Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae
Abstract
1. Introduction
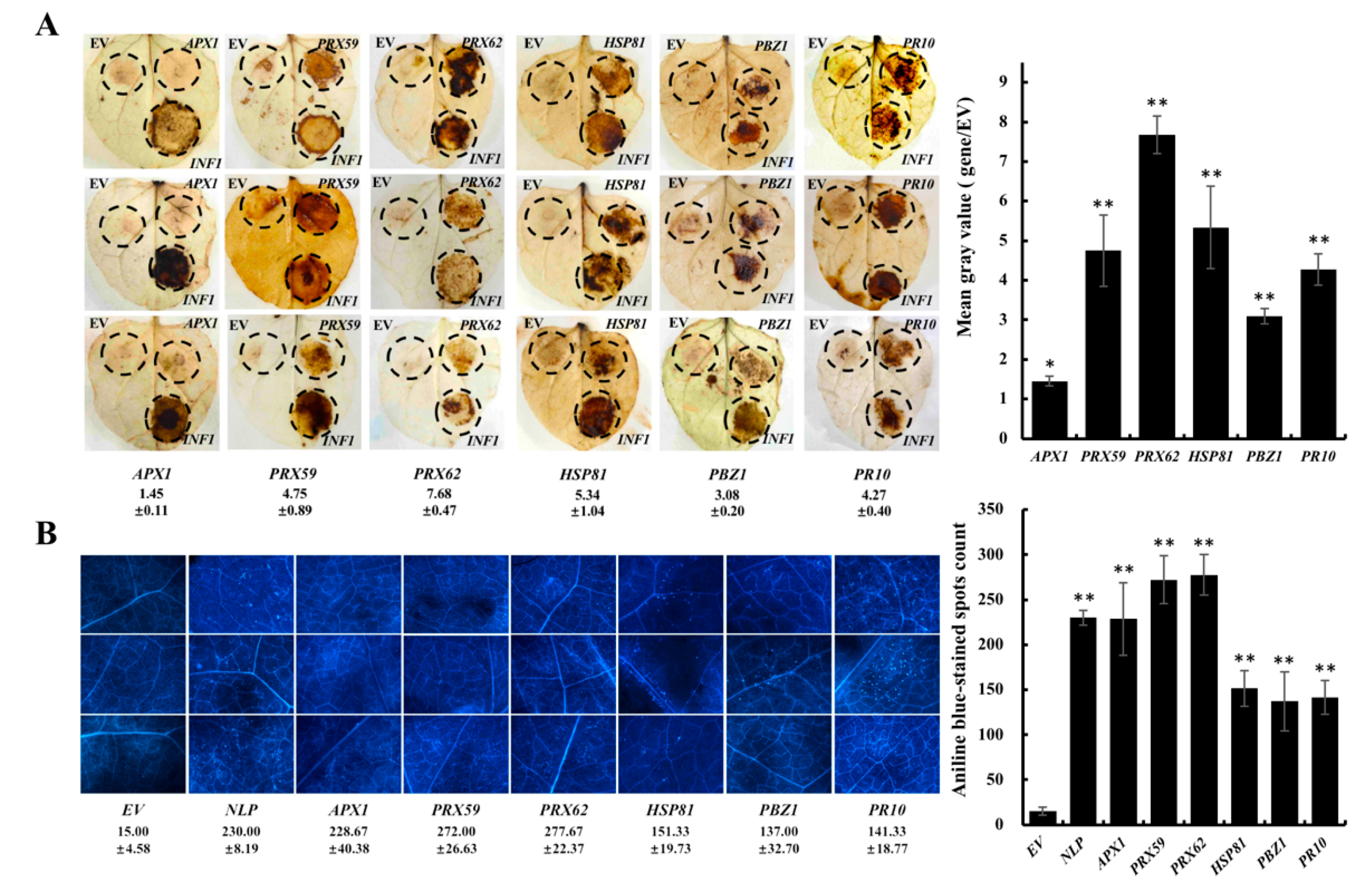
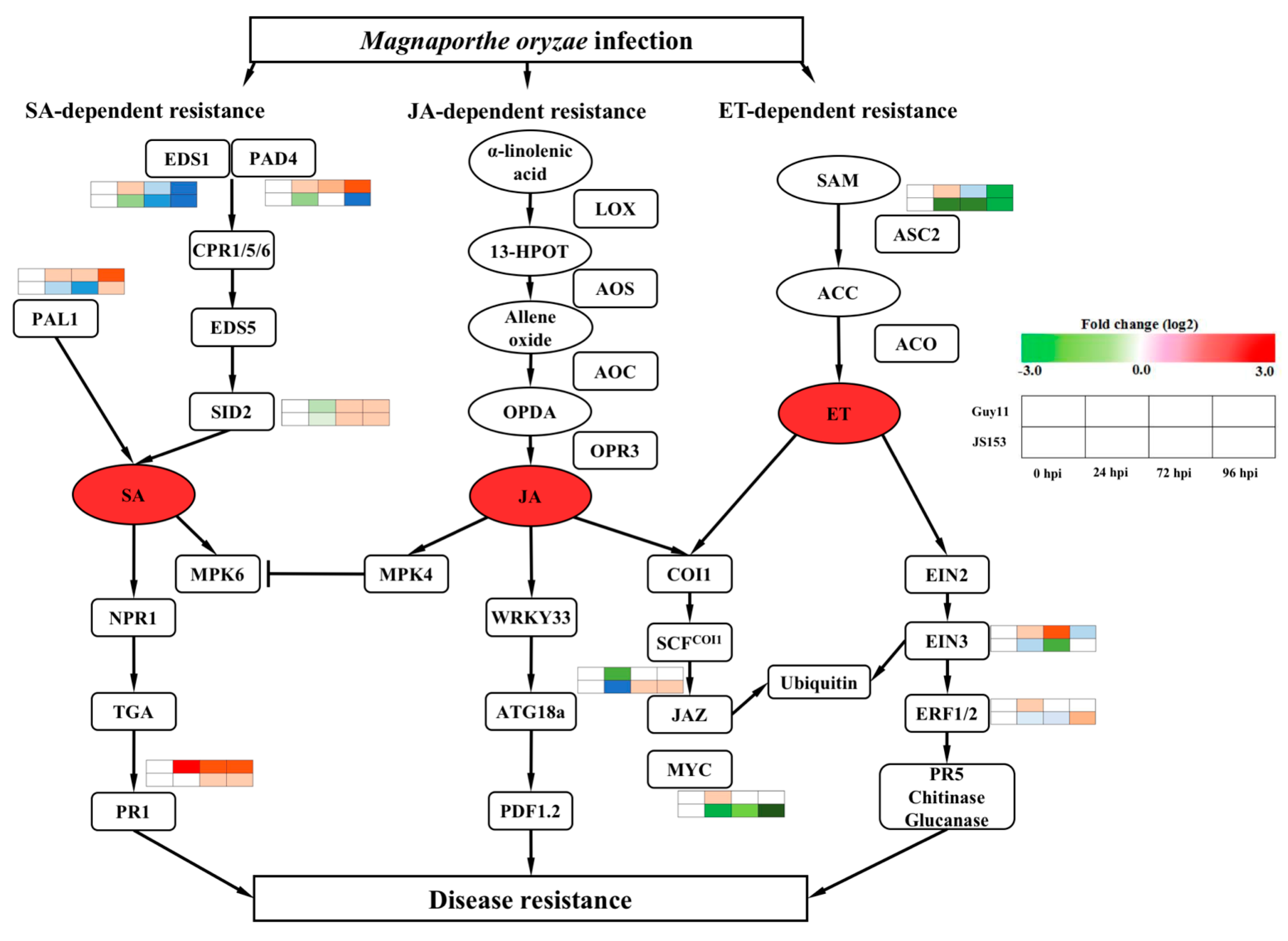
2. Results
3. Discussion
4. Materials and Methods
4.1. Plants and Inoculation
4.2. Protein Extraction
4.3. Protein Digestion, iTRAQ Labeling, and Strong Cation Exchange
4.4. Database Search and iTRAQ Quantification
4.5. Gene Ontology Analysis and Biological Processes Analysis
4.6. RT-PCR Analysis of the Small RNAs and Predicted Targets
4.7. Transient Expression Analysis in N. Benthamiana
4.8. DAB Staining
4.9. Aniline Blue Staining
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nalley, L.; Tsiboe, F.; Durand-Morat, A.; Shew, A.; Thoma, G. Economic and Environmental Impact of Rice Blast Pathogen (Magnaporthe oryzae) Alleviation in the United States. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Talbot, N. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Valent, B. Durable resistance to rice blast. Science 2017, 355, 906–907. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.; Dangl, J. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Ning, Y.; Ding, B.; Wang, X.; Wang, Z.; Wang, G. Recent Progress in Understanding PAMP- and Effector-Triggered Immunity against the Rice Blast Fungus Magnaporthe oryzae. Mole. Plant 2013, 6, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-Mediated Chitin Perception and Immune Signaling Requires Receptor-like Cytoplasmic Kinase 185 to Activate an MAPK Cascade in Rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Xue, Y.; Zhou, J.; Zhang, B.; Chang, H.; Takano, M. Phytochromes Regulate SA and JA Signaling Pathways in Rice and Are Required for Developmentally Controlled Resistance to Magnaporthe grisea. Mol. Plant 2011, 4, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Wang, Y.; Lee, K.; Park, Z.; Park, J.; Wu, J.; Kwon, S.; Lee, Y.; Agrawal, G.; Rakwal, R.; et al. In-depth insight into in vivo apoplastic secretome of rice–Magnaporthe oryzae interaction. J. Proteom. 2013, 78, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, Y.; Wang, Y.; Wu, J.; Park, Z.; Rakwal, R.; KumarAgrawal, G.; Lee, S.; Kang, K. Secretome analysis of differentially induced proteins in rice suspension-cultured cells triggered by rice blast fungus and elicitor. Proteomics 2009, 9, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Yang, C.; Li, L.; Jiang, L.; Wu, Y.; Wu, C.; Bu, Q.; Xia, G.; Liu, X.; Luo, Y.; et al. Rice Plasma Membrane Proteomics Reveals Magnaporthe oryzae Promotes Susceptibility by Sequential Activation of Host Hormone Signaling Pathways. Mol. Plant Microbe Interact. 2016, 29, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Franck, W.; Gokce, E.; Oh, Y.; Muddiman, D.; Dean, R. Temporal Analysis of the Magnaporthe oryzae Proteome During Conidial Germination and Cyclic AMP (cAMP)-mediated Appressorium Formation. Mol. Cell. Proteom. 2013, 12, 2249–2265. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.; Huang, Y.; Marchese, J.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Zieske, L. A perspective on the use of iTRAQ (TM) reagent technology for protein complex and profiling studies. J. Exp. Bot. 2006, 57, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae Induces the Expression of a MicroRNA to Suppress the Immune Response in Rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Li, Y.; Li, X.; Wei, L.-L.; Pan, Z.; Jiang, T.-T.; Chen, Z.-L.; Wang, C.; Cao, W.-M.; Zhang, X.; et al. Serum protein S100A9, SOD3, and MMP9 as new diagnostic biomarkers for pulmonary tuberculosis by iTRAQ-coupled two-dimensional LC-MS/MS. Proteomics 2015, 15, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, W.; Cao, J.; Meng, F.; Yu, Y.; Huang, J.; Jiang, L.; Liu, M.; Zhang, Z.; Chen, X.; et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 2017, 89, 338–353. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Munne-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.; Hwang, D.; Kang, S.; Kim, H.; Lee, B.; Lee, J.; Kang, K. Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 2004, 4, 3569–3578. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ekramoddoullah, A. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 2006, 68, 3–13. [Google Scholar] [CrossRef]
- Takahashi, A.; Kawasaki, T.; Henmi, K.; Shii, K.; Kodama, O.; Satoh, H.; Shimamoto, K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 1999, 17, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Qin, Y.; Han, J.; Zhang, L.; Xu, X.; Liu, X.; Wang, C. Expression analysis of innate immunity related genes in the true/field blast resistance gene-mediated defence response. Biotechnol. Biotechnol. Equip. 2014, 28, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.J.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stresses. Plant Physiol. 2000, 123, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Lu, Y.; Li, K.; Lin, L.; Zheng, H.; Yan, F.; Chen, J. Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol. Plant Pathol. 2014, 15, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Hwang, B. Pepper Heat Shock Protein 70a Interacts with the Type III Effector AvrBsT and Triggers Plant Cell Death and Immunity. Plant Physiol. 2015, 167. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.; Hu, J.; Liu, J.; Liu, K. A Heat Shock Protein Gene, CsHsp45.9, Involved in the Response to Diverse Stresses in Cucumber. Biochem. Genet. 2012, 50, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.H.; Dong, X.N. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Jing, M.; Ma, H.; Li, H.; Guo, B.; Zhang, X.; Ye, W.; Wang, H.; Wang, Q.; Wang, Y. Differential regulation of defense-related proteins in soybean during compatible and incompatible interactions between Phytophthora sojae and soybean by comparative proteomic analysis. Plant Cell Rep. 2015, 34, 1263–1280. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Mu, J.; Chen, L.; Feng, J.; Hu, J.; Li, L.; Zhou, J.; Zuo, J. S-Nitrosylation Positively Regulates Ascorbate Peroxidase Activity during Plant Stress Responses. Plant Physiol. 2015, 167, 1604–1753. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Kim, Y.; Lee, S.; Hong, J.; Hwang, B. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, C.; Brown, I.; Mansfield, J. Localized changes in peroxidase activity accompany hydrogen peroxide generation during the development of a nonhost hypersensitive reaction in lettuce. Plant Physiol. 1998, 118, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Diaz, F.; Orobio, R.; Chavarriaga, P.; Toro-Perea, N. Differential expression patterns among heat-shock protein genes and thermal responses in the whitefly Bemisia tabaci (MEAM 1). J. Therm. Biol. 2015, 52, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.; Chen, L.; Nakashima, A.; Hara, S.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 2007, 19, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.; Wang, Y.; Yu, S.; Choi, I.; Kim, Y.; Kim, W.; Agrawal, G.; Rakwal, R.; Kang, K. The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol. Cells 2011, 31, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kwon, S.; Wu, J.; Choi, J.; Lee, Y.; Agrawal, G.; Tamogami, S.; Rakwal, R.; Park, S.; Kim, B.; et al. Transcriptome Analysis of Early Responsive Genes in Rice during Magnaporthe oryzae Infection. Plant Pathol. J. 2014, 30, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.; Liu, G.; Zhou, B.; Bellizzi, M.; Zeng, L.; Dai, L.; Han, B.; Wang, G. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 2006, 172, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.; Katiyar-Agarwal, S.; Huang, H.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 Regulates Innate Immunity via miRNA393*-Mediated Silencing of a Golgi-Localized SNARE Gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Brown, S.; Patrick, E.; Brearley, C.; Turner, J. Enhanced transcription of the arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. Plant Cell 2003, 15, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Clay, N.; Adio, A.; Denoux, C.; Jander, G.; Ausubel, F. Glucosinolate Metabolites Required for an Arabidopsis Innate Immune Response. Science 2009, 323, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]



| Protein ID | Name | Annotation | Guy11 | JS153 | ||
|---|---|---|---|---|---|---|
| 24/0 hpi | 72/0 hpi | 24/0 hpi | 72/0 hpi | |||
| response to oxidative stress | ||||||
| Q652L6 | MDAR3 | cellular oxidant detoxification | 1.20 | 1.33 | 0.85 | 1.35 |
| B7E6Z4 | APX1 | hydrogen peroxide catabolic process | 1.29 | 1.29 | 0.80 | 1.29 |
| I1Q8M5 | TRXh1 | oxidoreductase activity | 1.11 | 1.35 | 0.95 | 1.47 |
| A0A0E0Q2V5 | CATB | hydrogen peroxide catabolic process | 1.17 | 1.81 | 0.88 | 1.86 |
| Q7XSU7 | PRX62 | hydrogen peroxide catabolic process | 1.57 | 3.44 | 0.98 | 2.67 |
| Q9ST80 | PRX59 | hydrogen peroxide catabolic process | 1.59 | 2.99 | 0.98 | 2.72 |
| Q5U1T0 | PRX13 | hydrogen peroxide catabolic process | 1.55 | 2.70 | 0.91 | 3.05 |
| Q5Z4D3 | PRX78 | hydrogen peroxide catabolic process | 1.44 | 1.71 | 0.75 | 1.72 |
| Q5Z7J2 | PRX86 | hydrogen peroxide catabolic process | 1.71 | 2.04 | 1.14 | 1.73 |
| Q654S1 | PRX12 | hydrogen peroxide catabolic process | 1.42 | 1.54 | 0.96 | 1.82 |
| Q6AVZ8 | PRX65 | hydrogen peroxide catabolic process | 1.26 | 1.41 | 0.78 | 1.35 |
| Q6ESJ0 | GPX3 | glutathione peroxidase activity | 1.15 | 1.36 | 0.93 | 1.58 |
| Q6EUS1 | PRX27 | hydrogen peroxide catabolic process | 1.35 | 1.56 | 0.84 | 1.79 |
| Q6Z4E4 | ALDH6B2 | methylmalonate-semialdehyde dehydrogenase activity | 1.27 | 1.66 | 0.87 | 1.82 |
| Q7XHB3 | PRX125 | hydrogen peroxide catabolic process | 1.39 | 1.77 | 1.02 | 1.75 |
| Q9FEV2 | riPHGPX | glutathione peroxidase activity | 1.36 | 1.75 | 0.8 | 1.54 |
| Q8W317 | NADH dehydrogenase | NADH dehydrogenase activity | 1.25 | 1.37 | 0.79 | 1.53 |
| A0A0E0PU51 | alkaline α-galactosidase | catalytic activity | 1.54 | 1.62 | 1.02 | 1.83 |
| response to biotic stimulus | ||||||
| Q7XPU1 | Harpin-induced 1 domain containing protein | signal transducer activity | 1.34 | 1.50 | 0.93 | 1.47 |
| Q40707 | PBZ1 | response to biotic stimulus, defense response | 1.14 | 9.22 | 0.63 | 2.77 |
| I1QJW3 | HSP81 | response to stress, ATP binding | 1.12 | 1.72 | 0.84 | 1.62 |
| Q75T45 | RSOsPR10 | pathogenesis-related protein | 1.58 | 8.25 | 0.66 | 2.68 |
| Q75L45 | OsRLCK178 | cell surface receptor signaling pathway | 1.49 | 1.55 | 1.02 | 1.66 |
| A0A0E0PMK8 | OsGDI1 | protein transport | 1.17 | 1.6 | 0.77 | 1.37 |
| Q945E9 | JIOsPR10 | response to biotic stimulus | 1.92 | 2.55 | 1.03 | 2.19 |
| response to cold, salt stress, water deprivation | ||||||
| Q8LHG8 | Os01g0542000 | isomerase activity | 1.35 | 1.53 | 0.87 | 1.56 |
| I1QGF2 | YchF1 | hydrolyzes ATP | 1.22 | 1.31 | 0.85 | 1.37 |
| Q7XXS0 | RMtATPd2 | mitochondrial membrane ATP synthase | 1.29 | 1.46 | 0.92 | 1.41 |
| Q7XUC9 | Histone H4 | transcription regulation, DNA repair, DNA replication | 0.96 | 1.44 | 0.91 | 1.32 |
| I1PYW0 | Os6PGDH1 | phosphogluconate dehydrogenase activity | 1.36 | 1.70 | 0.89 | 1.59 |
| I1PUR5 | UspA | response to stress | 3.26 | 1.18 | 2.17 | 1.38 |
| response to ion stress | ||||||
| Q5JK10 | Os01g0926300 | response to cadmium ion | 1.15 | 1.59 | 0.98 | 1.52 |
| Q6H734 | Os02g0198600 | ubiquitin binding | 1.35 | 1.53 | 0.87 | 1.50 |
| S4TZU3 | Os02g0621700 | magnesium ion binding | 1.23 | 1.49 | 0.79 | 1.31 |
| response to chemical | ||||||
| Q5W676 | HXK5 | fructose and glucose phosphorylating enzyme | 1.06 | 1.27 | 0.79 | 1.43 |
| Q2QYK6 | chalcone isomerase | chalcone isomerase activity | 1.15 | 1.4 | 0.94 | 1.59 |
| Q852M0 | GDH1 | glutamate dehydrogenase activity | 1.37 | 1.49 | 0.83 | 1.45 |
| Q8S718 | OsGSTU23 | glutathione transferase activity | 1.28 | 1.73 | 0.78 | 1.68 |
| response to physical stress | ||||||
| Q10LV7 | LOC_Os03g21560 | cellular response to light intensity | 1.20 | 1.36 | 0.81 | 1.44 |
| Q7XRB6 | Os04g0435700 | response to UV-B, photoreceptor activity | 1.39 | 1.46 | 0.75 | 1.46 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, S.; Nie, P.; Ding, S.; Zheng, L.; Chen, C.; Feng, R.; Wang, Z.; Wang, L.; Wang, J.; Fang, Z.; et al. Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae. Int. J. Mol. Sci. 2018, 19, 1950. https://doi.org/10.3390/ijms19071950
Lin S, Nie P, Ding S, Zheng L, Chen C, Feng R, Wang Z, Wang L, Wang J, Fang Z, et al. Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae. International Journal of Molecular Sciences. 2018; 19(7):1950. https://doi.org/10.3390/ijms19071950
Chicago/Turabian StyleLin, Siyuan, Pingping Nie, Shaochen Ding, Liyu Zheng, Chen Chen, Ruiying Feng, Zhaoyun Wang, Lin Wang, Jianan Wang, Ziwei Fang, and et al. 2018. "Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae" International Journal of Molecular Sciences 19, no. 7: 1950. https://doi.org/10.3390/ijms19071950
APA StyleLin, S., Nie, P., Ding, S., Zheng, L., Chen, C., Feng, R., Wang, Z., Wang, L., Wang, J., Fang, Z., Zhou, S., Ma, H., & Zhao, H. (2018). Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae. International Journal of Molecular Sciences, 19(7), 1950. https://doi.org/10.3390/ijms19071950





