Proposed Molecular and miRNA Classification of Gastric Cancer
Abstract
:1. Introduction
2. Histological and Molecular Classifications of GC
- these classifications are based on a highly complex methodology, which is not always available in every laboratory;
- they lack a prospective validation on a large scale;
- they have striking differences in epidemiology, underlying molecular mechanisms and prognosis;
- their prognostic power is decreased by limited follow-up of patients;
- none of them takes into account the active, non-malignant stromal cells
3. Integrated Molecular Signatures to Discriminate Intestinal and Diffuse Histological GC Subtypes
4. TCGA Classification of GC and Related Signaling Pathways
4.1. EBV-Related GC
4.2. GC with MSI
4.3. GC with CIN
4.4. Genomic Stable (GS) GC
4.5. Patient-Derived Preclinical Models of GC
4.6. Role of microRNAs in Signaling Pathways of GC
4.7. Clinical Implications of Tissue miRNAs in GC
5. Conclusions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrmann, R. Geschwulste des margens. In Handbuch spez pathol anat und histo; Henke, F., Lubarsch, O., Eds.; Springer: Berlin, Germany, 1926; pp. 864–871. [Google Scholar]
- Siewert, J.R.; Stein, H.J. Classification of adenocarcinoma of the oesophagogastric junction. Br. J. Surg. 1998, 85, 1457–1459. [Google Scholar] [CrossRef] [PubMed]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so called intestinal-type carcinoma: An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Lauwers, G.Y.; Carneiro, F.; Graham, D.Y. Gastric carcinoma. In WHO Classification of Tumours of the Digestive System, 4th ed.; Bosman, F.T., Carneiro, F., Hruban, R.H., Theise, N.D., Eds.; World Health Organization: Lyon, France, 2010; Volume 3, pp. 48–58, ISBN-13 9789283224327. [Google Scholar]
- Kajitani, T. The general rules for the gastric cancer study in surgery and pathology I: Clinical classification. Jpn. J. Surg. 1981, 1, 127–139. [Google Scholar]
- Tan, I.B.; Ivanova, T.; Lim, K.H.; Ong, C.W.; Deng, N.; Lee, J.; Tan, S.H.; Wu, J.; Lee, M.H.; Ooi, C.H.; et al. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology 2011, 141, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Cheong, J.H. Beyond precision surgery: Molecularly motivated precision care for gastric cancer. Eur. J. Surg. Oncol. 2017, 43, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Cho, Y.S.; Lee, G.K.; Lee, S.; Kim, K.W.; Jho, S.; Kim, H.M.; Hong, S.H.; Hwang, J.H.; Kim, S.Y.; et al. Genomic profile analysis of diffuse-type gastric cancers. Genome Biol. 2014, 15, R55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanabe, S.; Aoyagi, K.; Yokozaki, H.; Sasaki, H. Gene expression signatures for identifying diffuse-type gastric cancer associated with epithelial-mesenchymal transition. Int. J. Oncol. 2014, 44, 1955–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.; Bang, S.; Lee, S.; Kim, S.; Jung, Y.; Lee, C.; Choi, K.; Lee, S.G.; Lee, K.; Lee, Y.; et al. Expression Profiling and Subtype-Specific Expression of stomach cancer. Cancer Res. 2003, 63, 8248–8255. [Google Scholar] [PubMed]
- Min, L.; Zhao, Y.; Zhu, S.; Qiu, X.; Cheng, R.; Xing, J.; Shao, L.; Guo, S.; Zhang, S. Integrated Analysis Identifies Molecular Signatures and Specific Prognostic Factors for Different Gastric Cancer Subtypes. Transl. Oncol. 2017, 10, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Tan, I.B.; Das, K.; Deng, N.; Zouridis, H.; Pattison, S.; Chua, C.; Feng, Z.; Guan, Y.K.; Ooi, C.H.; et al. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology 2013, 145, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Benita, Y.; Cao, Z.; Giallourakis, C.; Li, C.; Gardet, A.; Xavier, R.J. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 2010, 115, 5376–5384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ooi, C.H.; Ivanova, T.; Wu, J.; Lee, M.; Tan, I.B.; Tao, J.; Ward, L.; Koo, J.H.; Gopalakrishnan, V.; Zhu, Y.; et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009, 5, e1000676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, G.; Wang, Q.; Lu, W.; Xu, M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget 2017, 10, 73826–73836. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki-Ushiku, A.; Kunita, A.; Fukayama, M. Update on Epstein–Barr virus and gastric cancer [review]. Int. J. Oncol. 2015, 46, 1421–1434. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Kaneda, A.; Fukayama, M. Epstein–Barr virus associated gastric carcinoma: Use of host cell machineries and somatic gene mutations. Pathobiology 2015, 82, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Fukayama, M.; Hino, R.; Uozaki, H. Epstein–Barr virus and gastric carcinoma: Virus–host interactions leading to carcinoma. Cancer Sci. 2008, 99, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Song, H.J.; Srivastava, A.; Lee, J.; Kim, Y.S.; Kim, K.M.; Ki Kang, W.; Kim, M.; Kim, S.; Park, C.K.; Kim, S. Host inflammatory response predicts survival of patients with Epstein–Barr virus-associated gastric carcinoma. Gastroenterology 2010, 139, 84–92.e2. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD- L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.; Gajewski, T.F.; Mackensen, A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion:implications for tumor immunotherapy. Cancer Immunol. Immunother. 2005, 54, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, M.; Jin, Y.; Li, B.; Wang, X.; Yan, S.; Xu, R.; Yang, D. Programmed cell death ligand 1 (PD-L1) expression on gastric cancer and its relationship with clinicopathologic factors. Int. J. Clin. Exp. Pathol. 2015, 8, 11084–11091. [Google Scholar] [PubMed]
- Qing, Y.; Li, Q.; Ren, T.; Xia, W.; Peng, Y.; Liu, G.L.; Luo, X.Y.; Dai, X.Y.; Zhou, S.F.; Wang, D. Upregulation of PD-L1 and APE1 is associated with tumorigenesis and poor prognosis of gastric cancer. Drug Des. Devel. Ther. 2015, 9, 901–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.W.; Nam, K.H.; Ahn, S.H.; Park, D.J.; Kim, H.H.; Kim, S.H.; Chang, H.; Lee, J.O.; Kim, Y.J.; Lee, H.S.; et al. Prognostic implications of immunosuppressive protein expression in tumors as well as immune cell infiltration within the tumor microenvironment in gastric cancer. Gastric Cancer 2016, 19, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.D.; Zahurak, M.; Murphy, A.; Cornish, T.; Cuka, N.; Abdelfatah, E.; Yang, S.; Duncan, M.; Ahuja, N.; Taube, J.M.; et al. Patterns of PD-L1 expression and CD8 T cell infiltration in gastric adenocarcinomas and associated immune stroma. Gut 2017, 66, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.X.; Wang, X.S.; Wang, Y.F.; Hu, X.C.; Yan, J.Q.; Zhang, Y.L.; Wang, W.; Yang, R.J.; Feng, Y.Y.; Gao, S.G.; et al. Prognostic significance of PD-L1 expression in patients with gastric cancer in East Asia: A meta-analysis. OncolTargets Ther. 2016, 9, 2649–2654. [Google Scholar] [CrossRef]
- Abe, H.; Kunita, A.; Yamashita, H.; Seto, Y.; Fukayama, M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1+ immune cells in Epstein-Barr virus-associated gastric cancer: The prognostic implications. Mod. Pathol. 2017, 30, 427–439. [Google Scholar] [CrossRef]
- Shankaran, V.; Muro, K.; Bang, Y.; Geva, R.; Catenacci, D.; Gupta, S.; Eder, J.P.; Berger, R.; Loboda, A.; Albright, A.; et al. Correlation of gene expression signatures and clinical outcomes in patients with advanced gastric cancer treated with pembrolizumab (MK-3475). J. Clin. Oncol. 2015, 33, 3026. [Google Scholar] [CrossRef]
- Bang, Y.; Im, S.; Lee, K.; Cho, J.; Song, E.; Lee, K.; Kim, Y.H.; Park, J.O.; Chun, H.G.; Zang, D.Y.; et al. Randomized, double-blind phase II trial with prospective classification by ATM protein level to evaluate the efficacy and tolerability of olaparib plus paclitaxel in patients with recurrent or metastatic gastric cancer. J. Clin. Oncol. 2015, 33, 3858–3865. [Google Scholar] [CrossRef] [PubMed]
- Fontana, E.; Smyth, E.C. Novel targets in the treatment of advanced gastric cancer: A perspective review. Ther. Adv. Med. Oncol. 2016, 8, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.F.; Zhou, Z.X.; Chen, J.H.; Yi, G.; Chen, H.G.; Ba, M.C.; Lin, S.Q.; Qi, Y.C. Up-regulation of PIK3CA promotes metastasis in gastric carcinoma. World J. Gastroenterol. 2010, 16, 4986–4991. [Google Scholar] [CrossRef] [PubMed]
- Tapia, O.; Riquelme, I.; Leal, P.; Sandoval, A.; Aedo, S.; Weber, H.; Letelier, P.; Bellolio, E.; Villaseca, M.; Garcia, P.; et al. The PI3K/AKT/mTOR pathway is activated in gastric cancer with potential prognostic and predictive significance. Virchows Arch. 2014, 465, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Jiang, L.; Xu, H.; Zhou, D.; Li, Z. Expression of PI3K/AKT pathway in gastric cancer and its blockade suppresses tumor growth and metastasis. Int. J. Immunopathol. Pharmacol. 2012, 25, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Cinti, C.; Vindigni, C.; Zamparelli, A.; Sala, D.; Epistolato, M.; Marrelli, D.; Cevenini, G.; Tosi, P. Activated Akt as an indicator of prognosis in gastric cancer. Virchows Arch. 2008, 453, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Sangawa, A.; Shintani, M.; Yamao, N.; Kamoshida, S. Phosphorylation status of Akt and caspase-9 in gastric and colorectal carcinomas. Int. J. Clin. Exp. Pathol. 2014, 7, 3312–3317. [Google Scholar] [PubMed]
- Welker, M.E.; Kulik, G. Recent syntheses of PI3K/Akt/mTOR signaling pathway inhibitors. Bioorg. Med. Chem. 2013, 2, 4063–4091. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Tsimberidou, A.M.; Garrido-Laguna, I.; Wang, X.; Luthra, R.; Hong, D.S.; Naing, A.; Falchook, G.S.; Moroney, J.W. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol. Cancer. Ther. 2011, 10, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.R.; Greenwood, H.; Dudley, P.; Crafter, C.; Yu, D.H.; Zhang, J.; Li, J.; Gao, B.; Ji, Q.; Maynard, J.; et al. Preclinical pharmacology of AZD5363, an inhibitor of AKT: Pharmacodynamics, antitumor activity, and correlation of monotherapy activity with genetic background. Mol. Cancer Ther. 2012, 11, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Li, V.; Wong, C.; Chan, T.; Chan, A.S.; Zhao, W.; Chu, K.M.; So, S.; Chen, X.; Yuen, S.T.; Leung, S.Y. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer 2005, 5, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.G.; Ai, Y.W.; Yu, L.L.; Zhou, X.D.; Liu, J.; Li, J.H.; Xu, X.M.; Liu, S.; Chen, J.; Liu, F.; et al. Phosphoinositide 3-kinase/Akt pathway plays an important role in chemoresistance of gastric cancer cells against etoposide and doxorubicin induced cell death. Int. J. Cancer 2008, 122, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Oki, E.; Kakeji, Y.; Tokunaga, E.; Nishida, K.; Koga, T.; Egashira, A.; Morita, M.; Maehara, Y. Impact of PTEN/AKT/ PI3K signal pathway on the chemotherapy for gastric cancer. J. Clin. Oncol. 2006, 24, 4034. [Google Scholar] [CrossRef]
- Im, S.; Lee, K.; Nam, E. Potential prognostic significance of p185HER2 overexpression with loss of PTEN expression in gastric carcinomas. Tumori 2005, 91, 513–521. [Google Scholar] [PubMed]
- Wu, H.; Huang, M.; Cao, P.; Wang, T.; Shu, Y.; Liu, P. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol. Ther. 2012, 13, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.J.; Dai, W.; O’Mara, M.L.; Abankwa, D.; Chhabra, Y.; Pelekanos, R.A.; Gardon, O.; Tunny, K.A.; Blucher, K.M.; Morton, C.J.; et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 2014, 344, 1249783. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.L.; Wadleigh, M.; Cools, J.; Ebert, B.L.; Wernig, G.; Huntly, B.J.; Boggon, T.J.; Wlodarska, I.; Clark, J.J.; Moore, S.; et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005, 7, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchert, M.; Burns, C.; Ernst, M. Targeting JAK kinase in solid tumors: Emerging opportunities and challenges. Oncogene 2016, 35, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, H.; Uppal, N.; Wagner, S.; Bendell, J.; Beck, J.; Wade, S.; Nemunaitis, J.J.; Stella, P.J.; Pipas, J.M.; Wainberg, Z.A.; et al. A randomized doubleblind phase 2 study of ruxolitinib (RUX) or placebo (PBO) with capecitabine (CAPE) as second-line therapy in patients (pts) with metastatic pancreatic cancer (mPC). J. Clin. Oncol. 2015, 32, 4000. [Google Scholar] [CrossRef]
- Pedrazzani, C.; Corso, G.; Velho, S.; Leite, M.; Pascale, V.; Bettarini, F.; Marrelli, D.; Seruca, R.; Roviello, F. Evidence of tumor micro satellite instability in gastric cancer with familial aggregation. Fam. Cancer 2009, 8, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Velho, S.; Fernandes, M.S.; Leite, M.; Figueiredo, C.; Seruca, R. Causes and consequences of microsatellite instability in gastric carcinogenesis. World J. Gastroenterol. 2014, 20, 16433–16442. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Rustgi, A.K. DNA mismatch repair and cancer. Gastroenterology 1995, 109, 1685–1699. [Google Scholar] [CrossRef]
- Smyth, E.C.; Wotherspoon, A.; Peckitt, C.; Nankivell, M.G.; Eltahir, Z.; Wilson, S.H.; de Castro, D.G.; Okines, A.F.C.; Langley, R.E.; Cunningham, D. Correlation between mismatch repair deficiency (MMRd), microsatellite instability (MSI) and survival in MAGIC. J. Clin. Oncol. 2016, 15, 4064. [Google Scholar] [CrossRef]
- Pinto, M.; Wu, Y.; Mensink, R.G.; Cirnes, L.; Seruca, R.; Hofstra, R.M. Somatic mutations in mismatch repair genes in sporadic gastric carcinomas are not a cause but a consequence of the mutator phenotype. Cancer Genet. Cytogenet. 2008, 180, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, Z.; Wang, Y.; Zhang, C.; Liu, Y.; Qu, X. Microsatellite instability and survival in gastric cancer: A systematic review and meta-analysis. Mol. Clin. Oncol. 2015, 3, 699–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, D.; Uram, J.; Wang, H.; Bartlett, B.; Kemberling, H.; Eyring, A.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Kim, W.H.; Chiaravalli, A.M.; Kim, K.M.; Corvalan, A.H.; Matsuo, K.; Yu, J.; Sung, J.J.; Herrera-Goepfert, R.; Meneses-Gonzalez, F.; et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: An international pooled analysis. Gut 2014, 63, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Bae, J.M.; An, J.Y.; Kwon, I.G.; Cho, I.; Shin, H.B.; Eiji, T.; Aburahmah, M.; Kim, H.I.; Cheong, J.H.; et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J. Surg. Oncol. 2014, 110, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Giam, M.; Rancati, G. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div. 2015, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Chia, N.Y.; Tan, P. Molecular classification of gastric cancer. Ann. Oncol. 2016, 27, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Aprile, G.; Giampieri, R.; Bonotto, M.; Bittoni, A.; Ongaro, E.; Cardellino, G.G.; Graziano, F.; Giuliani, F.; Fasola, G.; Cascinu, S.; et al. The challenge of targeted therapies for gastric cancer patients: The beginning of a long journey. Expert Opin. Investig. Drugs 2014, 23, 925–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xu, X.Y.; Zhou, P.H. Emerging molecular classifications and therapeutic implications for gastric cancer. Chin. J. Cancer 2016, 35, 49. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Yeoh, K.G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015, 149, 1153–1162.e3. [Google Scholar] [CrossRef] [PubMed]
- Gravalos, C.; Jimeno, A. HER2 in gastric cancer: A new prognostic factor and a novel therapeutic target. Ann. Oncol. 2008, 19, 1523–1529. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Hecht, J.R.; Bang, Y.J.; Qin, S.K.; Chung, H.C.; Xu, J.M.; Park, J.O.; Jeziorski, K.; Shparyk, Y.; Hoff, P.M.; Sombrero, A.; et al. Lapatinib in Combination with Capecitabine Plus Oxaliplatin in Human Epidermal Growth Factor Receptor 2-Positive Advanced or Metastatic Gastric, Esophageal, or Gastroesophageal Adenocarcinoma: TRIO-013/LOGiC—A Randomized Phase III Trial. J. Clin. Oncol. 2016, 34, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Cortés, J.; Kim, S.B.; Im, S.A.; Hegg, R.; Im, Y.H.; Roman, L.; Pdrini, J.L.; Pienkowki, T.; Kontt, A.; et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N. Engl. J. Med. 2012, 366, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.R.; Bang, Y.J.; Qin, S.K.; Chung, H.C.; Xu, J.M.; Park, J.O.; Jeziorski, K.; Shparyk, Y.; Hoff, P.M.; Sombrero, A.; et al. Lapatinib in combination with capecitabine plus oxaliplatin (CapeOx) in HER2 positive advanced or metastatic gastric (A/MGC), esophageal (EAC), or astroesophageal (GEJ) adenocarcinoma: The logic trial. J. Clin. Oncol. 2013, 31, LBA4001. [Google Scholar] [CrossRef]
- Satoh, T.; Xu, R.H.; Chung, H.C.; Sun, G.P.; Doi, T.; Xu, J.M.; Tsuji, A.; Omuro, Y.; Li, J.; Wang, J.W.; et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN-a randomized, phase III study. J. Clin. Oncol. 2014, 32, 2039–2049. [Google Scholar] [CrossRef] [PubMed]
- Deva, S.; Baird, R.; Cresti, N.; Garcia-Corbacho, J.; Hogarth, L.; Frenkel, E.; Kawaguchi, K.; Arimura, A.; Donaldson, K.; Posner, J.; et al. Phase I expansion of S-222611, a reversible inhibitor of EGFR and HER2, in advanced solid tumors, including patients with brain metastases. J. Clin. Oncol. 2015, 33, 2511. [Google Scholar] [CrossRef]
- Lee, J.Y.; Hong, M.; Kim, S.T.; Park, S.H.; Kang, W.K.; Kim, K.M.; Lee, J. The impact of concomitant genomic alterations on treatment outcome for trastuzumab therapy in HER2-positive gastric cancer. Sci. Rep. 2015, 5, 9289. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Liu, J.; Zhang, J.; Wu, M.; Guo, L.; Liao, W. Development of trastuzumab-resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci. Rep. 2015, 5, 11634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piro, G.; Carbone, C.; Cataldo, I.; Di Nicolantonio, F.; Giacopuzzi, S.; Aprile, G.; Simionato, F.; Boschi, F.; Zanotto, M.; Mina, M.M.; et al. An FGFR3 Autocrine Loop Sustains Acquired Resistance to Trastuzumab in Gastric Cancer Patients. Clin. Cancer Res. 2016, 22, 6164–6175. [Google Scholar] [CrossRef] [PubMed]
- Arienti, C.; Zanoni, M.; Pignatta, S.; Del Rio, A.; Carloni, S.; Tebaldi, M.; Tedaldi, G.; Tesei, A. Preclinical evidence of multiple mechanisms underlying trastuzumab resistance in gastric cancer. Oncotarget 2016, 7, 18424–18439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, C.D.; Brown, M.D.; Sacks, D.B. IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009, 583, 1817–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walch, A.; Seidl, S.; Hermannstädter, C.; Rauser, S.; Deplazes, J.; Langer, R.; von Weyhern, C.H.; Sarbia, M.; Busch, R.; Feith, M.; et al. Combined analysis of Rac1, IQGAP1, Tiam1 and E-cadherin expression in gastric cancer. Mod. Pathol. 2008, 21, 544–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoury, H.; Naujokas, M.A.; Zuo, D.; Sangwan, V.; Frigault, M.M.; Petkiewicz, S.; Dankort, D.L.; Muller, W.J.; Park, M. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol. Biol. Cell 2005, 16, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.T.; Kim, H.; Liska, D.; Gao, S.; Christensen, J.G.; Weiser, M.R. MET activation mediates resistance to lapatinib inhibition of HER2- amplified gastric cancer cells. Mol. Cancer Ther. 2012, 11, 660–669. [Google Scholar] [CrossRef] [PubMed]
- De Silva, N.; Schulz, L.; Paterson, A.; Qain, W.; Secrier, M.; Godfrey, E.; Cheow, H.; O’Donovan, M.; Lao-Sirieix, P.; Jobanputra, M.; et al. Molecular effects of Lapatinib in the treatment of HER2 overexpressing oesophago-gastric adenocarcinoma. Br. J. Cancer 2015, 113, 1305–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lordick, F.; Kang, Y.; Chung, H.; Salman, P.; Oh, S.; Bodoky, G.; Kurteva, G.; Volovat, C.; Moiseyenko, V.M.; Gorbunova, V.; et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): A randomised, open-label phase 3 Trial. Lancet Oncol. 2013, 14, 490–499. [Google Scholar] [CrossRef]
- Waddell, T.; Chau, I.; Cunningham, D.; Gonzalez, D.; Okines, A.F.; Okines, C.; Wotherspoon, A.; Saffert, C.; Middleton, G.; Wadsley, J.; et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): A randomised, open-label phase 3 trial. Lancet Oncol. 2013, 14, 481–489. [Google Scholar] [CrossRef]
- Dragovich, T.; Mccoy, S.; Fenoglio-Preiser, C.; Wang, J.; Benedetti, J.; Baker, A.F.; Hackett, C.B.; Urba, S.G.; Zaner, K.S.; Blanke, C.D.; et al. Phase II trial of erlotinib in gastroesophageal junction and gastric adenocarcinomas: SWOG 0127. J. Clin. Oncol. 2006, 24, 4922–4927. [Google Scholar] [CrossRef] [PubMed]
- Dutton, S.J.; Ferry, D.R.; Blazeby, J.M.; Abbas, H.; Dahle-Smith, A.; Mansoor, W.; Thompson, J.; Harrison, M.; Chatteriee, A.; Falk, S.; et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): A phase 3, multicentre, double-blind, placebocontrolled randomised trial. Lancet Oncol. 2014, 15, 894–904. [Google Scholar] [CrossRef]
- Ha, S.Y.; Lee, J.; Kang, S.Y.; Do, I.G.; Ahn, S.; Park, J.O; Kang, W.K.; Choi, M.G.; Sohn, T.S.; Bae, J.M.; et al. MET overexpression assessed by new interpretation method predicts gene amplification and poor survival in advanced gastric carcinomas. Mod. Pathol. 2013, 26, 1632–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scagliotti, G.V.; Novello, S.; Von Pawel, J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat. Rev. 2013, 39, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Tebbutt, N.; Davidenko, I.; Murad, A.; Al-Batran, S.; Ilson, D.; Tjulandin, S.; Gotovkin, E.; Karaszewska, B.; Bondarenko, I.; et al. Phase III, randomized, double-blind, multicenter, placebo (P)-controlled trial of rilotumumab (R) plus epirubicin, cisplatin and capecitabine (ECX) as first-line therapy in patients (pts) with advanced MET-positive (pos) gastric or gastroesophageal junction (G/GEJ) cancer: RILOMET-1 study. J. Clin. Oncol. 2015, 33, 4000. [Google Scholar] [CrossRef]
- Shah, M.; Bang, Y.; Lordick, F.; Tabernero, J.; Chen, M.; Hack, S.; Phan, S.; Shames, D.S.; Cunningham, D. Metgastric: A phase III study of onartuzumab plus mFOLFOX6 in patients with metastatic HER2-negative (HER2-) and METpositive (MET+) adenocarcinoma of the stomach or gastroesophageal junction (GEC). J. Clin. Oncol. 2015, 33, 4012. [Google Scholar] [CrossRef]
- Iveson, T.; Donehower, R.; Davidenko, I.; Tjulandin, S.; Deptala, A.; Harrison, M.; Nirni, S.; Lakshmaiah, K.; Thomas, A.; Jiang, Y.; et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: An open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet Oncol. 2014, 15, 1007–1018. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, S.J.; Zhang, Y.; Zhang, G.Q.; Zha, T.Z.; Feng, Y.Z.; Zhang, K. Clinicopathological and prognostic significance of galectin-1 and vascular endothelial growth factor expression in gastric cancer. World J. Gastroenterol. 2013, 19, 2073–2079. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, J.G.; Sohn, S.K.; Chae, Y.S.; Moon, J.H.; Kim, S.N.; Bae, H.I.; Chung, H.Y.; Yu, W. No association of vascular endothelial growth factor-A (VEGF-A) and VEGF-C expression with survival in patients with gastric cancer. Cancer Res. Treat. 2009, 41, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, K.; Ichikawa, D.; Soga, K.; Watanabe, K.; Kosuga, T.; Takeshita, H.; Konishi, H.; Morimura, R.; Tsujiura, M.; Komatsu, S.; et al. Clinical significance of vascular endothelial growth factors C and D and chemokine receptor CCR7 in gastric cancer. Anticancer Res. 2010, 30, 2361–2366. [Google Scholar] [PubMed]
- Gou, H.F.; Chen, X.C.; Zhu, J.; Jiang, M.; Yang, Y.; Cao, D.; Hou, M. Expressions of COX-2 and VEGF-C in gastric cancer: Correlations with lymphangiogenesis and prognostic implications. J. Exp. Clin. Canc. Res. 2011, 30, 14. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, A.; Shah, M.A.; Van Cutsem, E.; Rha, S.Y.; Sawaki, A.; Park, S.R.; Lim, H.Y.; Yamada, Y.; Wu, J.; Langer, B.; et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A randomized, double-blind, placebo-controlled phase III study. J. Clin. Oncol. 2011, 29, 3968–3976. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; de Haas, S.; Kang, Y.K.; Ohtsu, A.; Tebbutt, N.C.; Ming, X.J.; Peng Yong, W.; Langer, B.; Delmar, P.; Scherer, S.J.; et al. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: A biomarker evaluation from the AVAGAST randomized phase III trial. J. Clin. Oncol. 2012, 30, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebocontrolled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Li, J.; Qin, S.; Xu, J.; Guo, W.; Xiong, J.; Bai, Y.; Sun, G.; Yang, Y.; Wang, L.; Xu, N.; et al. Apatinib for chemotherapy-refractory advanced metastatic gastric cancer: Results from a randomized, placebocontrolled, parallel-arm, phase II trial. J. Clin. Oncol. 2013, 31, 3219–3225. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.Y.; Noh, S.H.; Cheong, J.H. Molecular Dimensions of Gastric Cancer: Translational and Clinical Perspectives. J. Pathol. Transl. Med. 2016, 50, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corso, G.; Marrelli, D.; Pascale, V.; Vindigni, C.; Roviello, F. Frequency of CDH1 germline mutations in gastric carcinoma coming from high- and low-risk areas: Metanalysis and systematic review of the literature. BMC Cancer 2012, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Shen, C.Y.; Wu, H.S.; Hsieh, T.Y.; Chan, D.C.; Chen, C.J.; Yu, J.C.; Yu, C.P.; Harn, H.J.; Chen, P.J.; et al. Mechanisms inactivating the gene for E-cadherin in sporadic gastric carcinomas. World J. Gastroenterol. 2006, 12, 2168–2173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, W.K.; Xing, R.; Wong, S.H.; Liu, Y.; Fang, X.; Zhang, Y.; Wang, M.; Wang, J.; Li, L.; et al. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res. 2016, 76, 1724–1732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weissman, B.; Knudsen, K.E. Hijacking the chromatin remodeling machinery: Impact of SWI/SNF perturbations in cancer. Cancer Res. 2009, 69, 8223–8230. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Chen, Y.B.; Pan, K.; Wang, W.; Chen, S.P.; Chen, J.G.; Zhao, J.J.; Lv, L.; Pan, Q.Z.; Li, Y.Q.; et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS ONE 2012, 7, e40364. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Marchioni, F.; Evelyn, C.R.; Sipes, N.; Zhou, X.; Seibel, W.; Wortman, M.; Zheng, Y. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc. Natl. Acad. Sci. USA 2013, 110, 3155–3160. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Marchioni, F.; Sipes, N.; Evelyn, C.R.; Jerabek-Willemsen, M.; Duhr, S.; Seibel, W.; Wortman, M.; Zheng, Y. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 2012, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Türeci, O.; Koslowski, M.; Helftenbein, G.; Castle, J.; Rohde, C.; Dhaene, K.; Seitz, G.; Sahin, U. Claudin-18 gene structure, regulation, and expression is evolutionary conserved in mammals. Gene 2011, 481, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Kausalya, J.P.; Sia, Y.Y.; Teo, A.S.; Lee, W.H.; Ong, A.G.; Zhang, Z.; Tan, J.H.; Li, G.; Bertrand, D.; et al. Recurrent Fusion Genes in Gastric Cancer: CLDN18-ARHGAP26 Induces Loss of Epithelial Integrity. Cell Rep. 2015, 12, 272–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Batran, S.E.; Schuler, M.H.; Zvirbule, Z.; Manikhas, G.; Lordick, F.; Rusyn, A.; Vynnyk, Y.; Vynnychenko, I.; Fadeeva, N.; Nechaeva, M.; et al. FAST: An international, multicenter, randomized, phase II trial of epirubicin, oxaliplatin, and capecitabine (EOX) with or without IMAB362, a first-in-class anti-CLDN18.2 antibody, as firstline therapy in patients with advanced CLDN18.2 gastric and gastroesophageal junction (GEJ) adenocarcinoma. J. Clin. Oncol. 2016, 34, LBA4001. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, AV.; Budinskà, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed]
- Hausser, H.J.; Brenner, R.E. Phenotypic instability of Saos-2 cells in long-term culture. Biochem. Biophys. Res. Commun. 2005, 333, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Calcagno, A.M.; Varma, S.; Marino, M.; Green, L.J.; Vora, M.I.; Patel, C.; Orina, J.N.; Eliseeva, T.A.; Singal, V.; et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 18708–18713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furukawa, T.; Kubota, T.; Watanabe, M.; Kitaijma, M.; Hoffman, R.M. Orthotopic transplantation of histologically intact clinical specimens of stomach cancer to nude mice: Correlation of metastatic sites in mouse and individual patient donors. Int. J. Cancer 1993, 53, 608–612. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Fu, X.; Kubota, T.; Watanabe, M.; Kitajima, M.; Hoffman, R.M. Nude mouse metastatic models of human stomach cancer constructed using orthotopic implantation of histologically intact tissue. Cancer Res. 1993, 53, 1204–1208. [Google Scholar] [PubMed]
- Zhang, L.; Yang, J.; Cai, J.; Song, X.; Deng, J.; Huang, H.; Chen, D.; Yang, M.; Wery, J.P.; Li, S.; et al. A subset of gastric cancers with EGFR amplification and overexpression respond to cetuximab therapy. Sci. Rep. 2013, 3, 2992. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tian, T.; Li, Z.; Tang, Z.; Wang, L.; Wu, J.; Li, Y.; Dong, B.; Li, Y.; Dong, B.; et al. Establishment and characterization of patient-derived tumor xenograft using gastroscopic biopsies in gastric cancer. Sci. Rep. 2015, 5, 8542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, W.M.; Teng, E.; Chong, H.S.; Lopez, K.A.; Tay, A.Y.; Salto-Tellez, M.; Shabbir, A.; So, J.B.; Shan, S.L. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014, 74, 2630–2641. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Cho, S.Y.; Kim, H.; Na, D.; Han, J.Y.; Chae, J.; Park, C.; Park, O.K.; Min, S.; Kang, J.; et al. Genomic alterations in BCL2L1 and DLC1 contribute to drug sensitivity in gastric cancer. Proc. Natl. Acad. Sci. USA 2015, 112, 12492–12497. [Google Scholar] [CrossRef] [PubMed]
- Dedhia, P.H.; Bertaux-Skeirik, N.; Zavros, Y.; Spence, J.R. Organoid models of human gastrointestinal development and disease. Gastroenterology 2016, 150, 1098–1112. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.R.; Spence, J.R. Gastrointestinal organoids: Understanding the molecular basis of the host-microbe interface. Cell Mol. Gastroenterol Hepatol. 2017, 3, 138–149. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rocjich, B.E.; Tsai, Y.H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stemcell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W.; et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadauld, L.D.; Garcia, S.; Natsoulis, G.; Bell, J.M.; Miotke, L.; Hopmans, E.S.; Xu, H.; Pai, R.K.; Palm, C.; Regan, J.F.; et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2 as a cancer driver in diffuse gastric cancer. Genome Biol. 2014, 15, 428. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yuen, S.T.; Xu, J.; Lee, S.P.; Yan, H.H.; Shi, S.T.; Siu, H.C.; Deng, S.; Chu, K.M.; Law, S.; et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014, 46, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Ruan, K.; Fang, X.; Ouyang, G. MicroRNAs: Novel regulators in the hallmarks of human cancer. Cancer Lett. 2009, 285, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Mak, T.W. Tumours and tremors: How PTEN regulation underlies both. Br. J. Cancer 2006, 94, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Z.; Han, L.; Zhang, A.-L.; Fu, Y.-C.; Yue, X.; Wang, G.-X.; Jia, Z.-F.; Pu, P.-Y.; Zhang, Q.-Y.; Kang, C.-S. MicroRNA221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10, 367. [Google Scholar] [CrossRef]
- Zhang, B.G.; Li, J.F.; Yu, B.Q.; Zhu, Z.G.; Liu, B.Y.; Yan, M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol. Rep. 2012, 27, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijina, T.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Iio, A.; Nakagawa, Y.; Naoe, T.; Tanigawa, N.; Akao, Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology 2009, 77, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Akiyama, Y.; Otsubo, T.; Shimada, S.; Yuasa, Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis 2010, 31, 777–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, N.; Liu, M.; Tang, Q.L.; Chen, X.; Liu, Z.; Bi, F. Effects of microRNA-29 family members on proliferation and invasion of gastric cancer cell lines. Chin. J. Cancer 2010, 29, 603–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.T., Jr.; McCubrey, J.A. The Raf/MEK/ERK signal transduction cascade as a target for chemotherapeutic intervention in leukemia. Leukemia 2002, 16, 486–507. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Xie, Y.; Zhang, H.; Wu, Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med. Oncol. 2012, 29, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Jin, H.; Guo, X.; Xia, L.; Chen, Z.; Bai, M.; Liu, J.; Shang, X.; Wu, K.; et al. MiR-206 inhibits gastric cancer proliferation in part by repressing CyclinD2. Cancer Lett. 2013, 332, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Xu, Y.; Zhang, W.; Deng, Y.; Si, M.; Du, Y.; Yao, H.; Liu, X.; Ke, Y.; Si, J.; et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010, 20, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Guo, L.; Ji, J.; Zhang, J.; Chen, X.; Cai, Q.; Li, J.; Gu, Q.; Liu, B.; Zhu, Z.; et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochem. Biophys. Res. Commun. 2010, 398, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Guo, J.; Miao, Y.; Jiang, Z.; Huan, R.; Zhang, Y.; Li, D.; Zhong, J. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin. Chim. Acta 2009, 400, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yu, J.; Han, T.S.; Park, S.Y.; Namkoong, B.; Kim, D.H.; Hur, K.; Yoo, M.W.; Lee, H.J.; Yang, H.K.; et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009, 37, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.L.; Peng, Z.; Yang, X.; Fan, K.J.; Ye, H.; Li, Z.H.; Wang, Y.; Xu, X.L.; Li, J.; Wang, Y.L.; et al. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int. J. Biol. Sci. 2011, 7, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, X.H.; Li, J.H.; Yang, J.S.; Zhang, E.B.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Ding, Y.; Li, S.Q.; et al. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol. Cancer Ther. 2012, 11, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.H.; Wu, C.W.; Li, A.F.; Chi, C.W.; Lin, W.C. miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 2008, 28, 907–911. [Google Scholar] [PubMed]
- Motoyama, K.; Inoue, H.; Mimori, K.; Tanaka, F.; Kojima, K.; Uetake, H.; Sugihara, K.; Mori, M. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int. J. Oncol. 2010, 36, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhao, Y.; Guo, G.; Li, W.; Zhu, E.D.; Luo, X.; Mao, X.H.; Zou, Q.M.; Yu, P.W.; Zuo, Q.F.; et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS ONE 2012, 7, e41629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Li, D.; Xiao, B.; Miao, Y.; Jiang, Z.; Zhuo, H. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med. Oncol. 2010, 27, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Luo, F.; Li, Q.; Xu, M.; Feng, D.; Zhang, G.; Wu, W. Identification of new aberrantly expressed miRNAs in intestinal-type gastric cancer and its clinical significance. Oncol. Rep. 2011, 26, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Katada, T.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitui, A.; Mori, Y.; Ogawa, R.; Harata, K.; Fujii, Y. microRNA expression profile in undifferentiated gastric cancer. Int. J. Oncol. 2009, 34, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Bandres, E.; Bitarte, N.; Arias, F.; Agorreta, J.; Fortes, P.; Agirre, X.; Zarate, R.; Diaz-Gonzalez, J.A.; Ramirez, N.; Sola, J.J.; et al. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin. Cancer Res. 2009, 15, 2281–2290. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Li, J.; Wang, Y.; Liu, C.; Jia, H.; Jiang, C.; Wang, Y.; Luo, M.; Zhao, H.; Dong, L.; et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis 2014, 35, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.S.; Yang, X.H.; Chen, X.; Wang, X.D.; Hua, J.; Zhou, D.L.; Zhou, B.; Song, Z.S. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014, 588, 2162–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishida, N.; Mimori, K.; Fabbri, M.; Yokobori, T.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin. Cancer Res. 2011, 17, 2725–2733. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, C.; Huang, B.; Li, H.; Zhang, R.; Huang, Y.; Wang, J. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur. J. Gastroenterol Hepatol. 2013, 25, 953–957. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, Y.; Zhang, Y.; Ding, J.; Wu, K.; Fan, D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut 2010, 59, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Zhang, H.; Liu, X.; Gong, T.; Li, M.; Sun, L.; Ji, G.; Shi, Y.; Han, Z.; et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol. Cancer Res. 2011, 9, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Kogo, R.; Mimori, K.; Tanaka, F.; Komune, S.; Mori, M. Clinical significance of miR-146a in gastric cancer cases. Clin. Cancer Res. 2011, 17, 4277–4284. [Google Scholar] [CrossRef] [PubMed]
- Bou Kheir, T.; Futoma-Kazmierczak, E.; Jacobsen, A.; Krogh, A.; Bardram, L.; Hother, C.; Gronbaek, K.; Federspiel, B.; Lund, A.H.; Friis-Hansen, L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol. Cancer 2011, 10, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, M.M.; Wang, C.S.; Tsai, C.Y.; Huang, H.W.; Chi, H.C.; Lin, Y.H.; Lu, P.H. Potential diagnostic, prognostic and therapeutic targets of microRNAs in human gastric cancer. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Guo, F.; Wang, Y.; Lv, Y.; Huo, B.; Wang, L.; Liu, W. MicroRna-200c regulates the sensitivity of chemotherapy of gastric cancer SGC7901/DDP cells by directly targeting RhoE. Pathol. Oncol. Res. 2014, 20, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Swiderski, P.M.; Kaplan, B.E.; Takao, M.; Yasui, A.; Shen, B.; Pfeifer, G.P. Processing of UV damage in vistro by FEN-1 proteins aas part of an alternative DNA excision repair pathway. Biochemistry 1999, 38, 4809–4817. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.B.; Li, Q.L.; Hu, J.F.; Zhang, Q.; Xie, J.P.; Deng, L. miR-124 inhibits growth and invasion of gastric cancer by targeting ROCK1. Asian Pac. J. Cncer Prev. 2014, 15, 6543–6546. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, Z.; Liu, Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol. Med. Rep. 2014, 10, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Pu, J.; Qi, T.; Qi, M.; Li, D.; Xiang, X.; Huang, K.; Tong, Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesi of gastric cancer cell. Mol. Cancer Res. 2013, 11, 182–193. [Google Scholar] [CrossRef] [PubMed]

| TCGA | EBV | MSI | CIN | GS |
| - Males >>> Females - Intestinal-type histology - Frequently located at fundus and body - JAK2 amplification - PIK3CA mutation (80% subtype) inactivating in the kinase domain (exon 20) - ARID1A (55%) mutations - Immune cell signaling enrichment | - >>>Females - Intestinal-type histology - An older age at diagnosis - Mutation in one of several different DNA mismatch repair genes (i.e., MLH1 or MSH2) - Lacks targetable amplifications | - Males >>> Females - Intestinal-type histology - Frequently located at EGJ - RTK-RAS amplifications (EGFR, ERRB2, ERRB3, VEGFA, FGFR2, MET, NRAS/KRAS, JAK2 and PIK3CA) - Amplification of cell cycle genes - TP53 mutations | - Males = Females - Distal location - Diffuse-type histology - An early age at diagnosis - Recurrent CDH1 inactivation, RHOA mutation, ARID1A mutation | |
| ACGR | MSS/TP53+ | MSI | MSS/TP53- | MSS/EMT |
| - Frequently EBV-positive - Intermediate prognosis - Mutations in ARID1A, APC, KRAS, PIK3CQA and SMAD4 | - Distal stomach - Intestinal-type histology - Early stage diagnosis - Favourable prognosis - Hypermutation | - TP53 mutation - Amplification of RTKs - Intermediate prognosis | - Diagnosed at younger age - Diffuse-type histology - Worse prognosis - Low number of mutations |
| Gene | Activity/Positivity | Molecular Alteration | Therapeutic Agents | Ref. |
|---|---|---|---|---|
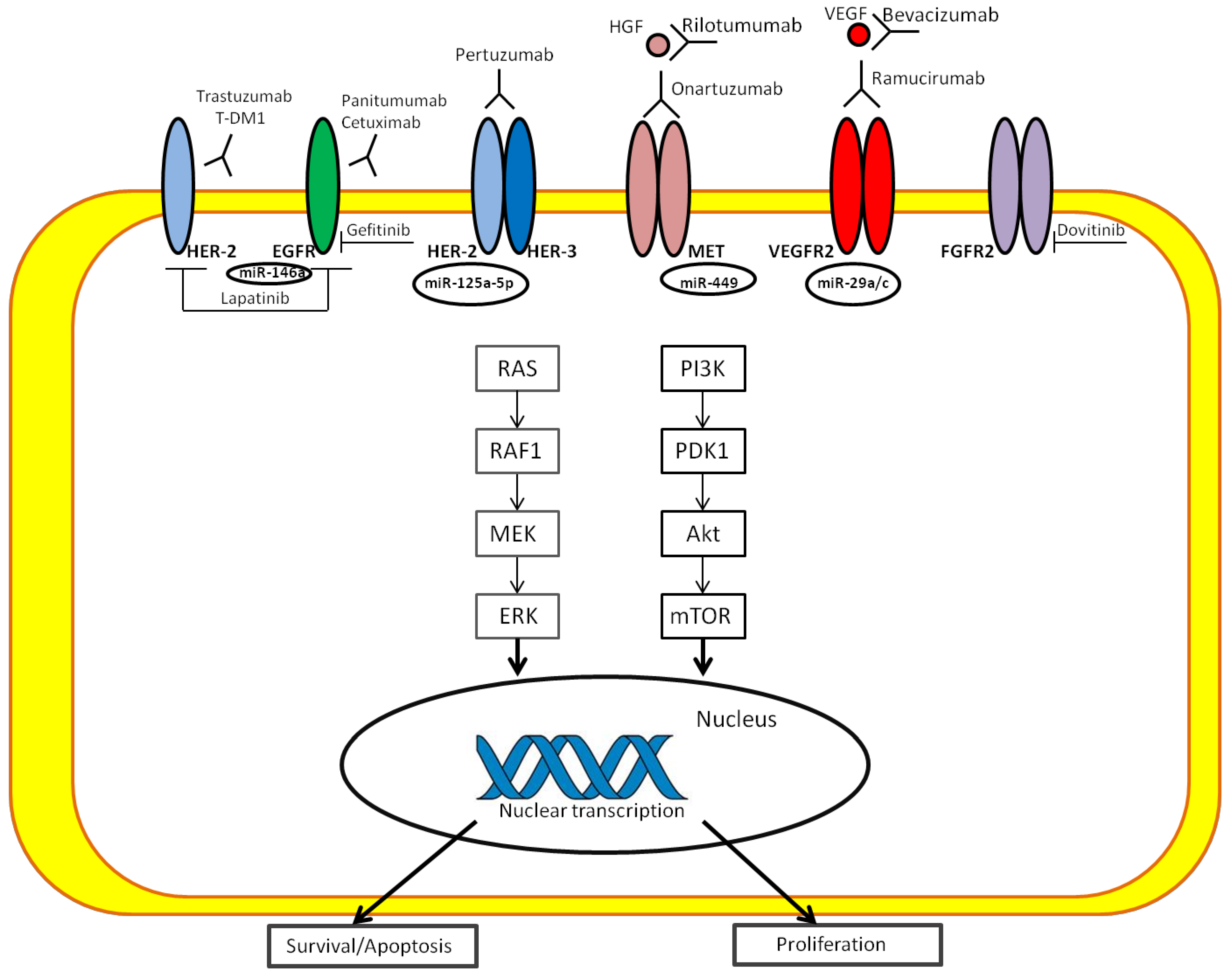
| HER2 | Member of the EGF RTK family Intestinal type (34%), diffuse type (6%) 24% in CIN, 12% in EBV and 7% in MSI subtypes | Amplification Overexpression | Trastuzumab + traditional chemotherapy (ToGA trial) Other anti-HER2 agents (lapatinib, pertuzumab and trastuzumab-emtansine) have not shown significant benefit; resistance is under investigation | [66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81] |
| EGFR | Member of the EGF RTK family; forms heterodimers with HER2 10% in the CIN molecular subtype | Amplification Overexpression | Panitumumab and cetuximab showed disappointing results in two large phase III trials; erlotinib and gefitinib were not effective | [82,83,84,85] |
| MET | RTK family; interacts with HGF 8% in the CIN molecular subtype | Amplification Overexpression | Rilotumumab was associated with significantly longer PFS and OS when added to chemotherapy in treatment-naive molecularly unselected patients with advanced GC; another anti-MET antibody, onartuzumab, did not show any advantage in combination with mFOLFOX | [86,87,88,89,90] |
| VEGF | Factors of angiogenesis 54–90% of GCs | Overexpression | Bevacizumab (AVAGAST trial) did not show increased OS Ramucirumab (RAINBOW trial) + paclitaxel confirmed OS advantage in a non-Asian population | [91,92,93,94,95,96,97,98] |
| FGFR | Fibroblast growth factor receptor family 9% CIN molecular subtype | Amplification | A phase II randomised trial is evaluating the activity of AZD4547, an inhibitor of FGFR 1–2 and 3, compared to paclitaxel in second-line treatment Other ongoing trials are testing dovitinib in FGFR2 amplified GC patients or in combination with docetaxel | [14,34] |
| KRAS | RAS GTPase; recruits the cytosolic protein RAF <5 GCs | Mutation codon 12–13 | No target therapies are currently approved for this alteration in GC | [99] |
| CDH1 | Tumour suppressor gene; encodes E-cadherin, a cell adhesion molecules 37% of the GS molecular subtype | Mutations, hypermethylation, downregulated expression | Treatments targeting EMT are under study | [100,101,102] |
| ARID1A | Tumour suppressor gene involved in chromatin remodelling 20% GS molecular subtype | Inactivating mutations | No target therapies are currently approved for this alteration in GC | [103,104] |
| RHOA | Rho GTPases are intracellular signaling molecules, regulating cytoskeleton organization, cell cycle and cell motility Diffuse type 30% GS molecular subtype | Mutations Interchromosomal translocation (between CLDN18 and ARHGAP26) | A recent trial tested IMAB362, a chimeric IgG1 antibody against CLDN18.2 showing clinical activity in patients with 2+/3+ immunostaining | [105,106,107,108,109] |
| Cons | Pros | |
|---|---|---|
| Cell line xenografts | - monodimensional - no tumour-microenvironment interaction - loss of architecture - genetic modifications | - rapid analysis of drug response - immortal cell lines allow unlimited source of material - low cost, low complexity |
| PDX models | - limited source of material - high failure rate of engraftment - long time for establishment - expensive - tissue must be rapidly processed | - reliable representation of tumour heterogeneity - includes microenvironment - can predict response to drugs |
| Organoids | - no tumour-microenvironment interaction | - high level of architectural and physiological similarity to native tissue - intermediate cost, easy to handle - large-scale drug screening |
| miRNAs | Role | Expression in Tissue | Note | Ref. |
|---|---|---|---|---|
| miR-21 | Diagnostic | Upregulated | Overexpressed miR-21 binds to PDCD4 and can inhibit protein expression; directly related to tumour size, depth of invasion, lymph node metastasis and vascular invasion | [145,146] |
| miR-21 miR-223 miR-218 | Diagnostic | Upregulated Downregulated | - | [147] |
| miR-31 | Diagnostic | Downregulated | - | [148] |
| miR-32 miR-182 miR-143 | Diagnostic | Upregulated | - | [149] |
| miR-106a | Diagnostic | Upregulated | Level of miR-106a is closely related to tumour size, differentiation degree, lymph node and distant metastasis | [141] |
| miR-20 miR-150b miR-451 | Prognostic | Upregulated Upregulated Downregulated | [150,151] | |
| miR-29 | Prognostic | Downregulated | This miRNA is associated with poor prognosis | [152] |
| miR-106b | Prognostic | Upregulated | This miRNA is associated with poor prognosis | [153] |
| miR-125a-5p | Prognostic | Downregulated | Multivariate analysis shows that its downregulation is an independent prognostic factor for survival | [154] |
| miR-206 | Prognostic | Downregulated | mRNA-206 is an independent prognostic factor in GC patients | [155] |
| miR-17-5p miR-21 miR-106a miR-106b miR-7a | Prognostic | Upregulated | - | [142,156] |
| miR-10b miR-21 miR-223 miR-338 let-7a miR-30a-5p miR-126 | Prognostic | - | These seven miRNAs are significantly related to recurrence-free periods and overall survival of patients; an overexpression of miR-223 in primary GC is associated with less survival without metastasis | [157,158] |
| miR-125b miR-199a miR-100 | Prognostic | Upregulated | These miRNAs are associated with progression of GC | [159] |
| Let-7g miR-433 miR-214 | Prognostic | Upregulated Upregulated Downregulated | Levels of these miRNAs are associated with tumour infiltration depth, lymph node metastasis and tumour stage. | [159] |
| miRNAs | Relative Expression | Target Gene | Cell Function | Ref. |
|---|---|---|---|---|
| miR-146a | Upregulated | EGFR | Invasion Migration | [160] |
| miR-449 | Upregulated | MET SIRT1 CDK6 | Cell proliferation Apoptosis Cell cycle | [161] |
| miR-29a/c | Downregulated | VEGF | Vascular cell Metastasis Growth | [162] |
| miR-181c | Upregulated | KRAS NOTCH4 | Cell proliferation | [134] |
| miR-221 miR-222 | Upregulated | CDKN1A CDKN1B CDKN1C | Cell Cycle | [142] |
| miR-200c | Upregulated | CDH RHO | Metastasis Chemoresistance | [163] |
| miR-150 | Upregulated | EGR2 | Apoptosis Cell proliferation | [150] |
| miR-382 | Upregulated | PTEN | Angiogenesis | [164] |
| miR-124 | Upregulated | ROCK1 | Cell proliferation Invasion | [165] |
| miR-125a-5p | Upregulated | ERBB2 E2F3 | Cell proliferation Metastasis Invasion Migration | [154,166] |
| miR-145 | Downregulated | ETS1 | Migration Invasion Angiogenesis | [167] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alessandrini, L.; Manchi, M.; De Re, V.; Dolcetti, R.; Canzonieri, V. Proposed Molecular and miRNA Classification of Gastric Cancer. Int. J. Mol. Sci. 2018, 19, 1683. https://doi.org/10.3390/ijms19061683
Alessandrini L, Manchi M, De Re V, Dolcetti R, Canzonieri V. Proposed Molecular and miRNA Classification of Gastric Cancer. International Journal of Molecular Sciences. 2018; 19(6):1683. https://doi.org/10.3390/ijms19061683
Chicago/Turabian StyleAlessandrini, Lara, Melissa Manchi, Valli De Re, Riccardo Dolcetti, and Vincenzo Canzonieri. 2018. "Proposed Molecular and miRNA Classification of Gastric Cancer" International Journal of Molecular Sciences 19, no. 6: 1683. https://doi.org/10.3390/ijms19061683
APA StyleAlessandrini, L., Manchi, M., De Re, V., Dolcetti, R., & Canzonieri, V. (2018). Proposed Molecular and miRNA Classification of Gastric Cancer. International Journal of Molecular Sciences, 19(6), 1683. https://doi.org/10.3390/ijms19061683









