Suppression of External NADPH Dehydrogenase—NDB1 in Arabidopsis thaliana Confers Improved Tolerance to Ammonium Toxicity via Efficient Glutathione/Redox Metabolism
Abstract
:1. Introduction
2. Results
2.1. Changes in Respiration and Pyridine Nucleotide Status in NDB1 Suppression Plants Cultured on NO3− or NH4+
2.2. Influence of NDB1 Suppression on ROS Content and Oxidative Damage in Tissues of Plants Fed NO3− or NH4+
2.3. Ascorbate Level and Ascorbate-Related Antioxidant Defense in NDB1 Knock-Down Plants in Response to NO3− or NH4+ Nutrition
2.4. Glutathione Metabolism in NDB1 Knock-Down Plants during NO3− or NH4+ Growth
2.5. The Effect of NDB1 Suppression on Redox-Related Enzymes in Plants Grown on NO3− or NH4+
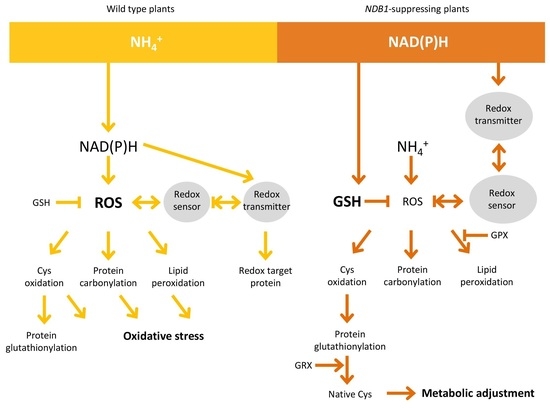
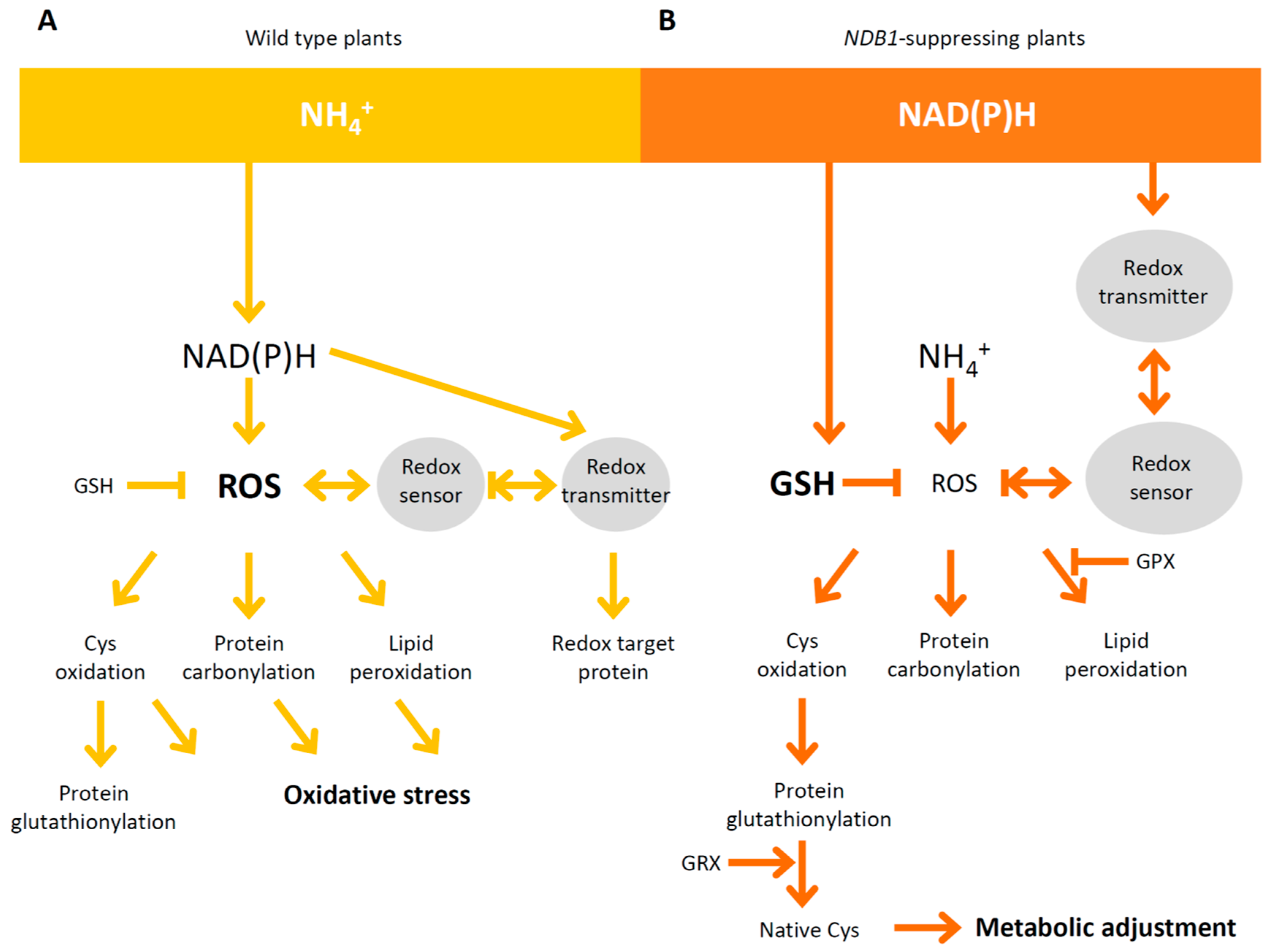
3. Discussion
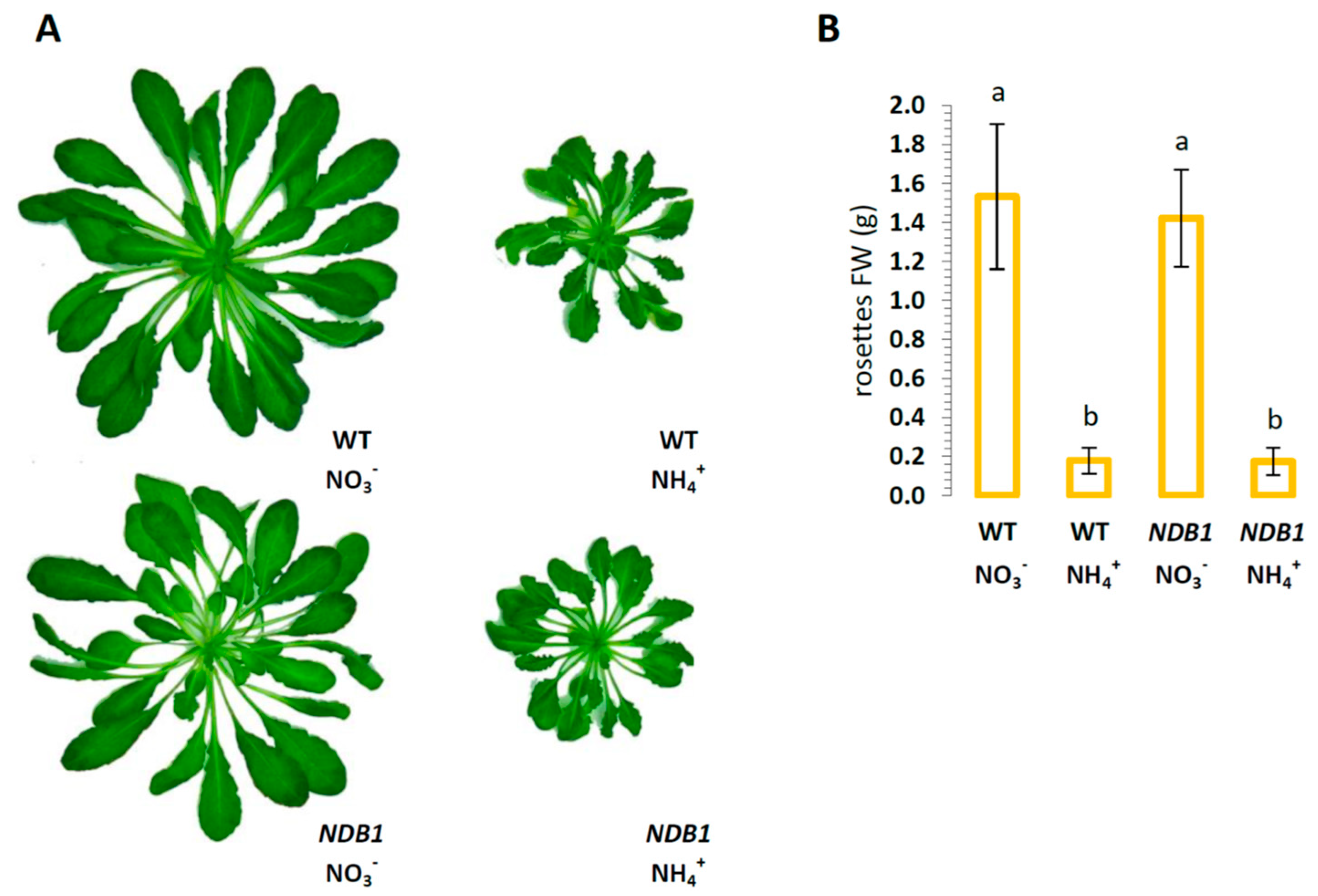
3.1. NDB1-Suppressed Line Does Not Show a Growth Phenotype or Over-Reduction under Ammonium Nutrition
3.2. Ammonium Nutrition Causes Less Oxidative Injury in NDB1 Knock-Down Plants
3.3. Improved Resistance of the NDB1 Knock-Down Line to Ammonium Stress Is Not Related to Ascorbate-Dependent Antioxidant Systems but May Be Attributable to a Glutathione-Dependent System
3.4. NDB1 Deletion Leads to Changes in the Expression of Genes Involved in the Redox Regulatory Network
4. Material and Methods
4.1. Plant Material and Growth Conditions
4.2. Phenotype Analysis
4.3. Respiratory Measurements
4.4. Quantitative RT–PCR Analyses
4.5. Analysis of Metabolites
4.6. Confocal Fluorescent Imaging of Intracellular ROS
4.7. Western Blotting Analyses
4.8. Enzyme Activity Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| APX | ascorbate peroxidase |
| AsA | reduced form of ascorbate |
| DHA | oxidized form of ascorbate (dehydroascorbate) |
| DHAR | dehydroascorbate reductase |
| GPX | glutathione peroxidase-like |
| GR | glutathione reductase |
| GRX | glutaredoxin |
| GSH | reduced form of glutathione |
| GSSG | oxidized form of glutathione (glutathione disulfide) |
| MDHAR | monodehydroascorbate reductase |
| NDB1 | external NADPH dehydrogenase |
| NTRC | NAPDH-dependent thioredoxin reductase C |
| Prx | peroxiredoxin |
| ROS | reactive oxygen species |
| SOD | superoxide dismutase |
| TRX | thioredoxin |
| UPOX | protein up-regulated by oxidative stress |
References
- Noctor, G. Metabolic signalling in defence and stress: The central roles of soluble redox couples. Plant Cell Environ. 2006, 29, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Gakière, B.; Fernie, A.R.; Pétriacq, P. More to NAD+ than meets the eye: A regulator of metabolic pools and gene expression in Arabidopsis. Free Radic. Biol. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; De Paepe, R.; Foyer, C.H. Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 2007, 12, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M. Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Biol. 2001, 52, 561–591. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.G.; Geisler, D.A.; Møller, I.M. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 2008, 8, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Michalecka, A.M.; Agius, S.C.; Møller, I.M.; Rasmusson, A.G. Identification of a mitochondrial external NADPH dehydrogenase by overexpression in transgenic Nicotiana sylvestris. Plant J. 2004, 37, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Elhafez, D.; Murcha, M.W.; Clifton, R.; Soole, K.L.; Day, D.A.; Whelan, J. Characterization of mitochondrial alternative NAD(P)H dehydrogenases in Arabidopsis: Intraorganelle location and expression. Plant Cell Physiol. 2006, 47, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Considine, M.J.; Holtzapffel, R.C.; Day, D.A.; Whelan, J.; Millar, A.H. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 2002, 129, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.G.; Wallström, S.V. Involvement of mitochondria in the control of plant cell NAD(P)H reduction levels. Biochem. Soc. Trans. 2010, 38, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Padmasree, K.; Padmavathi, L.; Raghavendra, A.S. Essentiality of mitochondrial oxidative metabolism for photosynthesis: Optimization of carbon assimilation and protection against photoinhibition. Crit. Rev. Biochem. Mol. Biol. 2002, 37, 71–119. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Yoshida, K. Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 2007, 8, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.M.; Rasmusson, A.G.; Fredlund, K.M. NAD(P)H-ubiquinone oxidoreductases in plant mitochondria. J. Bioenerg. Biomemb. 1993, 25, 377–384. [Google Scholar] [CrossRef]
- Potters, G.; Horemans, N.; Jansen, M.A. The cellular redox state in plant stress biology—A charging concept. Plant Physiol. Biochem. 2010, 48, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Araújo, W.L.; Fernie, A.R. Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 2011, 155, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Wallström, S.V.; Florez-Sarasa, I.; Araújo, W.L.; Aidemark, M.; Fernández-Fernández, M.; Fernie, A.R.; Ribas-Carbó, M.; Rasmusson, A.G. Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol. Plant 2014, 7, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Lelarge-Trouverie, C.; Mhamdi, A. The metabolomics of oxidative stress. Phytochemistry 2015, 112, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Diez, C.; Miguel, V.; Mennerich, D.; Kietzmann, T.; Sánchez-Pérez, P.; Cadenas, S.; Lamas, S. Antioxidant responses and cellular adjustments to oxidative stress. Redox Biol. 2015, 6, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Schertl, P.; Braun, H.P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, A.J. The integration of glutathione homeostasis and redox signaling. J. Plant Physiol. 2008, 165, 1390–1403. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.I.; Neukermans, J.; Marquez-Garcia, B.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Deponte, M. Glutathione catalysis and the reaction mechanisms of glutathione-dependent enzymes. Biochim. Biophys. Acta 2013, 1830, 3217–3266. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Skipsey, M.; Grundy, N.M.; Edwards, R. Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 2005, 138, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Halliwell, B. The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, F.; Jiménez, A.; Lázaro, J.J. What do the plant mitochondrial antioxidant and redox systems have to say under salinity, drought, and extreme temperature? In Reactive Oxygen Species and Oxidative Damage in Plants under Stress, 1st ed.; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 23–55. [Google Scholar]
- Geigenberger, P.; Thormählen, I.; Daloso, D.M.; Fernie, A.R. The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci. 2017, 22, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Rahantaniaina, M.S.; Tuzet, A.; Mhamdi, A.; Noctor, G. Missing links in understanding redox signaling via thiol/disulfide modulation: How is glutathione oxidized in plants? Front. Plant Sci. 2013, 4, 477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kirkham, M.B. Antioxidant responses to drought in sunflower and sorghum seedlings. New Phytol. 1996, 132, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Garmier, M.; Carroll, A.J.; Delannoy, E.; Vallet, C.; Day, D.A.; Small, I.D.; Millar, A.H. Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 2008, 148, 1324–1341. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Gutierrez, R.A. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr. Opin. Plant Biol. 2008, 11, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.J. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth. Res. 2015, 123, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Gerendás, J.; Bendixen, R.; Schinner, K.; Tabrizi, H.; Sattelmacher, B.; Hansen, U.P. Different Tolerance to Light Stress in NO3−-and NH4+-Grown Phaseolus vulgaris L. Plant Biol. 2000, 2, 558–570. [Google Scholar] [CrossRef]
- Guo, S.; Schinner, K.; Sattelmacher, B.; Hansen, U.P. Different apparent CO2 compensation points in nitrate-and ammonium-grown Phaseolus vulgaris and the relationship to non-photorespiratory CO2 evolution. Physiol. Plant. 2005, 123, 288–301. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. A re-evaluation of the ATP: NADPH budget during C3 photosynthesis: A contribution from nitrate assimilation and its associated respiratory activity? J. Exp. Bot. 1998, 49, 1895–1908. [Google Scholar] [CrossRef]
- Escobar, M.A.; Geisler, D.A.; Rasmusson, A.G. Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: Opposing effects of ammonium and nitrate. Plant J. 2006, 45, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.J.; Sukrapanna, S.S.; Warner, R.L. Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 1992, 99, 1294–1301. [Google Scholar] [CrossRef] [PubMed]
- Gerendás, J.; Zhu, Z.; Bendixen, R.; Ratcliffe, R.G.; Sattelmacher, B. Physiological and biochemical processes related to ammonium toxicity in higher plants. J. Plant Nutr. Soil Sci. 1997, 160, 239–251. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. Ecological significance and complexity of N-source preference in plants. Ann. Bot. 2013, 112, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Gieczewska, K.; Łukawska-Kuźma, K.; Rasmusson, A.G.; Gardeström, P.; Szal, B. Long-term ammonium nutrition of Arabidopsis increases the extrachloroplastic NAD(P)H/NAD(P)+ ratio and mitochondrial reactive oxygen species level in leaves but does not impair photosynthetic capacity. Plant Cell Environ. 2013, 36, 2034–2045. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Szal, B. The role of reactive oxygen species under ammonium nutrition. In Reactive Oxygen and Nitrogen Species Signaling and Communication in Plants; Gupta, K., Igamberdiev, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 23, pp. 133–153. [Google Scholar]
- Britto, D.T.; Kronzucker, H.J. NH4+ toxicity in higher plants: A critical review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef]
- Bittsánszky, A.; Pilinszky, K.; Gyulai, G.; Komives, T. Overcoming ammonium toxicity. Plant Sci. 2015, 231, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Esteban, R.; Ariz, I.; Cruz, C.; Moran, J.F. Review: Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 2016, 248, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Day, D.A.; Millar, A.H. Targets of stress-induced oxidative damage in plant mitochondria and their impact on cell carbon/nitrogen metabolism. J. Exp. Bot. 2004, 55, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Podgórska, A. The role of mitochondria in leaf nitrogen metabolism. Plant Cell Environ. 2012, 35, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G.; Hodges, M. Respiration and nitrogen assimilation: Targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1467–1482. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O.; Zhang, B.; Carrie, C.; Uggalla, V.; Paynter, E.; Giraud, E.; Whelan, J. Defining the mitochondrial stress response in Arabidopsis thaliana. Mol. Plant 2009, 2, 1310–1324. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Lee, H.; Xiong, L.; Zhu, J.K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Garmier, M.; Noctor, G.; Mathieu, C.; Chétrit, P.; Foyer, C.H.; De Paepe, R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance though altered signaling and diurnal regulation. Plant Cell 2003, 15, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E.H.; Tomaz, T.; Carroll, A.J.; Estavillo, G.; Delannoy, E.; Tanz, S.K.; Small, I.D.; Pogson, B.J.; Millar, A.H. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009, 151, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Łukawska, K.; Zdolińska, I.; Rychter, A.M. Chilling stress and mitochondrial genome rearrangement in the MSC16 cucumber mutant affect the alternative oxidase and antioxidant defense system to a similar extent. Physiol. Plant. 2009, 137, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, D.P.; Wang, Y.; McIntosh, L. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 1999, 96, 8271–8276. [Google Scholar] [CrossRef] [PubMed]
- Umbach, A.L.; Fiorani, F.; Siedow, J.N. Characterization of transformed Arabidopsis with altered oxidase levels and analysis of effect on reactive oxygen species in tissue. Plant Physiol. 2005, 139, 1806–1820. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Ho, L.H.; Clifton, R.; Carroll, A.; Estavillo, G.; Tan, Y.F.; Howell, K.A.; Ivanova, A.; Pogson, B.J.; Millar, A.H.; et al. The absence of alternative oxidase 1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 2008, 147, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Norberg, F.E.B.; Szilágyi, A.; De Paepe, R.; Åkerlund, H.E.; Rasmusson, A.G. The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol. 2008, 49, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.; Barthet, M.; Melino, V.; Smith, P.; Day, D.; Soole, K. Alterations in the mitochondrial alternative NAD(P)H dehydrogenase NDB4 lead to changes in mitochondrial electron transport chain composition, plant growth and response to oxidative stress. Plant Cell Physiol. 2011, 52, 1222–1237. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Nunes-Nesi, A.; Wallström, S.V.; Lager, I.; Michalecka, A.M.; Norberg, F.E.; Widell, S.; Fredlund, K.M.; Fernie, A.R.; Rasmusson, A.G. A redox-mediated modulation of stem bolting in transgenic Nicotiana sylvestris differentially expressing the external mitochondrial NADPH dehydrogenase. Plant Physiol. 2009, 150, 1248–1259. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B.; Stumpe, M.; Mauch, F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 2011, 233, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B. Compartment-specific importance of glutathione during abiotic and biotic stress. Front. Plant Sci. 2014, 5, 566. [Google Scholar] [CrossRef] [PubMed]
- Queval, G.; Thominet, D.; Vanacker, H.; Miginiac-Maslow, M.; Gakière, B.; Noctor, G. H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol. Plant 2009, 2, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Anjum, N.A.; Hasanuzzaman, M.; Gill, R.; Trivedi, D.K.; Ahmad, I.; Pereira, E.; Tuteja, N. Glutathione and glutathione reductase: A boon in disguise for plant abiotic stress defense operations. Plant Physiol. Biochem. 2013, 70, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Marty, L.; Siala, W.; Schwarzländer, M.; Fricker, M.D.; Wirtz, M.; Sweetlove, L.J.; Meyer, Y.; Meyer, A.J.; Reichheld, J.P.; Hell, R. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 9109–9114. [Google Scholar] [CrossRef] [PubMed]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, M.; Bedhomme, M.; Marchand, C.H.; Couturier, J.; Gao, X.H.; Rouhier, N.; Trost, P.; Lemaire, S.D. Glutaredoxin S12: Unique properties for redox signaling. Antioxid. Redox Sign. 2012, 16, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Patterson, K.; Walters, L.A.; Cooper, A.M.; Olvera, J.G.; Rosas, M.A.; Rasmusson, A.G.; Escobar, M.A. Nitrate-regulated glutaredoxins control Arabidopsis primary root growth. Plant Physiol. 2016, 170, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J. Redox signal integration: From stimulus to networks and genes. Physiol. Plant. 2008, 133, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Baier, M.; Dietz, K.-J. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999, 119, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Baier, M.; Horling, F.; Kahmann, U.; Harris, G.; Schürmann, P.; Dietz, K.-J. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. USA 2002, 99, 5738–5743. [Google Scholar] [CrossRef] [PubMed]
- Petersson, U.A.; Kieselbach, T.; García-Cerdán, J.G.; Schröder, W.P. The Prx Q protein of Arabidopsis thaliana is a member of the luminal chloroplast proteome. FEBS Lett. 2006, 580, 1873–3468. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Galliardt, H.; Jütte, P.; Schäper, S.; Dittmann, L.; Dietz, K.-J. The conformational bases for the two functionalities of 2-cysteine peroxiredoxins as peroxidase and chaperone. J. Exp. Bot. 2013, 64, 3483–3497. [Google Scholar] [CrossRef] [PubMed]
- Muthuramalingam, M.; Seidel, T.; Laxa, M.; Nunes de Miranda, S.M.; Gartner, F.; Stroher, E.; Kandlbinder, A.; Dietz, K.-J. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol. Plant 2009, 2, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.-J.; Horling, F.; König, J.; Baier, M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation, J. Exp. Bot. 2002, 53, 1321–1329. [Google Scholar] [CrossRef]
- Lamkemeyer, P.; Laxa, M.; Collin, V.; Li, W.; Finkemeier, I.; Schöttler, M.A.; Holtkamp, V.; Tognetti, V.B.; Issakidis-Bourguet, E.; Kandlbinder, A.; et al. Peroxiredoxin Q of Arabidopsis thaliana is attached to the thylakoids and functions in context of photosynthesis. Plant J. 2006, 45, 968–981. [Google Scholar] [CrossRef] [PubMed]
- Horling, F.; Lamkemeyer, P.; König, J.; Finkemeier, I.; Kandlbinder, A.; Baier, M.; Dietz, K.-J. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 2003, 131, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Gelhaye, E.; Gualberto, J.M.; Jordy, M.-N.; De Fay, E.; Hirasawa, M.; Duplessis, S.; Lemaire, S.D.; Frey, P.; Martin, F.; et al. Poplar Peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol. 2004, 134, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hisabori, T. Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proc. Natl. Acad. Sci. USA 2016, 113, 3967–3976. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ruiz, J.M.; Naranjo, B.; Ojeda, V.; Guinea, M.; Cejudo, F.J. NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proc. Natl. Acad. Sci. USA 2017, 114, 12069–12074. [Google Scholar] [CrossRef] [PubMed]
- Finkemeier, I.; Goodman, M.; Lamkemeyer, P.; Kandlbinder, A.; Sweetlove, L.J.; Dietz, K.J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 2005, 280, 12168–12180. [Google Scholar] [CrossRef] [PubMed]
- Horling, F.; König, J.; Dietz, K.-J. Type II peroxiredoxin C, a member of the peroxiredoxin family of Arabidopsis thaliana: Its expression and activity in comparison with other peroxiredoxins. Plant Physiol. Biochem. 2002, 40, 491–499. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Florez-Sarasa, I.; Ostaszewska, M.; Galle, A.; Flexas, J.; Rychter, A.M.; Ribas-Carbó, M. Changes of alternative oxidase activity, capacity and protein content in leaves of Cucumis sativus wild-type and MSC16 mutant grown under different light intensities. Physiol. Plant. 2009, 137, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Escobar, M.A.; Franklin, K.A.; Svensson, A.S.; Salter, M.G.; Whitelam, G.C.; Rasmusson, A.G. Light regulation of the Arabidopsis respiratory chain. Multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol. 2004, 136, 2710–2721. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Ostaszewska, M.; Gardeström, P.; Rasmusson, A.G.; Szal, B. In comparison with nitrate nutrition, ammonium nutrition increases growth of the frostbite1 Arabidopsis mutant. Plant Cell Environ. 2015, 38, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Dąbrowska, Z.; Malmberg, G.; Gardeström, P.; Rychter, A.M. Changes in energy status of leaf cells as the consequence of mitochondrial genome rearrangement. Planta 2008, 227, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Veljović-Jovanović, S.; Noctor, G.; Foyer, C.H. Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol. Biochem. 2002, 40, 501–507. [Google Scholar] [CrossRef]
- Okamura, M. An improved method for determination of L-ascorbic acid and L-dehydroascorbic acid in blood plasma. Clin. Chim. Acta 1980, 103, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Brehe, J.E.; Burch, H.B. Enzymatic assay for glutathione. Anal. Biochem. 1976, 74, 189–197. [Google Scholar] [CrossRef]
- Hodges, D.; DeLong, J.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Ostaszewska-Bugajska, M.; Rychter, A.M.; Juszczuk, I.M. Antioxidative and proteolytic systems protect mitochondria from oxidative damage in S-deficient Arabidopsis thaliana. J. Plant Physiol. 2015, 186–187, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Kehrer, I.; Kozlov, A.V.; Haller, M.; Redl, H.; Hermann, M.; Grimm, M.; Troppmair, J. Mitochondrial ROS production under cellular stress: Comparison of different detection methods. Anal. Bioanal. Chem. 2011, 400, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Hill, B.G.; Ramana, K.V.; Cai, J.; Bhatnagar, A.; Srivastava, S.K. Measurement and identification of S-glutathiolated proteins. In Methods in Enzymology; Cadenas, E., Packer, L., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 473, pp. 179–197. [Google Scholar]
- Podgórska, A.; Burian, M.; Rychter, A.M.; Rasmusson, A.G.; Szal, B. Short-term ammonium supply induces cellular defence to prevent oxidative stress in Arabidopsis leaves. Physiol. Plant. 2017, 160, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Asada, K. Inactivation of ascorbate peroxidase in spinach chloroplasts on dark addition of hydrogen peroxide: Its protection by ascorbate. Plant Cell Physiol. 1984, 25, 85–92. [Google Scholar] [CrossRef]
- Kuźniak, E.; Skłodowska, M. The effect of Botrytis cinerea infection on ascorbate-glutathione cycle in tomato leaves. Plant Sci. 1999, 148, 69–76. [Google Scholar] [CrossRef]
- Sehmer, L.; Fontaine, V.; Antoni, F.; Dizengremel, P. Effects of ozone and elevated atmospheric carbon dioxide on carbohydrate metabolism of spruce needles. Catabolic and detoxification pathways. Physiol. Plant. 1998, 102, 605–611. [Google Scholar] [CrossRef]
- Gutzat, R.; Mittelsten Scheid, O. Epigenetic responses to stress: Triple defense? Curr. Opin. Plant. Biol. 2012, 15, 568–573. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Rasool, B.; Davey, J.W.; Hancock, R.D. Cross-tolerance to biotic and abiotic stresses in plants: A focus on resistance to aphid infestation. J. Exp. Bot. 2016, 67, 2025–2037. [Google Scholar] [CrossRef] [PubMed]







| WT NO3− | WT NH4+ | NDB1 NO3− | NDB1 NH4+ | |
|---|---|---|---|---|
| Oxygen consumption (nmol O2 min−1 g−1 FW) | ||||
| Total respiration (Vt) | 40.54 a ± 9.93 | 93.16 c ± 3.67 | 41.38 a ± 5.99 | 72.92 b ± 2.96 |
| +SHAM (Vcyt) | 37.31 a ± 7.65 | 66.09 b ± 6.39 | 36.46 a ± 10.84 | 55.45 ab ± 4.62 |
| +KCN (Valt) | 5.94 a ± 3.11 | 38.98 b ± 10.58 | 5.82 a ± 1.38 | 46.47 b ± 2.74 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podgórska, A.; Ostaszewska-Bugajska, M.; Borysiuk, K.; Tarnowska, A.; Jakubiak, M.; Burian, M.; Rasmusson, A.G.; Szal, B. Suppression of External NADPH Dehydrogenase—NDB1 in Arabidopsis thaliana Confers Improved Tolerance to Ammonium Toxicity via Efficient Glutathione/Redox Metabolism. Int. J. Mol. Sci. 2018, 19, 1412. https://doi.org/10.3390/ijms19051412
Podgórska A, Ostaszewska-Bugajska M, Borysiuk K, Tarnowska A, Jakubiak M, Burian M, Rasmusson AG, Szal B. Suppression of External NADPH Dehydrogenase—NDB1 in Arabidopsis thaliana Confers Improved Tolerance to Ammonium Toxicity via Efficient Glutathione/Redox Metabolism. International Journal of Molecular Sciences. 2018; 19(5):1412. https://doi.org/10.3390/ijms19051412
Chicago/Turabian StylePodgórska, Anna, Monika Ostaszewska-Bugajska, Klaudia Borysiuk, Agata Tarnowska, Monika Jakubiak, Maria Burian, Allan G. Rasmusson, and Bożena Szal. 2018. "Suppression of External NADPH Dehydrogenase—NDB1 in Arabidopsis thaliana Confers Improved Tolerance to Ammonium Toxicity via Efficient Glutathione/Redox Metabolism" International Journal of Molecular Sciences 19, no. 5: 1412. https://doi.org/10.3390/ijms19051412
APA StylePodgórska, A., Ostaszewska-Bugajska, M., Borysiuk, K., Tarnowska, A., Jakubiak, M., Burian, M., Rasmusson, A. G., & Szal, B. (2018). Suppression of External NADPH Dehydrogenase—NDB1 in Arabidopsis thaliana Confers Improved Tolerance to Ammonium Toxicity via Efficient Glutathione/Redox Metabolism. International Journal of Molecular Sciences, 19(5), 1412. https://doi.org/10.3390/ijms19051412