediated Transient Expression in Plants
4.5. Analysis of the Subcellular Localization of VaHAESA and Its Effect on Pathogen Infection
4.6. Analysis of H2O2 and NO Levels in Transgenic Vitis vinifera Expressing VaHAESA
4.7. Screening and Identification of Transgenic Arabidopsis thaliana
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Casagrande, K.; Falginella, L.; Castellarin, S.D.; Testolin, R.; Di Gaspero, G. Defence responses in Rpv3-dependent resistance to grapevine downy mildew. Planta 2011, 234, 1097–1099. [Google Scholar] [CrossRef] [PubMed]
- Milli, A.; Cecconi, D.; Bortesi, L.; Persi, A.; Rinalducci, S.; Zamboni, A.; Zoccatelli, G.; Lovato, A.; Zolla, L.; Polverari, A. Proteomic analysis of the compatible interaction between Vitis vinifera and Plasmopara viticola. J. Proteom. 2012, 75, 1284–1302. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Gómezgómez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Robatzek, S.; Bittel, P.; Chinchilla, D.; Köchner, P.; Felix, G.; Shiu, S.H.; Boller, T. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 2007, 64, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Salamango, D.J.; Leslie, M.E.; Collins, C.A.; Heese, A. Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014, 164, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Robatzek, S.; Navarro, L.; Oakeley, E.J.; Jones, J.D.; Felix, G.; Boller, T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004, 428, 764. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Kunze, G.; Chinchilla, D.; Caniard, A.; Jones, J.D.; Boller, T.; Felix, G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006, 125, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, R.; Zhang, N.; Ma, F.; Jiao, Y.; Wang, Z. Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol. Biol. 2014, 86, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Qu, J.; Lu, J. Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol. Biochem. 2015, 95, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Fang, J.; Wang, C.; Yin, Y.; Sun, X.; Leng, X.; Song, C. Grapevine microRNAs responsive to exogenous gibberellin. BMC Genom. 2014, 15, 111. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.C. Receptor-like protein kinase genes of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2015, 3, 451–456. [Google Scholar] [CrossRef]
- Cho, S.K.; Larue, C.T.; Chevalier, D.; Wang, H.; Jinn, T.L.; Zhang, S.; Walker, J.C. From the Cover: Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 105, 15629–15634. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tör, M.; De Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Tintor, N.; Ross, A.; Kanehara, K.; Yamada, K.; Fan, L.; Kemmerling, B.; Nürnberger, T.; Tsuda, K.; Saijo, Y. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 6211–6216. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Shen, Q.; Qi, Y.; Yan, H.; Nie, H.; Chen, Y.; Zhao, T.; Katagiri, F.; Tang, D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell 2013, 25, 1143–1157. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Jinn, T.L.; Stone, J.M.; Walker, J.C. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Gene Dev. 2000, 14, 108–117. [Google Scholar] [PubMed]
- Butenko, M.A.; Wildhagen, M.; Albert, M.; Jehle, A.; Kalbacher, H.; Aalen, R.B.; Felix, G. Tools and Strategies to Match Peptide-Ligand Receptor Pairs. Plant Cell 2014, 26, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Julia, S.; Benjamin, B.; Mari, W.; Ulrich, H.; Hothorn, L.A.; Butenko, M.A.; Michael, H. Mechanistic insight into a peptide hormone signaling complex mediating floral organ abscission. eLife 2016, 5, e15075. [Google Scholar]
- Wong, F.P.; Burr, H.N.; Wilcox, W.F. Heterothallism in Plasmopara viticola. Plant Pathol. 2010, 50, 427–432. [Google Scholar] [CrossRef]
- Kemmerling, B.; Zipfel, C.; Chinchilla, D.; Felix, G.; Jones, J.D.G.; Robatzek, S.; Boller, T.; Nürnberger, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497. [Google Scholar]
- Böhm, H.; Albert, I.; Fan, L.; Reinhard, A.; Nürnberger, T. Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 2014, 20, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, L.; Macho, A.P.; Han, Z.; Hu, Z.; Zipfel, C.; Zhou, J.M.; Chai, J. Structural Basis for flg22-Induced Activation of the Arabidopsis FLS2-BAK1 Immune Complex. Science 2013, 342, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Schulze, B.; Mentzel, T.; Jehle, A.K.; Mueller, K.; Beeler, S.; Boller, T.; Felix, G.; Chinchilla, D. Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 2010, 285, 9444–9451. [Google Scholar] [CrossRef] [PubMed]
- Niederhuth, C.E.; Rahul, P.O.; Walker, J.C. Transcriptional profiling of the Arabidopsis abscission mutanthae hsl2by RNA-Seq. BMC Genom. 2013, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, L.; Boller, T. Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 2002, 7, 251–256. [Google Scholar] [CrossRef]
- Jonak, C.; Okrész, L.; Bögre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Betsuyaku, S.; Takahashi, F.; Kinoshita, A.; Miwa, H.; Shinozaki, K.; Fukuda, H.; Sawa, S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 2011, 52, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Ma, X.; Shan, L.; He, P. Big Roles of Small Kinases:The Complex Functions of Receptor-Like Cytoplasmic Kinases in Plant Immunity and Development. Chin. J. Plant Ecol. 2013, 55, 1188–1197. [Google Scholar]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Nühse, T.S.; Peck, S.C.; Hirt, H.; Boller, T. Microbial elicitors induce activation and dual phosphorylation of the Arabidopsis thaliana MAPK 6. J. Biol. Chem. 2000, 275, 7521–7526. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Dunning, F.M.; Pfund, C.; Weingarten, R.; Bent, A.F. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 2006, 18, 764. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant. 2010, 138, 414. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Wang, X.; Li, P.; Wang, H.; Ji, H.; Xie, J.; Qiu, Q.; Shen, D.; Dong, H. Plant Aquaporin AtPIP1;4 Links Apoplastic H2O2 Induction to Disease Immunity Pathways. Plant Physiol. 2016, 171. [Google Scholar] [CrossRef] [PubMed]
- Groß, F.; Durner, J.; Gaupels, F. Nitric oxide, antioxidants and prooxidants in plant defence responses. Front. Plant Sci. 2013, 4, 419. [Google Scholar] [CrossRef] [PubMed]
- Scheler, C.; Durner, J.; Astier, J. Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 2013, 16, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Raho, N.; Ramirez, L.; Lanteri, M.L.; Gonorazky, G.; Lamattina, L.; Have, A.T.; Laxalt, A.M. Phosphatidic acid production in chitosan-elicited tomato cells, via both phospholipase D and phospholipase C/diacylglycerol kinase, requires nitric oxide. J. Plant Physiol. 2011, 168, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Underwood, W. The Plant Cell Wall: A Dynamic Barrier Against Pathogen Invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Yin, L.; Lu, J. The mode of host resistance to Plasmopara viticola infection of grapevines. Phytopathology 2012, 102, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.L.; Jiao, W.; Pei, Z.; Hasi, G.; Yu, H.; Jiang, L.; Zhang, Y.L. Response of phytohormones and correlation of SAR signal pathway genes to the different resistance levels of grapevine against Plasmopara viticola infection. Plant Physiol. Biochem. 2016, 107, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Jürges, G.; Kassemeyer, H.H.; Dürrenberger, M.; Düggelin, M.; Nick, P. The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol. 2009, 11, 886–898. [Google Scholar] [PubMed]
- Iandolino, A.B.; Silva, F.G.D.; Lim, H.; Choi, H.; Williams, L.E.; Cook, D.R. High-Quality RNA, cDNA, and Derived EST Libraries From Grapevine (Vitis vinifera L.). Plant Mol. Biol. Rep. 2004, 22, 269–278. [Google Scholar] [CrossRef]
- Monteiro, F.; Sebastiana, M.; Pais, M.S.; Figueiredo, A. Reference Gene Selection and Validation for the Early Responses to Downy Mildew Infection in Susceptible and Resistant Vitis vinifera Cultivars. Minerva Stomatol. 2012, 56, 611. [Google Scholar] [CrossRef] [PubMed]
- Selim, M.; Legay, S.; Berkelmannlöhnertz, B.; Langen, G.; Kogel, K.H.; Evers, D. Identification of suitable reference genes for real-time RT-PCR normalization in the grapevine-downy mildew pathosystem. Plant Cell Rep. 2012, 31, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, X.; Wu, J.; Yin, L.; Zhang, Y.; Lu, J. Studying the Mechanism of Plasmopara viticola RxLR Effectors on Suppressing Plant Immunity. Front. Microbiol. 2016, 7, 709. [Google Scholar]
- Guan, X.; Zhao, H.; Xu, Y.; Wang, Y. Transient expression of glyoxal oxidase from the Chinese wild grape Vitis pseudoreticulata can suppress powdery mildew in a susceptible genotype. Protoplasma 2011, 248, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Keogh, R.C.; Deverall, B.J.; Mcleod, S. Comparison of histological and physiological responses to Phakopsora pachyrhizi resistant and susceptible soybean. Trans. Br. Mycol. Soc. 1980, 74, 329–333. [Google Scholar] [CrossRef]
- Díez-Navajas, A.M.; Greif, C.; Poutaraud, A.; Merdinoglu, D. Two simplified fluorescent staining techniques to observe infection structures of the oomycete Plasmopara viticola in grapevine leaf tissues. Micron 2007, 38, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]









© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhang, C.; Chao, N.; Lu, J.; Zhang, Y. Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola. Int. J. Mol. Sci. 2018, 19, 1204. https://doi.org/10.3390/ijms19041204
Liu S, Zhang C, Chao N, Lu J, Zhang Y. Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola. International Journal of Molecular Sciences. 2018; 19(4):1204. https://doi.org/10.3390/ijms19041204
Chicago/Turabian StyleLiu, Shaoli, Chi Zhang, Nan Chao, Jiang Lu, and Yali Zhang. 2018. "Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola" International Journal of Molecular Sciences 19, no. 4: 1204. https://doi.org/10.3390/ijms19041204
APA StyleLiu, S., Zhang, C., Chao, N., Lu, J., & Zhang, Y. (2018). Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola. International Journal of Molecular Sciences, 19(4), 1204. https://doi.org/10.3390/ijms19041204




