Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine
Abstract
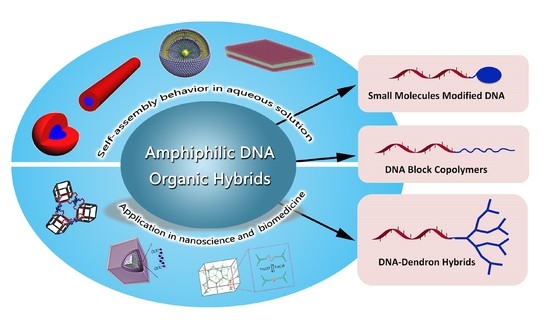
:1. Introduction
2. Small Molecules Modified DNA
2.1. Self-Assembly of Small Hydrophobic Molecule Modified DNA
2.1.1. Lipid-DNA Conjugates
2.1.2. Pyrene-Modified DNA
2.1.3. Perylenediimide-Modified DNA
2.1.4. Other Small Hydrophobic Molecule-DNA Conjugates
2.2. Application of Small Molecule Modified DNA
3. Amphiphilic DNA Block Copolymers
3.1. Self-Assembly of Amphiphilic DNA Block Copolymers
3.2. Application of Amphiphilic DNA Block Copolymers
4. Amphiphilic DNA-Dendron Hybrids
4.1. Self-Assembly of Amphiphilic DNA-Dendron Hybrids
4.2. Application of Amphiphilic DNA–Dendron Hybrids
5. Conclusions and Prospective
Author Contributions
Acknowledgments
Conflicts of Interest
Appendix A
| Category | Building Block | Assemblies | Application | Ref. |
|---|---|---|---|---|
| SMMD 1 | Lipid-DNA Conjugates | Vesicles, micelles, liposomes | Cargo release, drug delivery | [23,24,25,26,27,28,29,30,31,32,53,54,55] |
| SMMD 1 | Pyrene Modified DNA | Helical nanoribbons | Drug delivery | [35,36,37,38] |
| SMMD 1 | PDI 4 Modified DNA | Supramolecular polymers, fibers | Construct novel structural motifs | [40,41,42,43,44,45,46,47] |
| SMMD 1 | PpIX 5–DNA hybrids | 2D nanocages | ROS generation | [48,49] |
| DBCs 2 | DNA-b-PPO | Spherical micelles, nanofibers | Target drug delivery | [57,59,62,63] |
| DBCs 2 | PPO-dsDNA-PPO | Large spherical micelles | Construct novel structural motifs | [64] |
| DBCs 2 | DNA-b-PPO & AuNPs | Heterovesicles | Frame-guided assembly | [70] |
| DBCs 2 | ssDNA-b-PPO-b-ssDNA and Y-scafford DNA structure | 3D DNA network | Intramolecular collapse of PPO | [71] |
| DBCs 2 | DNA-b-PNIPAAm; | Spherical micelles | Drug delivery | [65] |
| DBCs 2 | DNA-b-PNIPAM-b-PMA | Spherical micelles and cylinders | Morphological control | [66] |
| DBCs 2 | DNA-b-PCL, PEG-b-PCL and the PCL homopolymer | Spherical micelles | High cellular uptake and effective antisense gene regulation | [68,69] |
| DBCs 2 | PMA-b-DNA and PBD-b-PEO | Giant polymersomes with DNA islands | Construct novel nanostructures | [72] |
| DBCs 2 | Sequence-defined DNA block copolymer on 3D DNA cage | DNA cage-ring structures, DNA–micelle cages | Targeted drug delivery and diagnostics | [73,74] |
| DDHs 3 | DNA–PDDB hybrid | Nanofibers | Drug delivery | [76,77] |
| DDHs 3 | DNA–PDMC hybrid | Spherical micelles and nanofibers | Carry Nile Red molecules and positioning AuNPs | [78] |
| DDHs 3 | DNA–DDOEG hybrid and gold nanoparticle | Heterovesicles | Biomimics and frame-guided assembly | [79] |
| DDHs 3 | DNA–DDOEG hybrid and 2D or 3D DNA Origami | 2D nanosheets, cuboid and dumbbell-shaped hetero-vesicles | Higher ordered nanostructures | [80,81,82] |
References
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, J.L.; Liu, M.H.; Liu, Y.; Yan, H. Spatially-interactive biomolecular networks organized by nucleic acid nanostructures. Acc. Chem. Res. 2012, 45, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Meyer, T.A.; Pan, V.; Dutta, P.K.; Ke, Y. The beauty and utility of DNA origami. Chem 2017, 2, 359–382. [Google Scholar] [CrossRef]
- Bathe, M.; Rothemund, P.W.K. DNA nanotechnology: A foundation for programmable nanoscale materials. MRS Bull. 2017, 42, 882–888. [Google Scholar] [CrossRef]
- Nummelin, S.; Kommeri, J.; Kostiainen, M.A.; Linko, V. Evolution of structural DNA nanotechnology. Adv. Mater. 2018, 30, 1703721. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Yurke, B.; Turberfield, A.J.; Mills, A.P.; Simmel, F.C.; Neumann, J.L. A DNA-fuelled molecular machine made of DNA. Nature 2000, 406, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhang, X.; Shen, Z.; Seeman, N.C. A robust DNA mechanical device controlled by hybridization topology. Nature 2002, 415, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, D. DNA nanomachines and their functional evolution. Chem. Commun. 2009, 2625–2636. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, Y.; Simmel, F.C. Nucleic acid based molecular devices. Angew. Chem. Int. Ed. 2011, 50, 3124–3156. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Ji, C.; Shi, J.; Pridgen, E.M.; Frieder, J.; Wu, J.; Farokhzad, O.C. DNA self-assembly of targeted near-infrared-responsive gold nanoparticles for cancer thermo-chemotherapy. Angew. Chem. Int. Ed. 2012, 51, 11853–11857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef] [PubMed]
- Sacca, B.; Niemeyer, C.M. Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 2011, 40, 5910–5921. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Fullhart, P.; Mirkin, C.A. Reversible and chemically programmable micelle assembly with DNA block-copolymer amphiphiles. Nano Lett. 2004, 4, 1055–1058. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, D.; Yang, Z. A brief review of methods for terminal functionalization of DNA. Methods 2014, 67, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Alemdaroglu, F.E.; Herrmann, A. DNA meets synthetic polymers—Highly versatile hybrid materials. Org. Biomol. Chem. 2007, 5, 1311–1320. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Herrmann, A. Nucleic acid amphiphiles: Synthesis and self-assembled nanostructures. Chem. Soc. Rev. 2011, 40, 5745–5755. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, T.; Herrmann, A. DNA block copolymers: Functional materials for nanoscience and biomedicine. Acc. Chem. Res. 2012, 45, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liang, F.; Liu, S. Self-assembly of DNA-based nanomaterials and potential application in drug delivery. Curr. Top. Med. Chem. 2017, 17, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Jin, X.; Mou, Q.; Zhang, C. Recent progress on DNA block copolymer. Chin. Chem. Lett. 2017, 28, 1822–1828. [Google Scholar] [CrossRef]
- Bandy, T.J.; Brewer, A.; Burns, J.R.; Marth, G.; Nguyen, T.; Stulz, E. DNA as supramolecular scaffold forfunctional molecules: Progress in DNA nanotechnology. Chem. Soc. Rev. 2011, 40, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Malinovskii, V.L.; Wenger, D.; Haner, R. Nucleic acid-guided assembly of aromatic chromophores. Chem. Soc. Rev. 2010, 39, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, Z.; Kang, H.; Wu, Y.; Sefan, K.; Tan, W. DNA-based micelles: Synthesis, micellar properties and size-dependent cell permeability. Chem. Eur. J. 2010, 16, 3791–3797. [Google Scholar] [CrossRef] [PubMed]
- Patwa, A.; Gissot, A.; Bestel, I.; Barthelemy, P. Hybrid lipid oligonucleotide conjugates: Synthesis, self-assemblies and biomedical applications. Chem. Soc. Rev. 2011, 40, 5844–5854. [Google Scholar] [CrossRef] [PubMed]
- Dentinger, P.M.; Simmons, B.A.; Cruz, E.; Sprague, M. DNA-mediated delivery of lipophilic molecules via hybridization to DNA-based vesicular aggregates. Langmuir 2006, 22, 2935–2937. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.P.; Chien, M.P.; Ku, T.H.; Rush, A.M.; Gianneschi, N.C. Smart lipids for programmable nanomaterials. Nano Lett. 2010, 10, 2690–2693. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, Y.; Zhang, Y.; Liu, D.; Yang, Z. The assembly of DNA amphiphiles at liquid crystal-aqueous interface. Nanomaterials 2016, 6, 229. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Liu, X.; Bai, H.; Wang, R.; Tan, J.; Peng, X.; Tan, W. Engineering stability-tunable DNA micelles using photocontrollable dissociation of an intermolecular G-quadruplex. ACS Nano 2017, 11, 12087–12093. [Google Scholar] [CrossRef] [PubMed]
- Cozzoli, L.; Gjonaj, L.; Stuart, M.C.A.; Poolman, B.; Roelfes, G. Responsive DNA G-quadruplex micelles. Chem. Commun. 2018, 54, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liu, D. Frame-guided assembly of amphiphiles. Chem. Eur. J. 2015, 21, 18018–18023. [Google Scholar] [CrossRef] [PubMed]
- Beales, P.A.; Geerts, N.; Inampudi, K.K.; Shigematsu, H.; Wilson, C.J.; Vanderlick, T.K. Reversible assembly of stacked membrane nanodiscs with reduced dimensionality and variable periodicity. J. Am. Chem. Soc. 2013, 135, 3335–3338. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang, Z.; Liu, D. Using small molecules to prepare vesicles with designable shapes and sizes via frame-guided assembly strategy. Small 2015, 11, 3768–3771. [Google Scholar] [CrossRef] [PubMed]
- Winiger, C.B.; Langenegger, S.M.; Khorev, O.; Häner, R. Influence of perylenediimide-pyrene supramolecular interactions on the stability of DNA-based hybrids: Importance of electrostatic complementarity. Beilstein J. Org. Chem. 2014, 10, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Krasheninina, O.A.; Novopashina, D.S.; Apartsin, E.K.; Venyaminova, A.G. Recent advances in nucleic acid targeting probes and supramolecular constructs based on pyrene-modified oligonucleotides. Molecules 2017, 22, 2108. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Häner, R. From ribbons to networks: Hierarchical organization of DNA-grafted supramolecular polymers. J. Am. Chem. Soc. 2015, 137, 14051–14054. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Häner, R. Pathway diversity in the self-Assembly of DNA-derived bioconjugates. Bioconjug. Chem. 2016, 27, 2755–2761. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Vybornyi, M.; Häner, R. Functional DNA-grafted supramolecular polymers—Chirality, cargo binding and hierarchical organization. Chem. Commun. 2017, 53, 5179–5181. [Google Scholar] [CrossRef] [PubMed]
- Winiger, C.B.; Langenegger, S.M.; Calzaferri, G.; Häner, R. Formation of two homo-chromophoric H-aggregates in DNA-assembled alternating dye stacks. Angew. Chem. Int. Ed. 2015, 54, 3643–3647. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Müllen, K.; Yin, M. Water-soluble perylenediimides: Design concepts and biological applications. Chem. Soc. Rev. 2016, 45, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Abdalla Moustafa, A.; Bayer, J.; Rädler Joachim, O.; Müllen, K. Synthesis and self-assembly of perylenediimide–oligonucleotide conjugates. Angew. Chem. Int. Ed. 2004, 43, 3967–3970. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Wagenknecht, H.-A. Perylene-3,4:9,10-tetracarboxylic acid bisimide dye as an artificial DNA base surrogate. Org. Lett. 2006, 8, 4191–4194. [Google Scholar] [CrossRef] [PubMed]
- Lewis, F.D.; Zhang, L.G.; Kelley, R.F.; McCamant, D.; Wasielewski, M.R. A Perylenedicarboxamide linker for DNA hairpins. Tetrahedron 2007, 63, 3457–3464. [Google Scholar] [CrossRef]
- Cordes, T.; Vogelsang, J.; Anaya, M.; Spagnuolo, C.; Gietl, A.; Summerer, W.; Herrmann, A.; Müllen, K.; Tinnefeld, P. Single-molecule redox blinking of perylene diimide derivatives in water. J. Am. Chem. Soc. 2010, 132, 2404–2409. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Guo, K.; Yu, J.; Sun, H.; Tang, J.; Shen, J.; Müllen, K.; Yang, W.; Yin, M. A unique perylene-based DNA intercalator: Localization in cell nuclei and inhibition of cancer cells and tumors. Small 2014, 10, 4087–4092. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, M.; Zheng, Y.; Long, H.; Zeidan, T.A.; Schatz, G.C.; Vura-Weis, J.; Wasielewski, M.R.; Zuo, X.B.; Tiede, D.M.; Lewis, F.D. Hydrophobic dimerization and thermal dissociation of perylenediimide-linked DNA hairpins. J. Am. Chem. Soc. 2009, 131, 5920–5929. [Google Scholar] [CrossRef] [PubMed]
- Neelakandan, P.P.; Pan, Z.; Hariharan, M.; Zheng, Y.; Weissman, H.; Rybtchinski, B.; Lewis, F.D. Hydrophobic self-assembly of a perylenediimide-linked DNA dumbbell into supramolecular polymers. J. Am. Chem. Soc. 2010, 132, 15808–15813. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Weissman, H.; Krieg, E.; Votaw, K.A.; McCullagh, M.; Rybtchinski, B.; Lewis, F.D. Self-assembly of perylenediimide–single-strand-DNA conjugates: Employing hydrophobic interactions and DNA base-pairing to create a diverse structural space. Chem. Eur. J. 2017, 23, 10328–10337. [Google Scholar] [CrossRef] [PubMed]
- Stulz, E. Nanoarchitectonics with porphyrin functionalized DNA. Acc. Chem. Res. 2017, 50, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Monisha, M.; Anindya, R.; Das, P. Self assembled nanocages from DNA–protoporphyrin hybrid molecules. RSC Adv. 2015, 5, 89025–89029. [Google Scholar] [CrossRef] [Green Version]
- Albert, S.K.; Sivakumar, I.; Golla, M.; Thelu, H.V.; Krishnan, N.; Varghese, R. DNA-decorated two-dimensional crystalline nanosheets. J. Am. Chem. Soc. 2017, 139, 17799–17802. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.K.; Thelu, H.V.P.; Golla, M.; Krishnan, N.; Varghese, R. Modular synthesis of supramolecular DNA amphiphiles through host-guest interactions and their self-assembly into DNA-decorated nanovesicles. Nanoscale 2017, 9, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Gissot, A.; Camplo, M.; Grinstaff, M.W.; Barthélémy, P. Nucleoside, nucleotide and oligonucleotide based amphiphiles: A successful marriage of nucleic acids with lipids. Org. Biomol. Chem. 2008, 6, 1324–1333. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Yang, J.; Liu, Q.; Willem de Vries, J.; Gruszka, A.; Rodriguez-Pulido, A.; Crielaard, B.J.; Kros, A.; Herrmann, A. Efficient fusion of liposomes by nucleobase quadruple-anchored DNA. Chem. Eur. J. 2017, 23, 9391–9396. [Google Scholar] [CrossRef] [PubMed]
- Ries, O.; Löffler, P.M.G.; Rabe, A.; Malavan, J.J.; Vogel, S. Efficient liposome fusion mediated by lipid-nucleic acid conjugates. Org. Biomol. Chem. 2017, 15, 8936–8945. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.; Minten, I.J.; Anaya, D.-M.; Musser, A.J.; Brasch, M.; Nolte, R.J.M.; Müllen, K.; Cornelissen, J.J.L.M.; Herrmann, A. Virus-like particles templated by DNA micelles: A general method for loading virus nanocarriers. J. Am. Chem. Soc. 2010, 132, 7834–7835. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Park, T.G. Novel polymer-DNA hybrid polymeric micelles composed of hydrophobic poly(d,l-lactic-co-glycolic acid) and hydrophilic oligonucleotides. Bioconjugate Chem. 2001, 12, 917–923. [Google Scholar] [CrossRef]
- Alemdaroglu, F.E.; Ding, K.; Berger, R.; Herrmann, A. DNA-templated synthesis in three dimensions: Introducing a micellar scaffold for organic reactions. Angew. Chem. Int. Ed. 2006, 45, 4206–4210. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.-P.; Rush, A.M.; Thompson, M.P.; Gianneschi, N.C. Programmable shape-shifting micelles. Angew. Chem. Int. Ed. 2010, 49, 5076–5080. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, L.; Liu, Y.; Yang, Z.; He, Y.-M.; Li, Z.; Fan, Q.-H.; Liu, D. pH-induced morphology-shifting of DNA-b-poly(propylene oxide) assemblies. Chem. Commun. 2012, 48, 9753–9755. [Google Scholar] [CrossRef] [PubMed]
- Kamps, A.C.; Cativo, M.H.M.; Chen, X.-J.; Park, S.-J. Self-assembly of DNA-coupled semiconducting block copolymers. Macromolecules 2014, 47, 3720–3726. [Google Scholar] [CrossRef]
- Kedracki, D.; Chekini, M.; Maroni, P.; Schlaad, H.; Nardin, C. Synthesis and self-assembly of a DNA molecular brush. Biomacromolecules 2014, 15, 3375–3382. [Google Scholar] [CrossRef] [PubMed]
- Alemdaroglu, F.E.; Alemdaroglu, N.C.; Langguth, P.; Herrmann, A. DNA block copolymer micelles—A combinatorial tool for cancer nanotechnology. Adv. Mater. 2008, 20, 899–902. [Google Scholar] [CrossRef]
- Alemdaroglu, F.E.; Wang, J.; Borsch, M.; Berger, R.; Herrmann, A. Enzymatic control of the size of DNA block copolymer nanoparticles. Angew. Chem. Int. Ed. 2008, 47, 974–976. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Song, Y.; Zhao, Z.; Zhang, S.; Yang, Z.; Li, Z.; Li, M.; Fan, Q.-H.; Liu, D. Preparation and self-assembly of supramolecular Coil–Rod–Coil triblock copolymer PPO–dsDNA–PPO. Macromolecules 2015, 48, 7550–7556. [Google Scholar] [CrossRef]
- Fujita, M.; Hiramine, H.; Pan, P.; Hikima, T.; Maeda, M. Effects of complementary DNA and salt on the thermoresponsiveness of Poly(N-isopropylacrylamide)-b-DNA. Langmuir 2016, 32, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-J.; Hu, X.; Park, S.-J. Multimodal shape transformation of dual-responsive DNA block copolymers. J. Am. Chem. Soc. 2016, 138, 14941–14947. [Google Scholar] [CrossRef] [PubMed]
- Alemdaroglu, F.E.; Alemdaroglu, N.C.; Langguth, P.; Herrmann, A. Cellular uptake of DNA block copolymer micelles with different shapes. Macromol. Rapid Commun. 2008, 29, 326–329. [Google Scholar] [CrossRef]
- Zhang, C.; Hao, L.; Calabrese, C.M.; Zhou, Y.; Choi, C.H.J.; Xing, H.; Mirkin, C.A. Biodegradable DNA-brush block copolymer spherical nucleic acids enable transfection agent-free intracellular gene regulation. Small 2015, 11, 5360–5368. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lu, X.; Jia, F.; Tan, X.; Sun, X.; Cao, X.; Wai, F.; Zhang, C.; Zhang, K. Precision tuning of DNA- and Poly(ethylene glycol)-based nanoparticles via coassembly for effective antisense gene regulation. Chem. Mater. 2017, 29, 9882–9886. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, C.; Dong, Y.; Yang, Z.; Fan, Q.-H.; Liu, D. Thermally triggered frame-guided assembly. Angew. Chem. Int. Ed. 2014, 53, 13468–13470. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, Z.; Chen, C.; Cao, T.; Li, C.; Shao, Y.; Zhang, Y.; Qiu, D.; Shi, Q.; Fan, Q.-H.; et al. Self-collapsing of single molecular polypropylene oxide (PPO) in a 3D DNA network. Small 2018, 14, 1703426. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Shi, Z.; Zhang, Y.; Chen, X.-J.; Han, S.-Y.; Baumgart, T.; Chenoweth, D.M.; Park, S.-J. DNA island formation on binary block copolymer vesicles. J. Am. Chem. Soc. 2016, 138, 10157–10162. [Google Scholar] [CrossRef] [PubMed]
- Chidchob, P.; Edwardson, T.G.W.; Serpell, C.J.; Sleiman, H.F. Synergy of two assembly languages in DNA nanostructures: Self-assembly of sequence-defined polymers on DNA cages. J. Am. Chem. Soc. 2016, 138, 4416–4425. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.; Liao, C.; Toader, V.; Barłóg, M.; Bazzi, H.S.; Li, J.; Sleiman, H.F. DNA-imprinted polymer nanoparticles with monodispersity and prescribed DNA-strand patterns. Nat. Chem. 2017, 10, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, K.M.M.; Aldaye, F.A.; Sleiman, H.F. Long-range assembly of DNA into nanofibers and highly ordered networks using a block copolymer approach. J. Am. Chem. Soc. 2010, 132, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Y.; Sun, Y.; Li, Z.; Yang, Z.; He, Y.-M.; Fan, Q.-H.; Liu, D. Amphiphilic DNA-dendron hybrid: A new building block for functional assemblies. Soft Matter 2011, 7, 7187–7190. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Y.; Yang, Z.; He, Y.-M.; Fan, Q.-H.; Liu, D. Reversibly controlled morphology transformation of an amphiphilic DNA-dendron hybrid. Chem. Commun. 2012, 48, 3715–3717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wu, F.; Yang, Z.; Liu, D.; Fan, Q. Synthesis and self-assembly of DNA-aliphatic polyether dendron hybrids. Acta Chim. Sin. 2013, 71, 549–554. [Google Scholar] [CrossRef]
- Dong, Y.; Sun, Y.; Wang, L.; Wang, D.; Zhou, T.; Yang, Z.; Chen, Z.; Wang, Q.; Fan, Q.; Liu, D. Frame-guided assembly of vesicles with programmed geometry and dimensions. Angew. Chem. Int. Ed. 2014, 53, 2607–2610. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wang, D.; Dong, Y.; Xin, L.; Sun, Y.; Yang, Z.; Liu, D. Preparation and self-folding of amphiphilic DNA origami. Small 2015, 11, 1161–1164. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang Yuhe, R.; Zhang, Y.; Wang, D.; Wei, X.; Banerjee, S.; Liu, Y.; Yang, Z.; Yan, H.; Liu, D. Cuboid vesicles formed by frame-guided assembly on DNA origami scaffolds. Angew. Chem. Int. Ed. 2016, 56, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, Y.; Dong, Y.; Wu, F.; Wang, D.; Xin, L.; Liu, D. Precisely controlled 2D free-floating nanosheets of amphiphilic molecules through frame-guided assembly. Adv. Mater. 2016, 28, 9819–9823. [Google Scholar] [CrossRef] [PubMed]
- Auvinen, H.; Zhang, H.; Nonappa; Kopilow, A.; Niemelä, E.H.; Nummelin, S.; Correia, A.; Santos, H.A.; Linko, V.; Kostiainen, M.A. Protein coating of DNA nanostructures for enhanced stability and immunocompatibility. Adv. Healthc. Mater. 2017, 6, 1700692. [Google Scholar] [CrossRef] [PubMed]
- Surana, S.; Shenoy, A.R.; Krishnan, Y. Designing DNA nanodevices for compatibilitywith the immunesystem of higher organisms. Nat. Nanotechnol. 2015, 10, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Jiang, Q.; Li, N.; Dai, L.; Liu, Q.; Song, L.; Wang, J.; Li, Y.; Tian, J.; Ding, B.; et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano 2014, 8, 6633–6643. [Google Scholar] [CrossRef] [PubMed]
- Mikkilä, J.; Eskelinen, A.-P.; Niemelä, E.H.; Linko, V.; Frilander, M.J.; Törmä, P.; Kostiainen, M.A. Virus-encapsulated DNA origami nanostructures for cellulardelivery. Nano Lett. 2014, 14, 2196–2200. [Google Scholar] [CrossRef] [PubMed]
- Perrault, S.D.; Shih, W.M. Virus-inspired membraneencapsulation of DNA nanostructures to achieve in vivo stability. ACS Nano 2014, 8, 5132–5140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, P.D.E.; Shen, Q.; Akpinar, B.; Davis, L.K.; Chung, K.K.H.; Baddeley, D.; Šarić, A.; Melia, T.J.; Hoogenboom, B.W.; Lin, C.; et al. A programmable DNA origami platform for organizing intrinsically disordered nucleoporins within nanopore confinement. ACS Nano 2018, 12, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Keyser, U.F. Enhancing nanopore sensing with DNA nanotechnology. Nat. Nanotechnol. 2016, 11, 106–108. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Du, T.; Liang, F.; Liu, S. Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine. Int. J. Mol. Sci. 2018, 19, 2283. https://doi.org/10.3390/ijms19082283
Zhao Z, Du T, Liang F, Liu S. Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine. International Journal of Molecular Sciences. 2018; 19(8):2283. https://doi.org/10.3390/ijms19082283
Chicago/Turabian StyleZhao, Zhiyong, Ting Du, Feng Liang, and Simin Liu. 2018. "Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine" International Journal of Molecular Sciences 19, no. 8: 2283. https://doi.org/10.3390/ijms19082283
APA StyleZhao, Z., Du, T., Liang, F., & Liu, S. (2018). Amphiphilic DNA Organic Hybrids: Functional Materials in Nanoscience and Potential Application in Biomedicine. International Journal of Molecular Sciences, 19(8), 2283. https://doi.org/10.3390/ijms19082283