Abstract
The administration of Everolimus (EVE), a mTOR inhibitor used in transplantation and cancer, is often associated with adverse effects including pulmonary fibrosis. Although the underlying mechanism is not fully clarified, this condition could be in part caused by epithelial to mesenchymal transition (EMT) of airway cells. To improve our knowledge, primary bronchial epithelial cells (BE63/3) were treated with EVE (5 and 100 nM) for 24 h. EMT markers (α-SMA, vimentin, fibronectin) were measured by RT-PCR. Transepithelial resistance was measured by Millicell-ERS ohmmeter. mRNA and microRNA profiling were performed by Illumina and Agilent kit, respectively. Only high dose EVE increased EMT markers and reduced the transepithelial resistance of BE63/3. Bioinformatics showed 125 de-regulated genes that, according to enrichment analysis, were implicated in collagen synthesis/metabolism. Connective tissue growth factor (CTGF) was one of the higher up-regulated mRNA. Five nM EVE was ineffective on the pro-fibrotic machinery. Additionally, 3 miRNAs resulted hyper-expressed after 100 nM EVE and able to regulate 31 of the genes selected by the transcriptomic analysis (including CTGF). RT-PCR and western blot for MMP12 and CTGF validated high-throughput results. Our results revealed a complex biological network implicated in EVE-related pulmonary fibrosis and underlined new potential disease biomarkers and therapeutic targets.
1. Introduction
Everolimus (EVE), marketed as Certican, is a pharmacological agent widely used in the anti-rejection therapy of solid organ transplantation and in the treatment of certain tumors (e.g., in advanced renal cell carcinoma, subependymal giant cell astrocytoma associated with tuberous sclerosis, pancreatic neuroendocrine tumors, breast cancer) [1]. Similar to Sirolimus and Tamsilorimus, it exerts its immunosuppressive activity by inhibiting mammalian target of rapamycin (mTOR), a phosphoinositide 3-kinase-related protein that controls cell cycle, protein synthesis, angiogenesis and autophagy [2]. These important multi-factorial biological/cellular effects allow this drug to avoid/minimize the onset of acute rejection episodes and to slow down the progression of chronic allograft lesions [3,4].
However, some authors have reported a high rate of discontinuation secondary to side effects after the introduction of this drug [5,6,7]. Among them, pneumonitis or interstitial lung disease with a range of pulmonary histopathologic changes (including alveolar hemorrhage, pulmonary alveolar proteinosis, focal fibrosis, bronchiolitis obliterans organizing pneumonia) have been largely reported in clinical records and they have been associated with worsened patients’ clinical outcomes and drug discontinuation [8,9,10,11,12,13,14,15,16]. The incidence of this complications is 2–11%, frequently reported between 1 and 51 months after the beginning of mTOR inhibitor therapy [17,18,19].
The pathogenic mechanism underlying lung toxicity is multi-factorial and epithelial to mesenchymal transition (EMT) of airway cells seems to have a pivotal role [20,21,22,23]. Our group has recently demonstrated that high doses of EVE are associated with a reprogramming of gene expression in several epithelial cell lines (airway, renal epithelial proximal tubular and hepatic cells) with a consequent loss of their phenotype (junctions and apical-basal polarity) and the acquisition of mesenchymal traits increasing the motility and enabling the development of an invasive and pro-fibrotic phenotype [24,25,26].
High dosage of EVE eliminating negative crosstalk from mTORC1/S6K, leads to activation of mTORC2 that enhances AKT phosphorylation at Ser473 and stimulates PI3K-AKT signaling that induces renal fibrosis [26,27,28,29,30].
The pro-fibrotic attitude of EVE has also been confirmed in vivo in renal transplant patients through the estimation of an arbitrary pulmonary fibrosis index score in renal transplant patients chronically treated with this drug. In this patients’ subset, high blood trough level of EVE was associated with a high rate of pulmonary signs of fibrosis [24].
However, although the aforementioned studies and the large clinical evidences, the complete biological machinery involved in this condition has not been completely clarified.
Therefore, we employed, for the first time, a highthroughput approach combining a transcriptomic with a miRNome analysis to study the capability of EVE to induce pro-fibrotic changes in primary bronchial epithelial cells.
All together our results could represent a step forward in the comprehension of the mTOR-I associated biological machinery and in the identification of new targets for therapeutic interventions.
2. Results
2.1. High Dosage Everolimus (EVE) Induced Epithelial to Mesenchymal Transition (EMT) of BE63/3 (Primary Bronchial Epithelial Cells)
To confirm our previous results obtained in immortalized bronchial and pulmonary cell lines [24], we decided to measure by Real Time-PCR the expression level of alpha smooth muscle actin (α-SMA), vimentin (VIM), and fibronectin (FN) in BE63/3 treated for 24 h with 2 different dosages of EVE (5 and 100 nM) chosen according to literature evidences [31,32,33,34] and previous experiments performed by our research group in different cell lines [24,25,26].
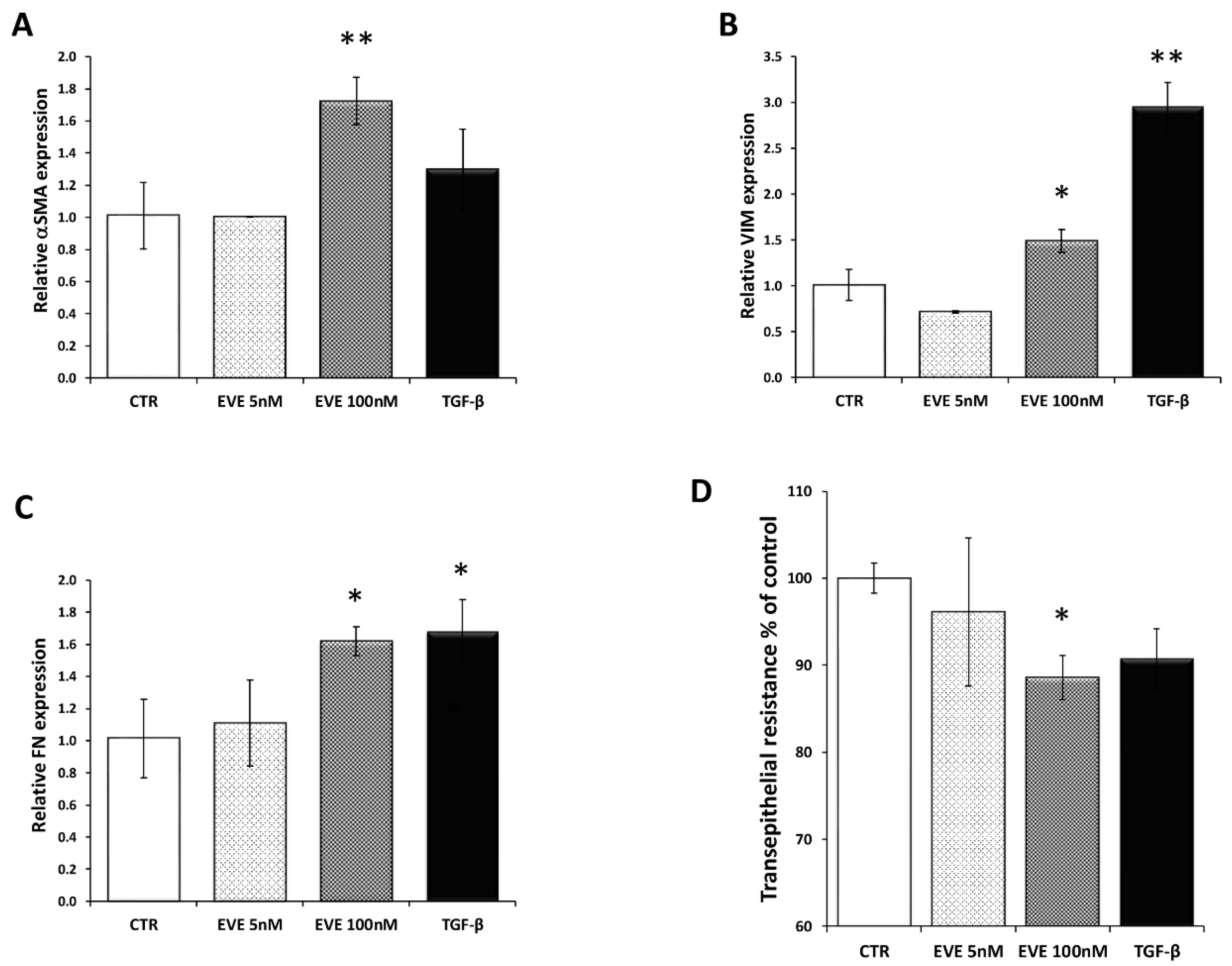
Only high dose of EVE (100 nM), similarly to TGF-β (20 ng/mL), increased the mRNA level of the EMT-related markers (Figure 1A–C). Moreover E-cadherin resulted downregulated although it did not reach a statistically significant level (Figure S1). Contrarily, 5 nM EVE was ineffective (Figure 1A–C).
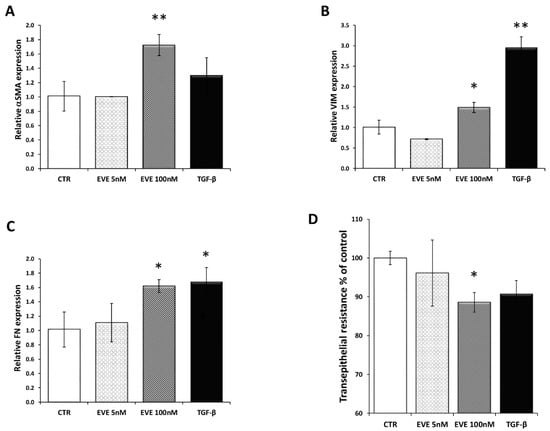
Figure 1.
Gene expression of epithelial to mesenchymal transition (EMT) related markers. Relative (A) alpha smooth muscle actin (α-SMA), (B) fibronectin (FN) and (C) vimentin (VIM) expression evaluated by Real-time PCR in BE 63/3 cells treated or untreated with Everolimus (EVE) (5 and 100 nM) or TGF-β (20 ng/mL); expression values were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Mean ± S.D. (error bars) of three separate experiments performed in triplicate. * p < 0.05, ** p < 0.01 vs. control (CTR). (D) Histogram represents transepithelial resistance as percentage change with respect to control cells. * p < 0.05 vs. CTR.
Additionally, high dosage of EVE was also able to reduce the transepithelial resistance (TER) evaluated by a Millicell-ERS ohmmeter indicating dysfunctional tight junctions (Figure 1D).
2.2. Transcriptomic Analysis Revealed That High Dosage of EVE Up-Regulated Genes Involved in Collagen Synthesis and Metabolism
Gene expression profiling evaluated by transcriptomic analysis revealed that in vitro treatment of BE63/3 cells with 100 nM EVE for 24 h deregulated 147 probe sets (corresponding to 125 genes): 60/147 probe sets (47 genes) resulted up-regulated while 87/147 probe sets (corresponding to 78 genes) were down-regulated (≥1.5-fold change) in EVE-treated cells compared with control (CTR) (Table 1). According to enrichment analysis, selected genes belonged to 44 pathways (Table 2) and 5 of them were involved in collagen synthesis/metabolism and regulation of stress fiber assembly. Interestingly, connective tissue growth factor (CTGF) was a representative gene in all these pro-fibrotic pathways.

Table 1.
List of the differentially expressed probe sets after treatment with 100 nM EVE.

Table 2.
List of pathways differentially regulated after 100 nM EVE.
Instead, low dosage EVE (5 nM) was able to change the expression level of only 33 probe sets (24 genes): 25/33 probe sets (20 genes) were hyper-expressed and 4 probe sets (4 genes) down-regulated after treatment (Table 3). None of the selected pathways was associated with the pro-fibrotic cellular machinery (Table 4).

Table 3.
List of probe sets differentially expressed after treatment with 5 nM EVE.

Table 4.
List of pathways differentially regulated after treatment with 5 nM EVE.
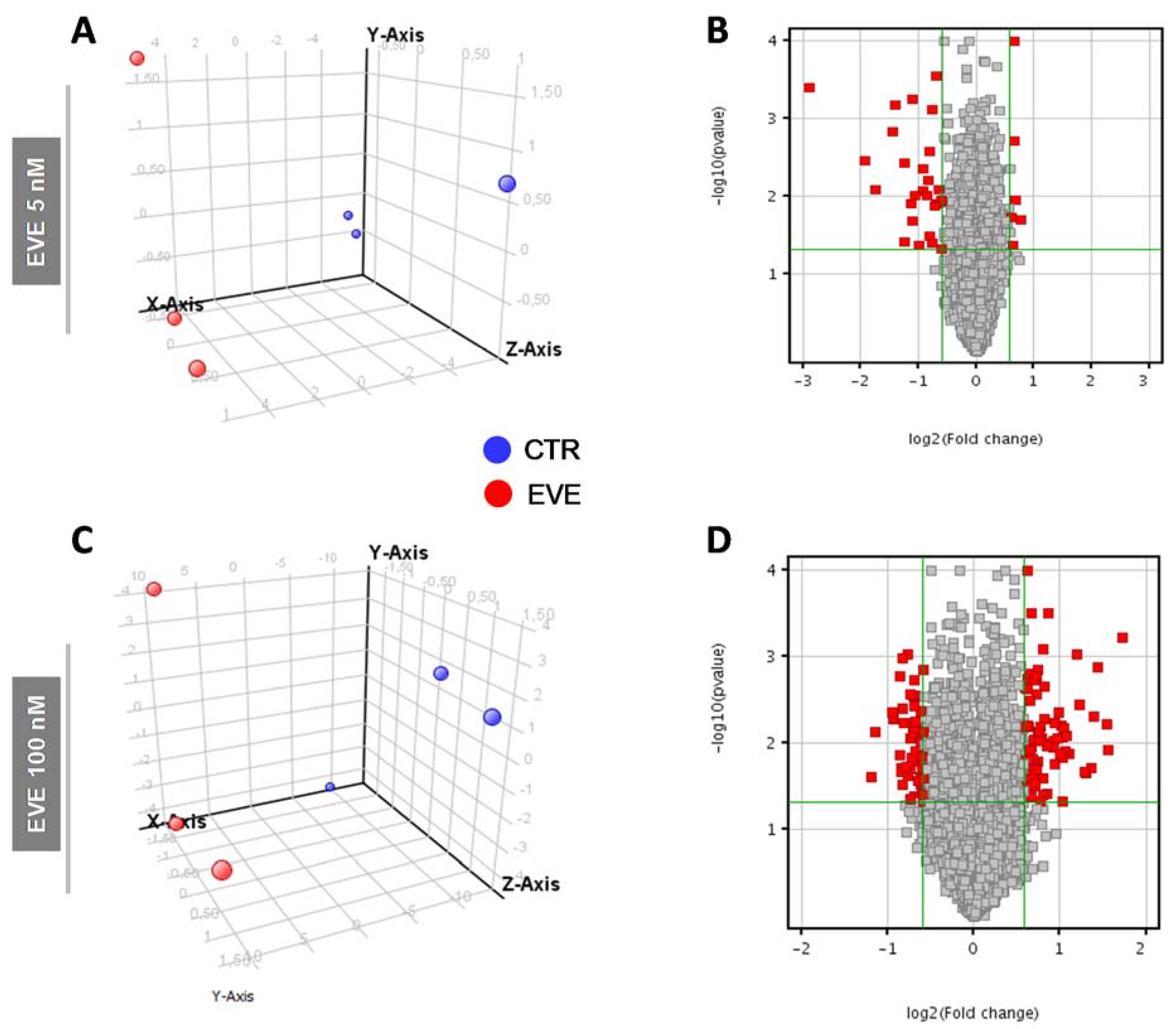
Principal component analysis (PCA) and volcano plot showed the degree of separation of untreated versus treated cells at both EVE dosages (Figure 2).
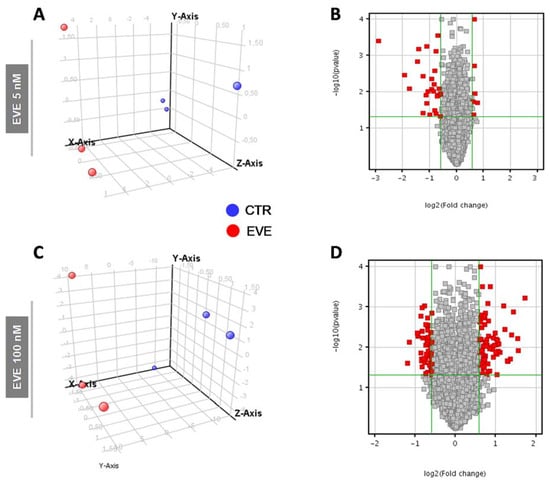
Figure 2.
Principal Component Analysis (PCA) and Volcano Plot discriminating BE63/3 CTR from EVE treated cells. PCA plots were built using the expression level of all differentially expressed genes obtained from mRNA expression profiling after treatment with (A) 5 nM and (C) 100 nM EVE. Volcano Plot based on fold change (Log2) and p value (−Log10) of all genes identified in BE63/3 after treatment with (B) 5 nM and (D) 100 nM EVE. In both graphs red circles indicate the genes that showed statistically significant change.
2.3. MiRNome Analysis Identified Specific MicroRNAs Deregulated by EVE
To gain insights into the mechanism leading to EMT induced by EVE and to discover possible regulatory miRNAs of this effect, we performed a miRNome analysis by miRNA Complete Labeling and Hybridization kit. Statistical analysis identified three miRNAs up-regulated after high dosage (100 nM) (Table 5) and four after treatment with EVE at low dosage (5 nM) (Table 6). Among these, miR-8485 was the most up-regulated miRNA (more than 4-fold changes in both treatments).

Table 5.
List of microRNAs differentially regulated after treatment with 100 nM EVE.

Table 6.
List of microRNAs differentially regulated after treatment with 5 nM EVE.
By matching mRNA and miRNA expression data, we found that 31 genes were specific target of the three identified miRNAs (Table 7).

Table 7.
miRNA/mRNA pairs matched on the basis of mRNA and miRNA profiling results.
2.4. Gene Expression and Protein Analysis for Matrix Metalloproteinase 12 (MMP12) and Connective Tissue Growth Factor (CTGF) Validated High-Throughput Results
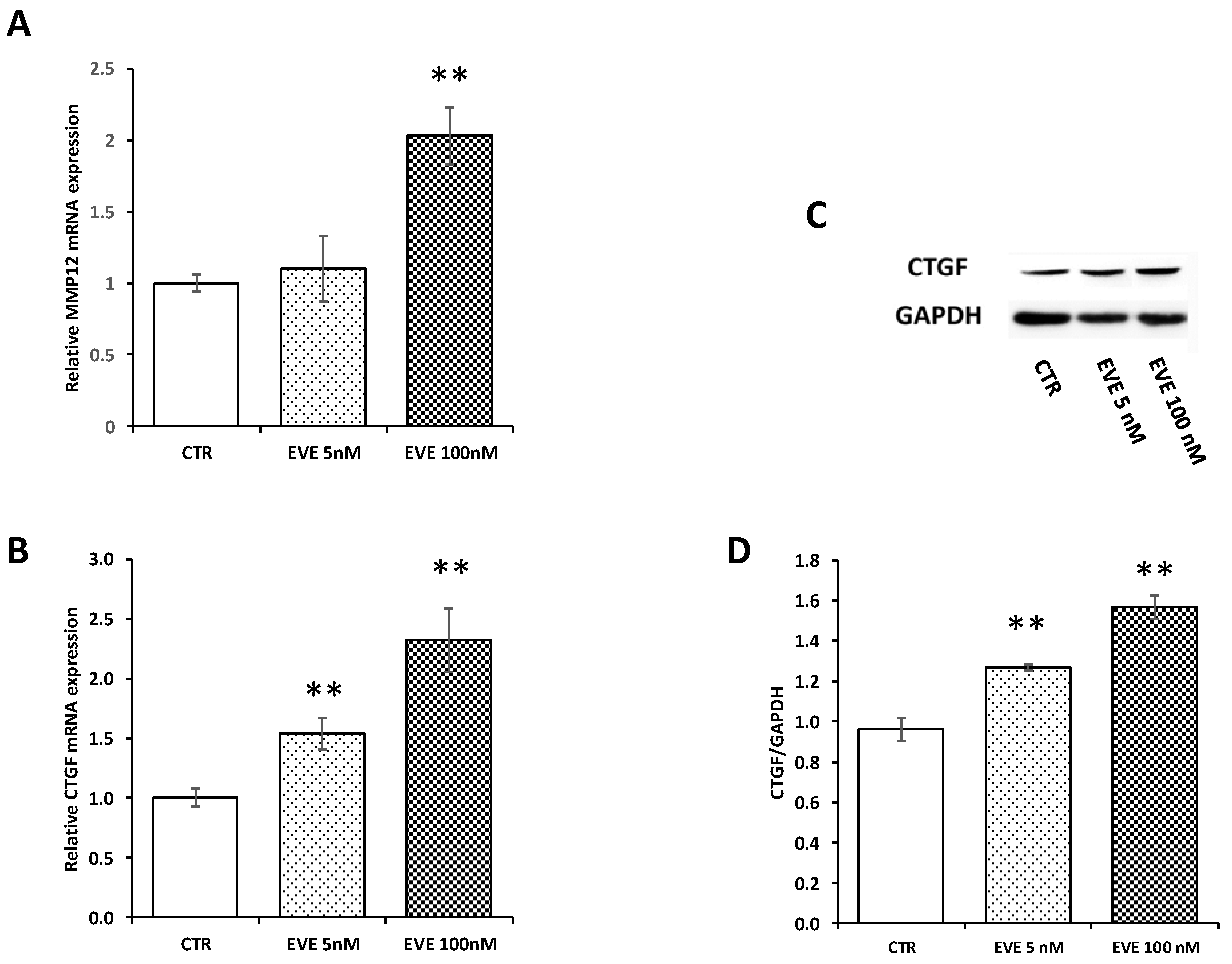
In order to validate microarray results, we measured by Real-Time PCR the level of mRNA expression of MMP12 and CTGF. Both transcripts were up-regulated after treatment with 100 nM EVE. Contrarily 5 nM EVE had no effect (Figure 3A,B). In addition, western blot analysis of CTGF confirmed gene expression results at protein level (Figure 3C,D).
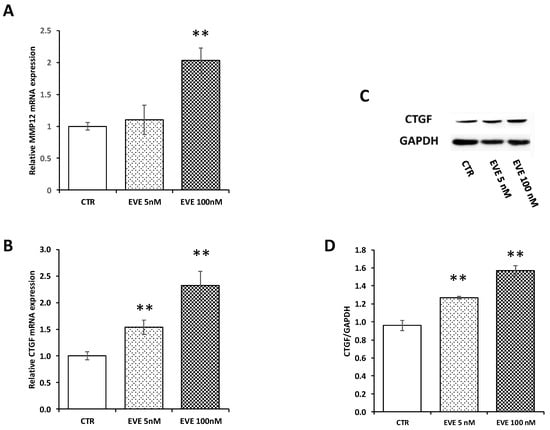
Figure 3.
Gene expression of MMP12 and connective tissue growth factor (CTGF). mRNA level of (A) MMP12 and (B) CTGF evaluated by real-time PCR in BE63/3 cells treated or not with EVE (5 and 100 nM). Data were normalized to GAPDH expression. Mean ± SD (error bars) of two separate experiments performed in triplicate. ** p < 0.001, * p < 0.05 vs. CTR. (C) Representative western blotting experiments for CTGF. (D) Histogram represents the mean ± SD of CTGF protein level. GAPDH was included as loading control. ** p < 0.001 vs. CTR.
2.5. Validation of Transcriptomic Results in an Additional Primary Cell Line (BE121/3)
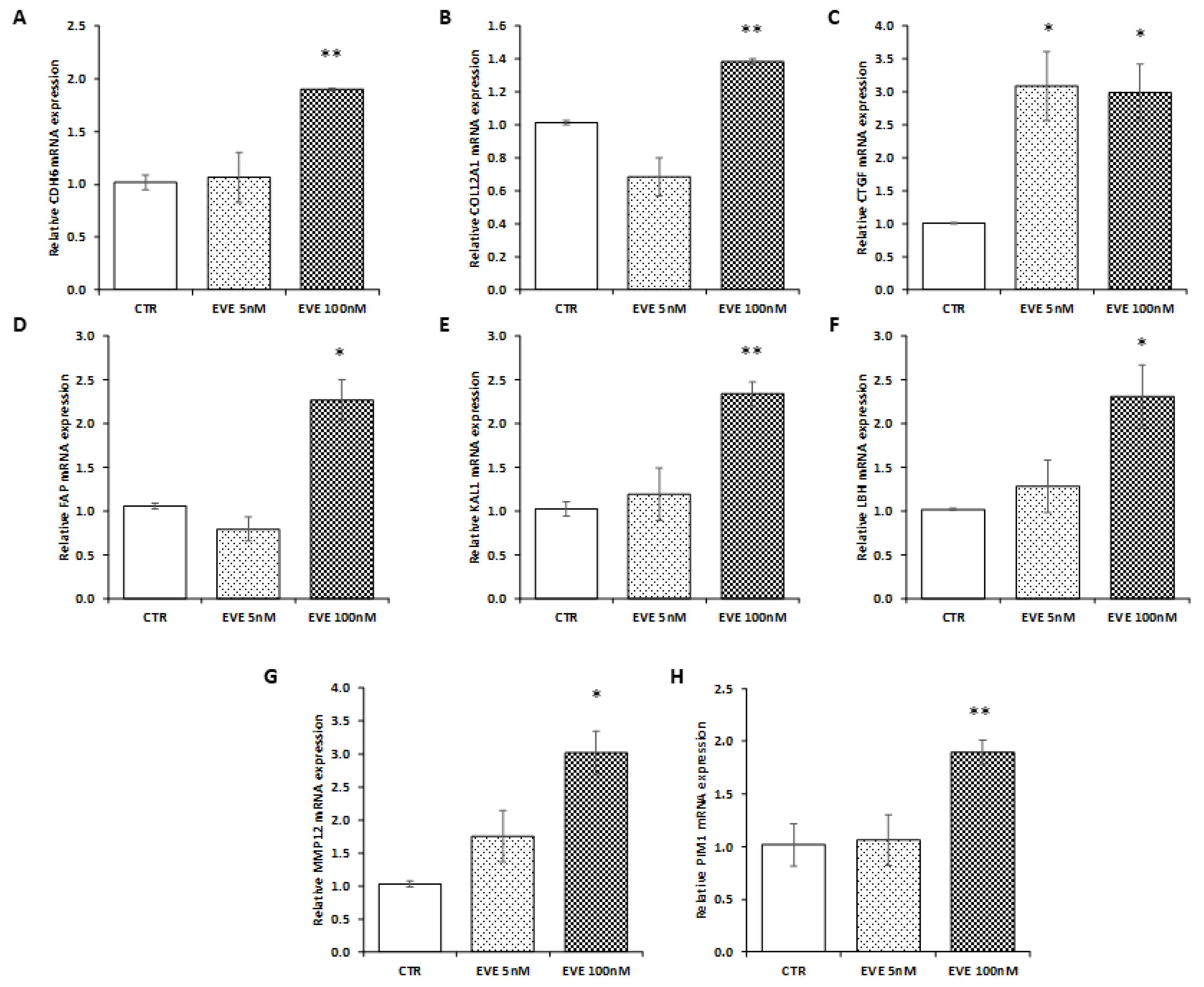
To confirm transcriptomic results, we decided to measure the expression level of 8 selected genes (involved in EMT) up-regulated after high dosage EVE in a new primary bronchial epithelial cell line. As showed in Figure 4, results were in line with those obtained in BE63/3 (Figure 4).
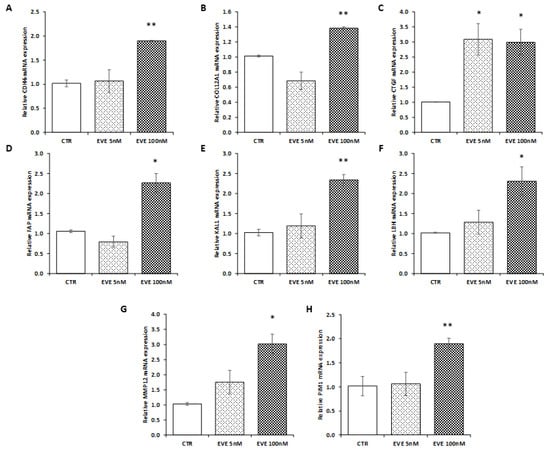
Figure 4.
Gene expression in BE121/3. mRNA level of (A) CDH6, (B) COL12A1, (C) CTGF, (D) FAP, (E) KAL1, (F) LBH, (G) MMP12, (H) PIM1 evaluated by real-time PCR in BE121/3 cells treated or not with EVE (5 and 100 nM). Data were normalized to GAPDH expression. Mean ± SD (error bars) of two separate experiments performed in triplicate. ** p < 0.001, * p < 0.05 vs. CTR.
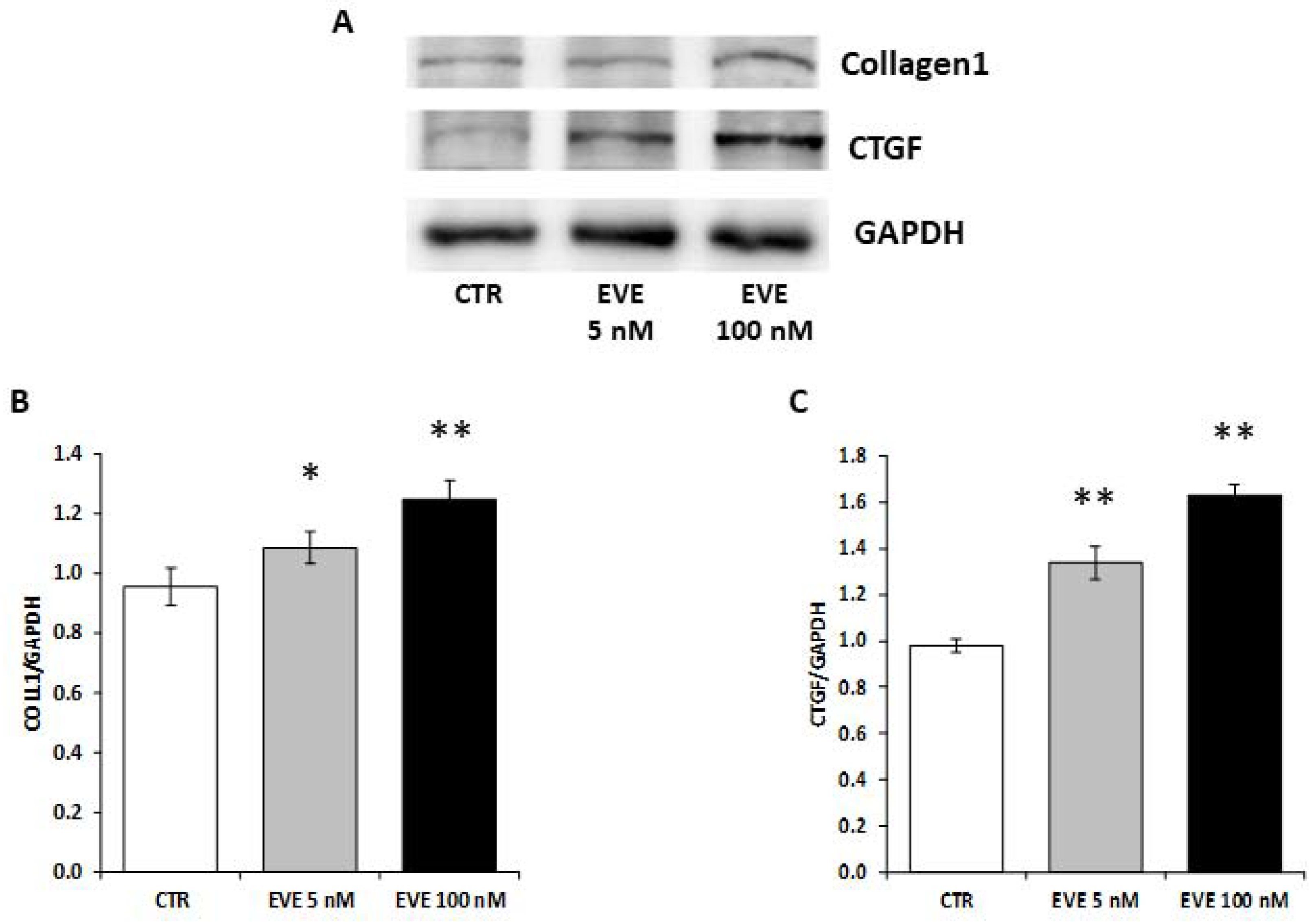
2.6. High Dosage EVE Up-Regulated CTGF and Collagen1 in Fibroblasts and Hepatic Stellate Cells
To validate the pro-fibrotic effect of high dosage EVE we measured the expression level of collagen1 and CTGF in NIH/3T3 (mouse embryo fibroblast cell line) treated with EVE.
Interestingly, also in fibroblasts high dosage EVE up-regulated the protein levels of collagen1 and CTGF (Figure 5).
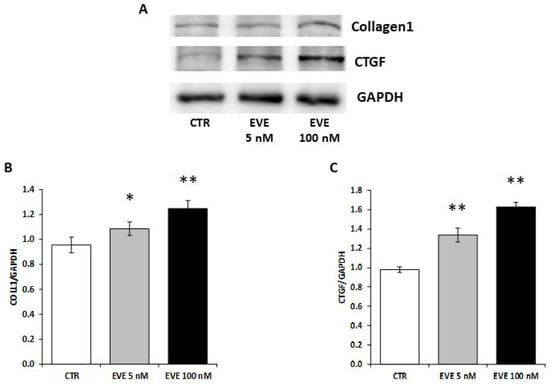
Figure 5.
Protein levels of collagen1 and CTGF in NIH/3T3 cells. (A) Representative western blotting experiments for collagen1 and CTGF. Histograms represent the mean ± SD of (B) collagen1 and (C) CTGF protein levels. GAPDH was included as loading control. ** p < 0.001, * p < 0.05 vs. CTR.
Also, in hepatic stellate cells high dosage EVE induced the up-regulation of CTGF and collagen1 (Figure S2).
3. Discussion
Pulmonary fibrosis is a potential serious adverse effect following administration of mTOR-I in patients undergoing solid organ transplantation or receiving anti-cancer therapies. It is generally accepted that pulmonary disease is related to mTOR-I therapy, whether the following conditions are present: (1). The symptoms of pulmonary disease occur after initiation of mTOR-I therapy; (2). Infection, other pulmonary diseases or toxicity associated with other drugs are excluded; (3). mTOR-I minimization or discontinuation lead to resolution of the symptoms. In fact, the dose-dependent effect was proved by the observation of this disease particularly in patients receiving high doses of mTOR-I.
Pulmonary manifestations in these patients are numerous and include several clinical/histological phenotypes (e.g., focal pulmonary fibrosis, bronchiolitis obliterans with organizing pneumonia) [8,9,35,36].
This multi-factorial and heterogeneous clinical condition is often responsible for drug discontinuation and it requires long and expensive clinical evaluations and treatments (e.g., antibiotics, corticosteroids, immunosuppressive drugs) [14] with the involvement of a multidisciplinary team of experts (e.g., pulmonologists, infectivologists, nephrologists).
The etiopathogenic mechanism of pulmonary toxicity associated with mTOR-I therapy is not known and several in vivo and in vitro studies have tried to define the underlying mechanisms. It has been proposed a T cell-mediated autoimmune response induced when pulmonary cryptic antigens are exposed, leading to lymphocytic alveolitis and interstitial pneumonitis [15]. Other possible pathogenic mechanisms could be a delayed-type hypersensitivity reaction [9] or pulmonary inflammation as a direct effect of mTOR-I to stimulate cells of the innate immune system to produce proinflammatory cytokines [37,38].
Additionally, Ussavarungsi et al. have reported that sirolimus may induce granulomatous interstitial inflammation and proposed a mechanism of T-cell mediated hypersensitivity reaction triggered by circulating antigens or immune complexes in the lungs [39].
Moreover, several authors have emphasized the pathogenetic role of the EMT of bronchial epithelial cells in these important Everolimus (EVE)-related adverse events [20,21,22,23].
To obtain more insights, we decided to employ, for the first time, innovative high throughput technologies, to identify new elements involved in the biological/cellular reprogramming induced by high dose of mTOR-I and leading to fibrosis.
In vitro experiments using classical bio-molecular strategies, confirmed, in primary bronchial epithelial cell lines, our previous results demonstrating the ability of high dosages EVE to induce EMT. In particular, 100 nM EVE caused the up-regulation of EMT-related genes (α-SMA, VIM, FN) and reduced the trans-epithelial resistance to the same levels induced by TGF-β. Then, high doses of this drug significantly changed the expression level of 125 genes (47 up- and 78 down-regulated).
Several of the selected genes were target of miR-8485, the top significant and up-regulated microRNA (miRNA) by EVE 100 nM. Other 2 miRNAs were identified after the same treatment: miR-937-5p and miR-5194. Except for miR-8485, at our knowledge, none of them has been previously associated with fibrosis or supposed to be regulatory of genes implicated in this process. It’s unquestionable that further studies are warranted to confirm the involvement of these miRNAs in EVE induced EMT since all identified miRNAs were up-regulated demonstrating their possible role as enhancer of fibrotic machinery. This could be in line with recent findings suggesting that miRNA-mediated down-regulation is not a one-way process and some miRNAs could up-regulate gene expression in specific cell types and conditions with distinct transcripts and proteins [40,41]. It is noteworthy that these miRNAs are up-regulated also after treatment with 5 nM EVE. Many reasons could be responsible of this effect. In particular, the expression of these miRNAs could be regulated by several factors and networks (some of them also unrelated to mTOR-I treatment). Additional studies are needed to clarify the role of miRNA in EVE-mediated pro-fibrotic effect.
Moreover, analyzing the results of the transcriptomic analysis and the hypothetic targets of miR-8485, we found that connective tissue growth factor (CTGF), a protein secreted into the extracellular environment where it interacts with distinct cell surface receptors, growth factors and extra-cellular matrix [42,43] was one of the top scored genes. Gene expression by RT-PCR and protein analysis by western blotting confirmed the result obtained by microarray.
It is well known that CTGF modulates the activities of TGF-β or vascular endothelial growth factor (VEGF), with consequent pro-fibrotic and angiogenetic effects [44,45,46,47]. However, the overexpression of CTGF in fibroblast of mice caused tissue fibrosis in vivo [48] without involving the canonical TGF-β pathway. This is in line with several reports that demonstrated a mTOR-I dose-related induction of CTGF at gene and protein levels in vitro and in vivo [49,50,51,52].
Moreover, Xu et al. have demonstrated that rapamycin, an analogue of EVE, exerted a profibrotic effect in lung epithelial cells as well as in lung fibroblasts via up-regulation of CTGF expression and PI3K/AKT pathway [50,51]. Similarly, Mikaelian et al. using a combination of RNAi and pharmacological approaches showed that inhibition of mTOR triggers EMT in mammalian epithelial cells by a mechanism TGF-β independent [53]. In the transplant context it has been described a synergistic fibrotic effect of sirolimus with cyclosporine in kidney also mediated by the up-regulation of CTGF [54,55].
Another interested gene up-regulated by EVE, selected by microarray and validated by RT-PCR, was metalloproteinase 12 (MMP12), a member of the zinc-dependent endopeptidases family able to proteolyze all components of the extracellular matrix [56,57] by degrading collagen, other extracellular filaments, cytokines, growth factors and their receptors. MMP12 has a pivotal role in TGF-β mediated pulmonary fibrosis [58,59].
Interestingly, other identified genes by transcriptomic analysis and target of miR-8485 (Table 7) were Kallmann syndrome-1 gene (KAL1, fold change: 1.705), Limb-bud and heart (LBH, fold change: 1.808) and insulin receptor substrates 2 (IRS2, fold change: 1.646) that resulted up-regulated after 100 nM EVE treatment and Protocadherin 7 (PCDH7, fold change: −1.625) down-regulated by similar treatment. All of them have been described in literature as directly or indirectly involved in the EMT.
KAL1, codes for anosmin-1, a cell adhesion protein in extracellular matrix induced by TGF-β [60,61]. IRS2 expression appears to repress the expression of E-cadherin [62], marker of epithelial cells deregulated during EMT.
LBH is a transcription cofactor with both transcriptional activator and corepressor functions. LBH is a direct Wnt/β-catenin target gene and is induced by TGF-β [63,64]. Wnt/β-catenin signaling activation occurs in cells during EMT [65] and treated with mTOR-I.
Protocadherin 7 is an integral membrane protein having a role in cell–cell recognition and adhesion. Down-regulation of PCDH7 gene was correlated with E-cadherin inhibition [66].
All these findings, although speculatively interesting, need to be validated in vivo. Our study is an hypothesis generating study that should be considered a starting point for bio-molecular study involving transplanted patients or animal models.
Nevertheless, after 21 days in culture, most of the cells were not ciliated and we cannot exclude that differentiation state may have affected the response to EVE (Figure S3).
However, our results suggested that high concentrations of EVE, through the activation of a multi-factorial biological/cellular machinery, may lead to pulmonary fibrosis and underlined potential pathogenetic, diagnostic biomarkers and targets for future pharmacological interventions to introduce in the “day by day” clinical practice. Finally, at a clinical point of view, we confirm that, whenever possible, the dose of EVE should be the minimized in patients with early signs of lung toxicity.
4. Materials and Methods
4.1. Cell Culture Treatment
Primary wild-type bronchial epithelial cells (BE63/3 and BE121/3) were obtained from “Servizio Colture Primarie” of the Italian Cystic Fibrosis Research Foundation (ICFRF) and cultured following the supplier instructions [67]. The protocols to isolate, culture, store, and study bronchial epithelial cells from patients undergoing lung transplant was approved by the Ethical Committee of Gaslini Institute (ethical approval number IGG:192 date of approval: 9/24/2010) under the supervision of the Italian Ministry of Health. Cells were grown on rat tail collagen-coated tissue culture plates in serum-free LHC9/RPMI 1640 medium at 37 °C and 5% CO2.
After 4–5 passages, cells were seeded on Transwell porous inserts. After 24 h from seeding, the medium was switched to DMEM/F12 supplemented with 2% Ultroser G, 2 mM l-glutammine, 100 U/mL penicillin, 100 μg/mL streptomycin.
Exchange of culture medium is repeated every day on both sides of permeable supports up to 5 days. Then the apical culture medium was removed, and the medium was added only in the basolateral side (air-liquid interface) favoring a differentiation of the epithelium (Figure S3). After 11 days the epithelium was treated with EVE (5 nM and 100 nM) and TGF-β (20 ng/mL), an EMT inducer, for 24 h. “The timing of cell culture for gene expression and western blot experiments (17 days) was based on clear instructions supplied by the “Servizio Colture Primarie” of the ICFRF in order to reach the differentiation of epithelium”. Although the in vitro model cannot completely represent the in vivo pharmacokinetic/effect of this drug, we can postulate that 5 nM EVE corresponds to a trough level of approximately 5 ng/mL (drug level frequently reached in the immunosuppressive maintenance therapy of solid organ transplantation), while 100 nM may correspond to very high dosages (trough level more than 50 ng/mL) that patients could reach in anticancer therapy.
NIH/3T3 fibroblasts, purchased from American Type Culture Collection (Manassas, VA, USA) were maintained at 37 °C in DMEM supplemented with 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. Cells were treated with or without 5 and 100 nM Everolimus for 24 h.
4.2. RNA Extraction and Gene Expression Profiling
Trizol reagent (Invitrogen) was used to extract total RNA and then, yield and purity were checked using a Nanodrop spectrophotometer.
Gene expression data were produced using the HumanHT-12 v3 Expression BeadChip (Release 38, Illumina, San Diego, CA, USA). Five hundred ng total RNA from BE63/3 was used to synthesize biotin-labeled cRNA using the Illumina®TotalPrep™ RNA amplification kit (Applied Biosystems, Foster City, CA, USA). Quality of labelled cRNA was assessed by NanoDrop® ND-100 spectrophotometer and the Agilent 2100 Bioanalyzer. Then, 750 ng biotinylated cRNA was used for hybridization to illumina microarrays that were then scanned with the HiScanSQ.
4.3. Pathway Analysis
The Ingenuity Pathway Analysis software (IPA, Ingenuity System, Redwood City, CA, USA) was used to assess biological relationships among differentially regulated genes. The reference gene selection was performed by own software written in Java program language. The canonical pathways generated by IPA are the most significant for the uploaded data set. Fischer’s exact test with false discovery rate (FDR) option was used to calculate the significance of the canonical pathway.
4.4. MicroRNA Expression Profiling
Fluorescently-labeled miRNAs were generated using the miRNA Complete Labeling and Hybridization kit (Agilent Technologies, Santa Clara, CA, USA), with a sample input of 100 ng of total RNA from BE63/3 and hybridized for 20 h at 55 °C on the Agilent 8 × 60 K Human miRNA Microarray slide (Agilent Technologies), based on miRBase database (Release 21.0). Following hybridization, the slides were washed and scanned using the High-Resolution Microarray C Scanner (Agilent Technologies). The image files were processed using the Agilent Feature Extraction software (v10.7.3): the microarray grid was correctly placed; inlier pixels were identified, and outlier pixels were rejected.
4.5. Real-Time PCR
Five hundred ng total RNA from each sample was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR amplification reactions were performed in duplicate via SYBR Green chemistry on CFX-connect (Bio-Rad, Hercules, CA, USA) and SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad). Primers for α-SMA, VIM, FN, MMP12, CTGF, CDH6, COL12A1, FAP, KAL1, LBH, PIM1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were obtained from Qiagen (QuantiTect Primer Assay, Hilden, Germany).
The comparative Ct method (ΔΔCt) was used to quantify gene expression and the relative quantification was calculated as 2−ΔΔCt. Melting curve analysis was employed to exclude non-specific amplification products.
4.6. Western Blot
Equal amounts of proteins were resolved in 10% SDS-PAGE and electrotransferred to nitrocellulose membranes. Non-specific binding was blocked for 1 h at room temperature with non-fat milk (5%) in TBST buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20). Membranes were exposed to primary antibodies directed against GAPDH (Santa Cruz sc-25778), CTGF (NovusBio, Littleton, CO, USA) and collagen1 (ORIGENE TA309096) (overnight at 4 °C) and incubated with a secondary peroxidase-conjugated antibody for 1 h at room temperature. The signal was detected with SuperSignals West Pico Chemiluminescent substrate solution (Pierce) according to the manufacturer’s instructions.
4.7. Transepithelial Resistance (TER)
Millicell-ERS ohmmeter with electrodes (Millipore) was used to measure TER (alternating current applied between the electrodes: ±20 μA and frequency: 12.5 Hz). The resistance of the monolayer multiplied by the effective surface area was used to obtain the electrical resistance of the monolayer (Ω cm2). Once stable resistances were obtained, different culture media (control, EVE 5 nM, EVE 100 nM, TGF-β 20 ng/mL) were tested. After the addition of test solutions, measurements were taken at 24 h.
4.8. Statistical Analysis
For transcriptomics statistical analyses were carried out by Genespring GX 11.0 software (Agilent Technologies). Gene probe sets were filtered based on the FDR method of Benjamini–Hochberg and fold-change. Only genes that were significantly (adjusted-p value < 0.05 and fold-change > 1.5) modulated were considered for further analysis.
In the miRNome analysis, after normalization (Quantile method), unpaired t-test (p-value cut-off: 0.05 and fold-change cut-off: 2.0, after Benjamini–Hochberg multiple testing correction) was employed to identify most differentially expressed probes.
For the statistical analysis of RT-PCR and western-blot, differences between control and treated cell were compared using Student’s t-test. A p-value < 0.05 was set as statistically significant.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/4/1250/s1.
Acknowledgments
This study was funded by grants from the Italian Cystic Fibrosis (CF) Research Foundation (FFC#28/2014, Delegazione FFC di Torino, Lodi/Latina, Italy) and from the Fondazione Cariverona 2015. This study was performed in the LURM (Laboratorio Universitario di Ricerca Medica) Research Center, University of Verona, Verona, Italy.
Author Contributions
Gianluigi Zaza, Simona Granata, Valentina Masola conceived and designed the experiments; Simona Granata, Valentina Masola, Gloria Santoro, Nadia Antonucci, Fabio Sallustio, Paola Pontrelli, Matteo Accetturo, Paola Tomei performed the experiments; Gianluigi Zaza, Simona Granata, Antonio Lupo, Pierluigi Carratù analyzed the data; Gianluigi Zaza and Simona Granata wrote the manuscript. All co-authors revised and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fasolo, A.; Sessa, C. Targeting mTOR pathways in human malignancies. Curr. Pharm. Des. 2012, 18, 2766–2777. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.; Hartmann, E.; Cibrik, D.; Cooper, M.; Shaw, L.M. Optimal everolimus concentration is associated with risk reduction for acute rejection in de novo renal transplant recipients. Transplantation 2010, 90, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, J.; Citterio, F.; Favi, E.; Salerno, M.P.; Tondolo, V.; Spagnoletti, G.; Renna, R.; Castagneto, M. Higher incidence of acute rejection in renal transplant recipients with low everolimus exposure. Transplant. Proc. 2007, 39, 1823–1826. [Google Scholar] [CrossRef] [PubMed]
- Zaza, G.; Tomei, P.; Ria, P.; Granata, S.; Boschiero, L.; Lupo, A. Systemic and nonrenal adverse effects occurring in renal transplant patients treated with mTOR inhibitors. Clin. Dev. Immunol. 2013, 2013, 403280. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Qazi, Y.; Wellen, J.R. Strategies for the management of adverse events associated with mTOR inhibitors. Transplant. Rev. 2014, 28, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Engelen, M.A.; Welp, H.A.; Gunia, S.; Amler, S.; Klarner, M.P.; Dell’aquila, A.M.; Stypmann, J. Prospective study of everolimus with calcineurin inhibitor-free immunosuppression after heart transplantation: Results at four years. Ann. Thorac. Surg. 2014, 97, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Champion, L.; Stern, M.; Israël-Biet, D.; Mamzer-Bruneel, M.-F.; Peraldi, M.-N.; Kreis, H.; Porcher, R.; Morelon, E. Sirolimus-associated pneumonitis: 24 cases in renal transplant recipients. Ann. Intern. Med. 2006, 144, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.T.; Pham, P.C.; Danovitch, G.M.; Ross, D.J.; Gritsch, H.A.; Kendrick, E.A.; Singer, J.; Shah, T.; Wilkinson, A.H. Sirolimus-associated pulmonary toxicity. Transplantation 2004, 77, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Weiner, S.M.; Sellin, L.; Vonend, O.; Schenker, P.; Buchner, N.J.; Flecken, M.; Viebahn, R.; Rump, L.C. Pneumonitis associated with sirolimus: Clinical characteristics, risk factors and outcome—A single-centre experience and review of the literature. Nephrol. Dial. Transplant. 2007, 22, 3631–3637. [Google Scholar] [CrossRef] [PubMed]
- West, M.L. Bronchiolitis obliterans and organizing pneumonia in renal transplant recipients. Transplantation 2000, 69, 1531. [Google Scholar] [CrossRef]
- Feagans, J.; Victor, D.; Moehlen, M.; Florman, S.S.; Regenstein, F.; Balart, L.A.; Joshi, S.; Killackey, M.T.; Slakey, D.P.; Paramesh, A.S. Interstitial pneumonitis in the transplant patient: Consider sirolimus-associated pulmonary toxicity. J. La. State Med. Soc. 2009, 161, 166–172. [Google Scholar] [PubMed]
- Molas-Ferrer, G.; Soy-Muner, D.; Anglada-Martínez, H.; Riu-Viladoms, G.; Estefanell-Tejero, A.; Ribas-Sala, J. Interstitial pneumonitis as an adverse reaction to mTOR inhibitors. Nefrologia 2013, 33, 297–300. [Google Scholar] [PubMed]
- Lopez, P.; Kohler, S.; Dimri, S. Interstitial lung disease associated with mTOR inhibitors in solid organ transplant recipients: Results from a large phase III clinical trial program of everolimus and review of the literature. J. Transplant. 2014, 2014, 305931. [Google Scholar] [CrossRef] [PubMed]
- Morelon, E.; Stern, M.; Israël-Biet, D.; Correas, J.M.; Danel, C.; Mamzer-Bruneel, M.F.; Peraldi, M.N.; Kreis, H. Characteristics of sirolimus-associated interstitial pneumonitis in renal transplant patients. Transplantation 2001, 72, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Hasni, K.; Slusher, J.; Siddiqui, W.; Matsumura, D.; Malek, B.; Heifets, M.; Ahmed, Z. Bronchiolitis obliterans organizing pneumonia in renal transplant patients. Dial. Transplant. 2010, 39, 449–451. [Google Scholar] [CrossRef]
- Errasti, P.; Izquierdo, D.; Martín, P.; Errasti, M.; Slon, F.; Romero, A.; Lavilla, F.J. Pneumonitis associated with mammalian target of rapamycin inhibitors in renal transplant recipients: A single-center experience. Transplant. Proc. 2010, 42, 3053–3054. [Google Scholar] [CrossRef] [PubMed]
- Alexandru, S.; Ortiz, A.; Baldovi, S.; Milicua, J.M.; Ruíz-Escribano, E.; Egido, J.; Plaza, J.J. Severe everolimus-associated pneumonitis in a renal transplant recipient. Nephrol. Dial. Transplant. 2008, 23, 3353–3355. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Moreno, A.; Ridao, N.; García-Ledesma, P.; Calvo, N.; Pérez-Flores, I.; Marques, M.; Barrientos, A.; Sánchez-Fructuoso, A.I. Sirolimus and everolimus induced pneumonitis in adult renal allograft recipients: Experience in a center. Transplant. Proc. 2009, 41, 2163–2165. [Google Scholar] [CrossRef] [PubMed]
- Kage, H.; Borok, Z. EMT and interstitial lung disease: A mysterious relationship. Curr. Opin. Pulm. Med. 2012, 18, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, J.C.; Thannickal, V.J. Epithelial-mesenchymal interactions in pulmonary fibrosis. Semin. Respir. Crit. Care Med. 2006, 27, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Strieter, R.M.; Mehrad, B. New mechanisms of pulmonary fibrosis. Chest 2009, 136, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Felton, V.M.; Inge, L.J.; Willis, B.C.; Bremner, R.M.; Smith, M.A. Immunosuppression-induced bronchial epithelial-mesenchymal transition: A potential contributor to obliterative bronchiolitis. J. Thorac. Cardiovasc. Surg. 2011, 141, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Tomei, P.; Masola, V.; Granata, S.; Bellin, G.; Carratù, P.; Ficial, M.; Ventura, V.A.; Onisto, M.; Resta, O.; Gambaro, G.; et al. Everolimus-induced epithelial to mesenchymal transition (EMT) in bronchial/pulmonary cells: When the dosage does matter in transplantation. J. Nephrol. 2016, 29, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Carraro, A.; Zaza, G.; Bellin, G.; Montin, U.; Violi, P.; Lupo, A.; Tedeschi, U. Epithelial to mesenchymal transition in the liver field: The double face of Everolimus in vitro. BMC Gastroenterol. 2015, 15, 118. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Zaza, G.; Granata, S.; Gambaro, G.; Onisto, M.; Lupo, A. Everolimus-induced epithelial to mesenchymal transition in immortalized human renal proximal tubular epithelial cells: Key role of heparanase. J. Transl. Med. 2013, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Breuleux, M.; Klopfenstein, M.; Stephan, C.; Doughty, C.A.; Barys, L.; Maira, S.M.; Kwiatkowski, D.; Lane, H.A. Increased AKT S473 phosphorylation after mTORC1 inhibition is rictor dependent and does not predict tumor cell response to PI3 K/mTOR inhibition. Mol. Cancer Ther. 2009, 8, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Harkavy, B.; Shen, N.; Grohar, P.; Helman, L.J. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 2007, 26, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, P.T.; Hay, N. The two TORCs and Akt. Dev. Cell 2007, 12, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Ma, L.; Teruya-Feldstein, J.; Rojo, F.; Salmena, L.; Alimonti, A.; Egia, A.; Sasaki, A.T.; Thomas, G.; Kozma, S.C.; et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J. Clin. Investig. 2008, 118, 3065–3074. [Google Scholar] [CrossRef] [PubMed]
- Witzig, T.E.; Reeder, C.; Han, J.J.; LaPlant, B.; Stenson, M.; Tun, H.W.; Macon, W.; Ansell, S.M.; Habermann, T.M.; Inwards, D.J.; et al. The mTORC1 inhibitor everolimus has antitumor activity in vitro and produces tumor responses in patients with relapsed T-cell lymphoma. Blood 2015, 126, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhong, Y.; Jackson, A.L.; Clark, L.H.; Kilgore, J.; Zhang, L.; Han, J.; Sheng, X.; Gilliam, T.P.; Gehrig, P.A.; et al. Everolimus exhibits anti-tumorigenic activity in obesity-induced ovarian cancer. Oncotarget 2016, 7, 20338–20356. [Google Scholar] [CrossRef] [PubMed]
- Yunokawa, M.; Koizumi, F.; Kitamura, Y.; Katanasaka, Y.; Okamoto, N.; Kodaira, M.; Yonemori, K.; Shimizu, C.; Ando, M.; Masutomi, K.; et al. Efficacy of everolimus, a novel mTOR inhibitor, against basal-like triple-negative breast cancer cells. Cancer Sci. 2012, 103, 1665–1671. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Kubasch, M.L.; Göbel, A.; Hadji, P.; Chen, D.; Rauner, M.; Stölzel, F.; Hofbauer, L.C.; Rachner, T.D. Concurrent antitumor and bone-protective effects of everolimus in osteotropic breast cancer. Breast Cancer Res. 2017, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Vandewiele, B.; Vandecasteele, S.J.; Vanwalleghem, L.; De Vriese, A.S. Diffuse alveolar hemorrhage induced by everolimus. Chest 2010, 137, 456–459. [Google Scholar] [CrossRef] [PubMed]
- Vlahakis, N.E.; Rickman, O.B.; Morgenthaler, T. Sirolimus-associated diffuse alveolar hemorrhage. Mayo Clin. Proc. 2004, 79, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Cravedi, P.; Ruggenenti, P.; Remuzzi, G. Sirolimus for calcineurin inhibitors in organ transplantation: Contra. Kidney Int. 2010, 78, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.; Heit, A.; Dreher, S.; Eisenächer, K.; Mages, J.; Haas, T.; Krug, A.; Janssen, K.P.; Kirschning, C.J.; Wagner, H. Mammalian target of rapamycin (mTOR) orchestrates the defense program of innate immune cells. Eur. J. Immunol. 2008, 38, 2981–2992. [Google Scholar] [CrossRef] [PubMed]
- Ussavarungsi, K.; Elsanjak, A.; Laski, M.; Raj, R.; Nugent, K. Sirolimus induced granulomatous interstitial pneumonitis. Respir. Med. Case Rep. 2012, 7, 8–11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vasudevan, S.; Steitz, J.A. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef] [PubMed]
- Duncan, M.R.; Frazier, K.S.; Abramson, S.; Williams, S.; Klapper, H.; Huang, X.; Grotendorst, G.R. Connective tissue growth factor mediates transforming growth factor β-induced collagen synthesis: Down-regulation by cAMP. FASEB J. 1999, 13, 1774–1786. [Google Scholar] [CrossRef] [PubMed]
- Cicha, I.; Goppelt-Struebe, M. Connective tissue growth factor: Context-dependent functions and mechanisms of regulation. Biofactors 2009, 35, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.H.; Yamauchi, K.; Uzuki, M.; Nakanishi, T.; Takigawa, M.; Inoue, H.; Sawai, T. Type II alveolar epithelial cells and interstitial fibroblasts express connective tissue growth factor in IPF. Eur. Respir. J. 2001, 17, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Lipson, K.E.; Wong, C.; Teng, Y.; Spong, S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenes. Tissue Repair 2012, 5, S24. [Google Scholar] [CrossRef] [PubMed]
- Grotendorst, G.R. Connective tissue growth factor: A mediator of TGF-beta action on fibroblasts. Cytokine Growth Factor Rev. 1997, 8, 171–179. [Google Scholar] [CrossRef]
- Nishida, T.; Kondo, S.; Maeda, A.; Kubota, S.; Lyons, K.M.; Takigawa, M. CCN family 2/connective tissue growth factor (CCN2/CTGF) regulates the expression of Vegf through Hif-1α expression in a chondrocytic cell line, HCS-2/8, under hypoxic condition. Bone 2009, 44, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Sonnylal, S.; Shi-Wen, X.; Leoni, P.; Naff, K.; van Pelt, C.S.; Nakamura, H.; Leask, A.; Abraham, D.; Bou-Gharios, G.; de Crombrugghe, B. Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheumatol. 2010, 62, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Balah, A.; Ezzate, O. The mTOR inhibitor rapamycin induces CTGF and TIMP-1 expression in rat kidney: Implication of TGF-β/SMAD signaling cascade. Eur. J. Pharm. Med. Res. 2017, 4, 49–56. [Google Scholar]
- Xu, X.; Dai, H.; Geng, J.; Wan, X.; Huang, X.; Li, F.; Jiang, D.; Wang, C. Rapamycin increases CCN2 expression of lung fibroblasts via phosphoinositide 3-kinase. Lab. Investig. 2015, 95, 846–859. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, X.; Wan, X.; Geng, J.; Li, F.; Yang, T.; Dai, H. Rapamycin regulates connective tissue growth factor expression of lung epithelial cells via phosphoinositide 3-kinase. Exp. Biol. Med. 2013, 238, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Finckenberg, P.; Inkinen, K.; Ahonen, J.; Merasto, S.; Louhelainen, M.; Vapaatalo, H.; Müller, D.; Ganten, D.; Luft, F.; Mervaala, E. Angiotensin II induces connective tissue growth factor gene expression via calcineurin-dependent pathways. Am. J. Pathol. 2003, 163, 355–366. [Google Scholar] [CrossRef]
- Mikaelian, I.; Malek, M.; Gadet, R.; Viallet, J.; Garcia, A.; Girard-Gagnepain, A.; Hesling, C.; Gillet, G.; Gonzalo, P.; Rimokh, R.; et al. Genetic and pharmacologic inhibition of mTORC1 promotes EMT by a TGF-β-independent mechanism. Cancer Res. 2013, 73, 6621–6631. [Google Scholar] [CrossRef] [PubMed]
- Shihab, F.S.; Bennett, W.M.; Yi, H.; Andoh, T.F. Effect of cyclosporine and sirolimus on the expression of connective tissue growth factor in rat experimental chronic nephrotoxicity. Am. J. Nephrol. 2006, 26, 400–407. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, S.; Slattery, C.; Ryan, M.P.; McMorrow, T. Sirolimus enhances cyclosporine a-induced cytotoxicity in human renal glomerular mesangial cells. J. Transplant. 2012, 2012, 980910. [Google Scholar] [CrossRef] [PubMed]
- Catania, J.M.; Chen, G.; Parrish, A.R. Role of matrix metalloproteinases in renal pathophysiologies. Am. J. Physiol. Renal. Physiol. 2007, 292, F905–F911. [Google Scholar] [CrossRef] [PubMed]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Matute-Bello, G.; Wurfel, M.M.; Lee, J.S.; Park, D.R.; Frevert, C.W.; Madtes, D.K.; Shapiro, S.D.; Martin, T.R. Essential role of MMP-12 in Fas-induced lung fibrosis. Am. J. Respir. Cell Mol. Biol. 2007, 37, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.R.; Cho, S.J.; Lee, C.G.; Homer, R.J.; Elias, J.A. Transforming growth factor (TGF)-β1 stimulates pulmonary fibrosis and inflammation via a Bax-dependent, Bid-activated pathway that involves matrix metalloproteinase-12. J. Biol. Chem. 2007, 282, 7723–7732. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kanda, M.; Sugimoto, H.; Shimizu, D.; Sueoka, S.; Takami, H.; Ezaka, K.; Hashimoto, R.; Okamura, Y.; Iwata, N.; et al. Translational implication of Kallmann syndrome-1 gene expression in hepatocellular carcinoma. Int. J. Oncol. 2015, 46, 2546–2554. [Google Scholar] [CrossRef] [PubMed]
- Raju, R.; Jian, B.; Hooks, J.J.; Nagineni, C.N. Transforming growth factor-β regulates the expression of anosmin (KAL-1) in human retinal pigment epithelial cells. Cytokine 2013, 61, 724–727. [Google Scholar] [CrossRef] [PubMed]
- Carew, R.M.; Browne, M.B.; Hickey, F.B.; Brazil, D.P. Insulin receptor substrate 2 and FoxO3a signalling are involved in E-cadherin expression and transforming growth factor-β1-induced repression in kidney epithelial cells. FEBS J. 2011, 278, 3370–3380. [Google Scholar] [CrossRef] [PubMed]
- Rieger, M.E.; Sims, A.H.; Coats, E.R.; Clarke, R.B.; Briegel, K.J. The embryonic transcription cofactor LBH is a direct target of the Wnt signaling pathway in epithelial development and in aggressive basal subtype breast cancers. Mol. Cell Biol. 2010, 30, 4267–4279. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Guan, X.; Lv, J.; Li, X.; Wang, Y.; Li, L. Limb-bud and Heart (LBH) functions as a tumor suppressor of nasopharyngeal carcinoma by inducing G1/S cell cycle arrest. Sci. Rep. 2015, 5, 7626. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.P.; Flozak, A.S.; Russell, S.; Wei, J.; Jain, M.; Mutlu, G.M.; Budinger, G.R.; Feghali-Bostwick, C.A.; Varga, J.; Gottardi, C.J. Nuclear β-catenin is increased in systemic sclerosis pulmonary fibrosis and promotes lung fibroblast migration and proliferation. Am. J. Respir. Cell Mol. Biol. 2011, 45, 915–922. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Ma, R.R.; He, J.Y.; Zhang, H.; Liu, X.L.; Guo, X.Y.; Gao, P. Protocadherin 7 inhibits cell migration and invasion through E-cadherin in gastric cancer. Tumour Biol. 2017, 39, 1010428317697551. [Google Scholar] [CrossRef] [PubMed]
- Galietta, L.J.; Lantero, S.; Gazzolo, A.; Sacco, O.; Romano, L.; Rossi, G.A.; Zegarra-Moran, O. An improved method to obtain highly differentiated monolayers of human bronchial epithelial cells. In Vitro Cell. Dev. Biol. Anim. 1998, 34, 478–481. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).