Large-Scale Functional Analysis of CRP-Mediated Feed-Forward Loops
Abstract
:1. Introduction
2. Results
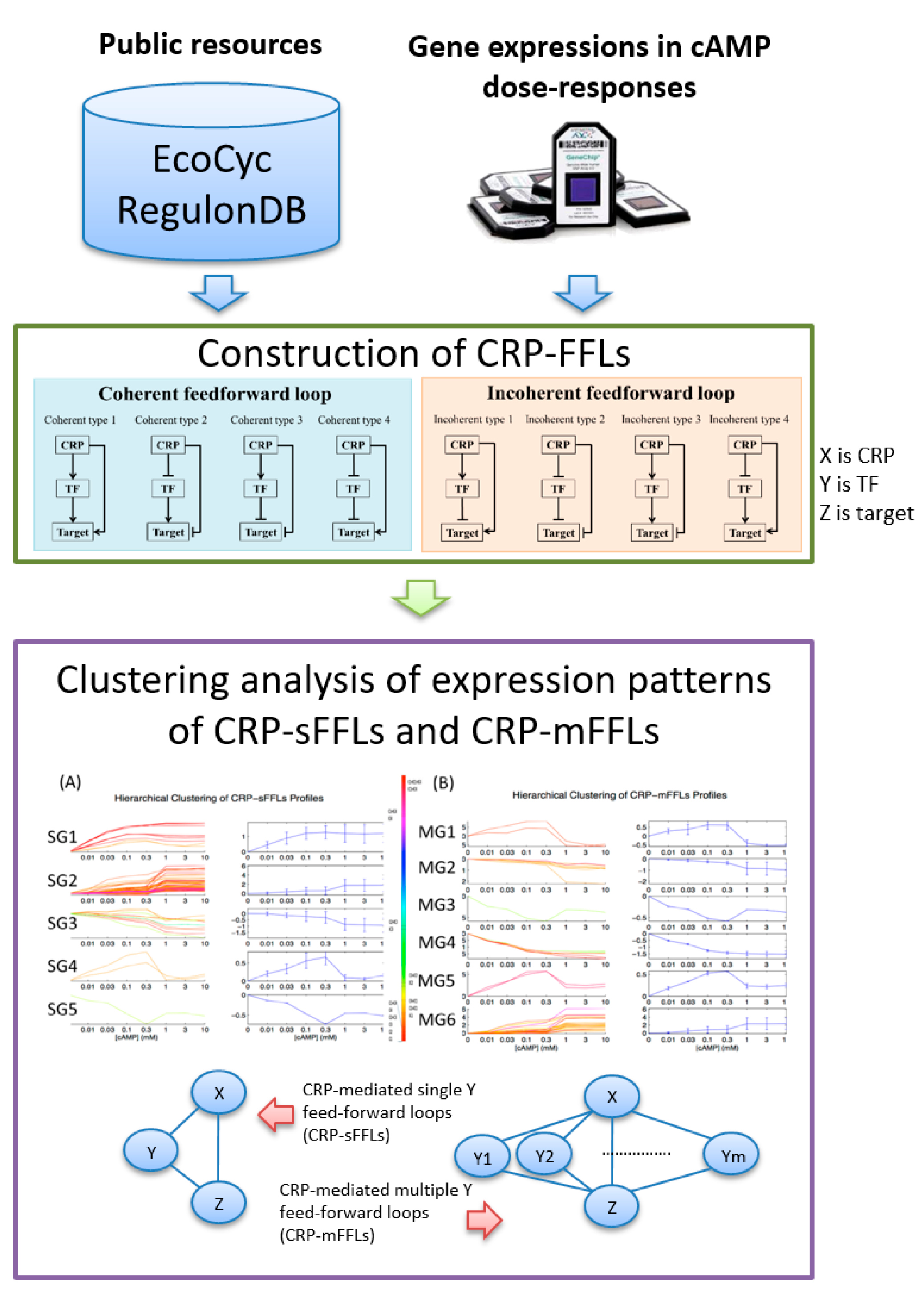
2.1. CRP-Mediated FFLs in E. coli
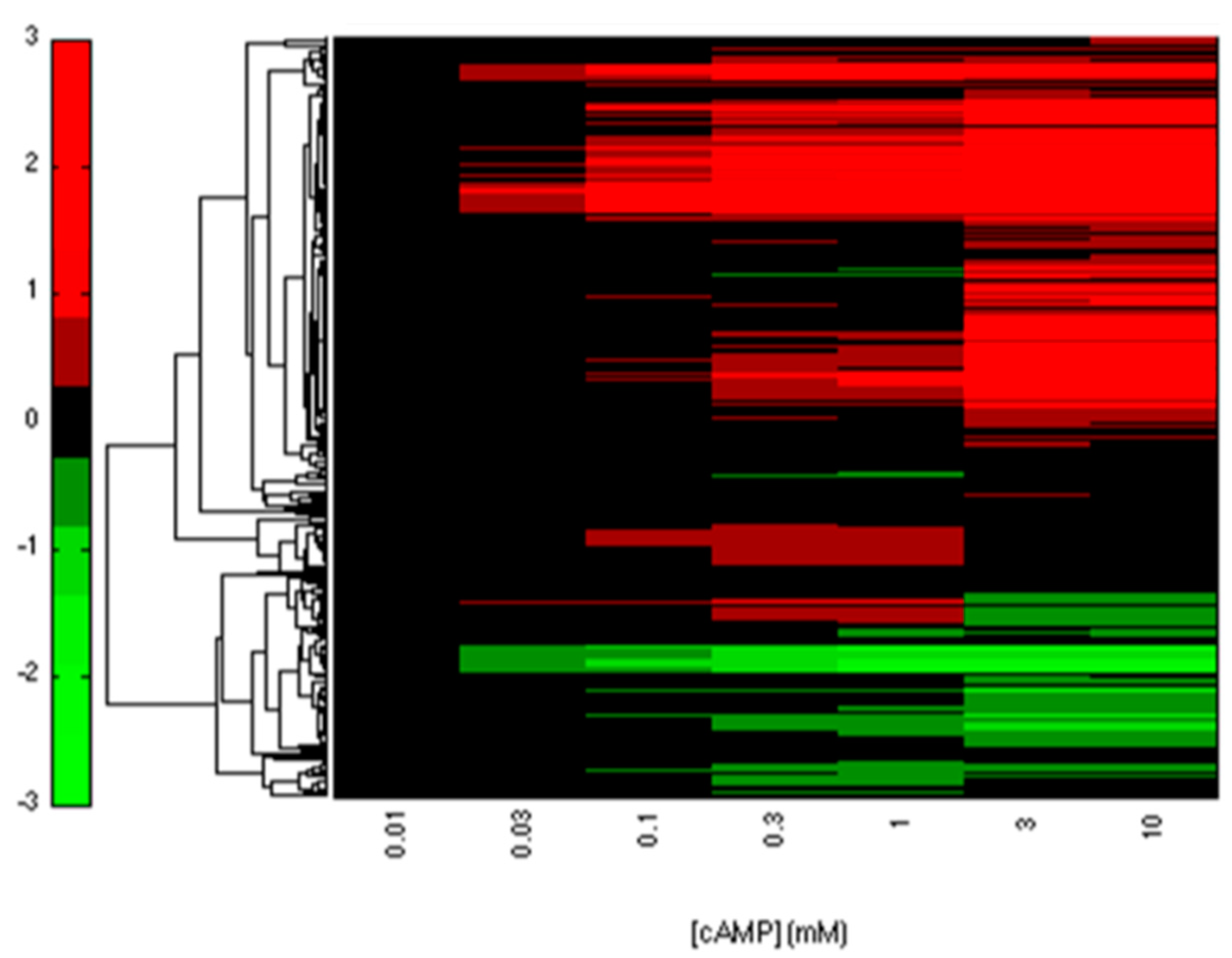
2.2. CRP-FFLs Characterized by Distinct cAMP Dose–Responses
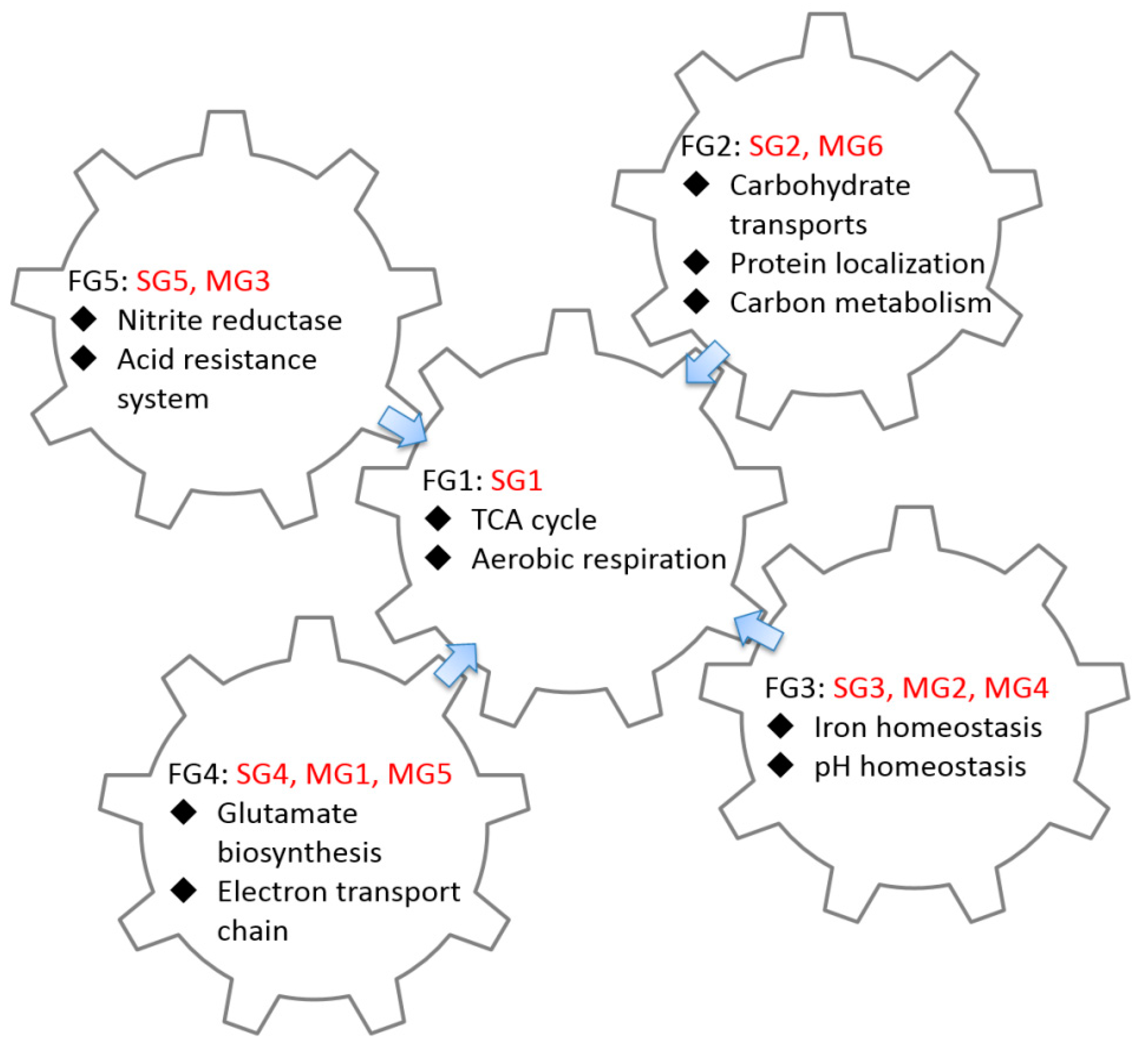
2.3. GO Enrichment Analysis of FFLs in CRP Regulatory Network
3. Discussion
3.1. Integration of Different FFL Types Diversifies the Output Response
3.2. Functional Analysis of FFLs in CRP-Regulated Networks
4. Application of FFLs
5. Materials and Methods
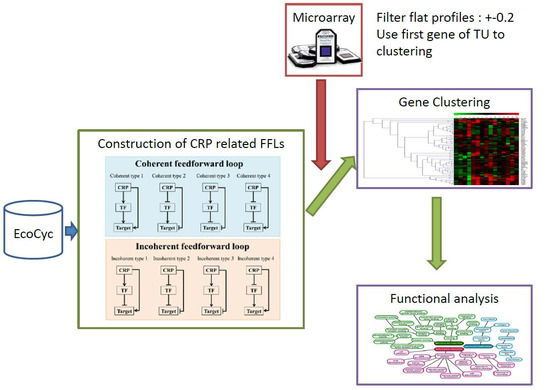
5.1. Construction of CRP-FFLs
5.2. Identification of CRP-Regulated Genes Using cAMP Dose–Response Microarray
5.3. Analysis of Microarray Data
5.4. Hierarchical Clustering and Functional Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Freyre-Gonzalez, J.A.; Trevino-Quintanilla, L.G. Analyzing Regulatory Networks in Bacteria. Net. Educ. 2010, 3, 24. [Google Scholar]
- Van Hijum, S.A.; Medema, M.H.; Kuipers, O.P. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol. Mol. Biol. Rev. MMBR 2009, 73, 481–509. [Google Scholar] [CrossRef] [PubMed]
- Keseler, I.M.; Mackie, A.; Santos-Zavaleta, A.; Billington, R.; Bonavides-Martinez, C.; Caspi, R.; Fulcher, C.; Gama-Castro, S.; Kothari, A.; Krummenacker, M.; et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017, 45, D543–D550. [Google Scholar] [CrossRef] [PubMed]
- Gama-Castro, S.; Salgado, H.; Santos-Zavaleta, A.; Ledezma-Tejeida, D.; Muniz-Rascado, L.; Garcia-Sotelo, J.S.; Alquicira-Hernandez, K.; Martinez-Flores, I.; Pannier, L.; Castro-Mondragon, J.A.; et al. RegulonDB version 9.0: High-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016, 44, D133–D143. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Cruz, C.F.; Gama-Castro, S.; Mejia-Almonte, C.; Castillo-Villalba, M.P.; Muniz-Rascado, L.J.; Collado-Vides, J. First steps in automatic summarization of transcription factor properties for RegulonDB: Classification of sentences about structural domains and regulated processes. Database 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Madan Babu, M.; Teichmann, S.A. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 2003, 31, 1234–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janga, S.C.; Salgado, H.; Martinez-Antonio, A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009, 37, 3680–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.W.; Kumar, B.; Ditges, U.; Gunzer, F.; Buer, J.; Zeng, A.P. An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nucleic Acids Res. 2004, 32, 6643–6649. [Google Scholar] [CrossRef] [PubMed]
- Beisel, C.L.; Storz, G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell 2011, 41, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Mangan, S.; Alon, U. Structure and function of the feed-forward loop network motif. Proc. Nat. Acad. Sci. USA 2003, 100, 11980–11985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangan, S.; Zaslaver, A.; Alon, U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J. Mol. Biol. 2003, 334, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Alon, U. Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 2007, 8, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Kashtan, N.; Itzkovitz, S.; Milo, R.; Alon, U. Efficient sampling algorithm for estimating subgraph concentrations and detecting network motifs. Bioinformatics 2004, 20, 1746–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urrios, A.; Gonzalez-Flo, E.; Canadell, D.; de Nadal, E.; Macia, J.; Posas, F. Plug-and-Play Multicellular Circuits with Time-Dependent Dynamic Responses. ACS Synth. Biol. 2018, 7, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Nampoothiri, S.S.; Fayaz, S.M.; Rajanikant, G.K. A Novel Five-Node Feed-Forward Loop Unravels miRNA-Gene-TF Regulatory Relationships in Ischemic Stroke. Mol. Neurobiol. 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Antoneli, F.; Golubitsky, M.; Stewart, I. Homeostasis in a feed forward loop gene regulatory motif. J. Theor. Biol. 2018, 445, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Fujita, N.; Yamamoto, K.; Ishihama, A. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS ONE 2011, 6, e20081. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Constantinidou, C.; Hobman, J.L.; Minchin, S.D. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 2004, 32, 5874–5893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hantke, K.; Winkler, K.; Schultz, J.E. Escherichia coli exports cyclic AMP via TolC. J. Bacteriol. 2011, 193, 1086–1089. [Google Scholar] [CrossRef] [PubMed]
- Hecker, M.; Lambeck, S.; Toepfer, S.; van Someren, E.; Guthke, R. Gene regulatory network inference: Data integration in dynamic models—A review. Biosystems 2009, 96, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Buetti-Dinh, A.; Ungricht, R.; Kelemen, J.Z.; Shetty, C.; Ratna, P.; Becskei, A. Control and signal processing by transcriptional interference. Mol. Syst. Biol. 2009, 5, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.P.; Lin, H.H.; Yang, C.D.; Huang, S.H.; Tseng, C.P. Regulatory role of cAMP receptor protein over Escherichia coli fumarase genes. J. Microbiol. 2012, 50, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Valgepea, K.; Adamberg, K.; Nahku, R.; Lahtvee, P.J.; Arike, L.; Vilu, R. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst. Biol. 2010, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Nam, T.W.; Park, Y.H.; Jeong, H.J.; Ryu, S.; Seok, Y.J. Glucose repression of the Escherichia coli sdhCDAB operon, revisited: Regulation by the CRP*cAMP complex. Nucleic Acids Res 2005, 33, 6712–6722. [Google Scholar] [CrossRef] [PubMed]
- Gorke, B.; Stulke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef] [PubMed]
- Stulke, J.; Hillen, W. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 1999, 2, 195–201. [Google Scholar] [CrossRef]
- Zhang, Z.; Gosset, G.; Barabote, R.; Gonzalez, C.S.; Cuevas, W.A.; Saier, M.H., Jr. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 2005, 187, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO: TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]


| Type | Number of CRP-FFLs | Number of TF | Number of Gene | Genes |
|---|---|---|---|---|
| Coh1 | 49 | 13 | 49 | araF, araG, dctA, fecA, fecB, fecC, fecD, fecE, fucA, fucI, fucK, fucO, fucP, fucR, fumC, hyfH, idnD, idnK, idnO, lamB, malE, malF, malG, malK, malM, malS, marB, ompF, prpB, prpC, prpD, prpE, rhaR, sdhA, sdhB, sdhC, sdhD, srlA, srlB, srlD, srlE, sucA, sucB, sucC, sucD, xylA, xylF, xylG, xylH |
| Coh2 | 15 | 3 | 10 | gadA, gadB, gadB, gadC, gadC, gadE, gadE, gadX, gltD, gltF, mdtE, mdtE, mdtF, mdtF, proP |
| Coh3 | 2 | 1 | 2 | gltD, gltF |
| Coh4 | 26 | 3 | 26 | acs, actP, aldB, fadL, flhC, flhD, glcC, guaA, guaB, hlyE, hupB, mglA, mglC, mtlA, mtlD, mtlR, nanA, nanE, nanK, nanT, nmpC, xylF, xylG, xylH, yiaK, yjcH |
| InCoh1 | 87 | 22 | 73 | cdd, chbB, cirA, cyoD, cyoD, cyoE, cyoE, cytR, entD, entH, fecA, fecB, fecC, fecD, fecE, fepA, fiu, flhC, flhD, fumC, galK, galK, galP, galP, galS, glcC, glpD, glpF, glpK, glpT, gntK, gntP, gntP, gntU, grcA, grcA, lsrA, lsrB, lsrC, lsrD, lsrF, lsrG, lsrK, lsrR, malI, malX, manX, manY, manZ, marB, mglA, mglA, mglA, mglB, mglB, mglB, mglC, mglC, mglC, mtlA, mtlD, mtlR, nagB, nagE, nupC, nupG, prpR, rbsA, rbsB, rbsC, rbsD, rbsK, rbsR, srlA, srlB, srlD, srlE, tsx, udp, uidA, uidB, uidC, uxuA, uxuA, uxuB, uxuB, xylA |
| InCoh2 | 8 | 3 | 8 | bhsA, gadA, gadB, gadC, gadX, nirB, osmY, yiaJ |
| InCoh3 | 1 | 1 | 1 | araJ |
| InCoh4 | 14 | 4 | 14 | csgD, csgE, csgF, csgG, cyoD, cyoE, exuT, glpB, grcA, malE, malF, malG, marB, nrdB |
| Functional Group ID | Group ID | GO Term | p-Value | Number of Gene | Summary of Function |
|---|---|---|---|---|---|
| FG1 | SG1 | GO:0006099 tricarboxylic acid cycle | 5.02 × 10−12 | 8 |
|
| GO:0009060 aerobic respiration | 8.00 × 10−11 | 8 | |||
| GO:0045333 cellular respiration | 3.13 × 10−9 | 8 | |||
| GO:0015980 energy derivation by oxidation of organic compounds | 2.18 × 10−8 | 8 | |||
| GO:0006091 generation of precursor metabolites and energy | 2.25 × 10−7 | 8 | |||
| GO:0055114 oxidation-reduction process | 7.12 × 10−3 | 8 | |||
| FG2 | SG2 MG6 | GO:0008643 carbohydrate transport | 3.84 × 10−26 | 37 |
|
| GO:0034219 carbohydrate transmembrane transport | 6.99 × 10−18 | 27 | |||
| GO:0071702 organic substance transport | 4.69 × 10−12 | 44 | |||
| GO:0006810 transport | 2.93 × 10−8 | 49 | |||
| GO:0051234 establishment of localization | 3.75 × 10−8 | 49 | |||
| GO:0015749 monosaccharide transport | 5.37 × 10−8 | 14 | |||
| GO:0044765 single-organism transport | 2.23 × 10−7 | 46 | |||
| GO:0051179 localization | 4.36 × 10−7 | 49 | |||
| GO:0015768 maltose transport | 4.45 × 10−5 | 6 | |||
| GO:0009401phosphoenolpyruvate-dependent sugar phosphotransferase system | 1.03 × 10−4 | 10 | |||
| GO:0055085 transmembrane transport | 1.20 × 10−4 | 36 | |||
| GO:0015750 pentose transport | 2.71 × 10−4 | 7 | |||
| GO:0044724 single-organism carbohydrate catabolic process | 6.79 × 10−4 | 21 | |||
| GO:0015766 disaccharide transport | 7.07 × 10−4 | 7 | |||
| GO:0042956 maltodextrin transport | 7.40 × 10−4 | 5 | |||
| GO:0015772 oligosaccharide transport | 1.61 × 10−3 | 7 | |||
| GO:0016052 carbohydrate catabolic process | 2.08 × 10−3 | 21 | |||
| GO:0006004 fucose metabolic process | 3.19 × 10−3 | 6 | |||
| GO:0019521 D-gluconate metabolic process | 7.58 × 10−3 | 6 | |||
| GO:0044275 cellular carbohydrate catabolic process | 8.56 × 10−3 | 11 | |||
| FG3 | SG3 MG2 MG4 | GO:0055080 cation homeostasis | 7.34 × 10−6 | 8 |
|
| GO:0050801 ion homeostasis | 9.10 × 10−6 | 8 | |||
| GO:0048878 chemical homeostasis | 1.37 × 10−5 | 8 | |||
| GO:0042592 homeostatic process | 3.33 × 10−4 | 8 | |||
| GO:0055072 iron ion homeostasis | 7.44 × 10−3 | 5 | |||
| GO:0045852 pH elevation | 9.50 × 10−3 | 3 | |||
| GO:0051454 intracellular pH elevation | 9.50 × 10−3 | 3 | |||
| FG4 | SG4 MG1 MG5 | No significant GO term |
| ||
| FG5 | SG5 MG3 | No significant GO term |
| ||
| Functional Group ID | Type | Group ID | Number of CRP-FFLs (Gene) | Coh1 | Coh2 | Coh3 | Coh4 | InCoh1 | InCoh2 | InCoh3 | InCoh4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FG1 | CRP-sFFLs | SG1 | 10 (10) | V | V | ||||||
| FG2 | SG2 | 78 (78) | V | V | V | V | |||||
| FG3 | SG3 | 10 (10) | V | V | V | V | V | V | |||
| FG4 | SG4 | 2 (2) | V | ||||||||
| FG5 | SG5 | 1 (1) | V | ||||||||
| FG4 | CRP-mFFLs | MG1 | 4 (2) | V | V | ||||||
| FG3 | MG2 | 15 (7) | V | V | V | ||||||
| FG5 | MG3 | 2 (1) | V | V | |||||||
| FG3 | MG4 | 14 (6) | V | V | |||||||
| FG4 | MG5 | 6 (2) | V | V | |||||||
| FG2 | MG6 | 60 (27) | V | V | V | V |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-D.; Huang, H.-Y.; Shrestha, S.; Chen, Y.-H.; Huang, H.-D.; Tseng, C.-P. Large-Scale Functional Analysis of CRP-Mediated Feed-Forward Loops. Int. J. Mol. Sci. 2018, 19, 2335. https://doi.org/10.3390/ijms19082335
Yang C-D, Huang H-Y, Shrestha S, Chen Y-H, Huang H-D, Tseng C-P. Large-Scale Functional Analysis of CRP-Mediated Feed-Forward Loops. International Journal of Molecular Sciences. 2018; 19(8):2335. https://doi.org/10.3390/ijms19082335
Chicago/Turabian StyleYang, Chi-Dung, Hsi-Yuan Huang, Sirjana Shrestha, Yen-Hua Chen, Hsien-Da Huang, and Ching-Ping Tseng. 2018. "Large-Scale Functional Analysis of CRP-Mediated Feed-Forward Loops" International Journal of Molecular Sciences 19, no. 8: 2335. https://doi.org/10.3390/ijms19082335
APA StyleYang, C.-D., Huang, H.-Y., Shrestha, S., Chen, Y.-H., Huang, H.-D., & Tseng, C.-P. (2018). Large-Scale Functional Analysis of CRP-Mediated Feed-Forward Loops. International Journal of Molecular Sciences, 19(8), 2335. https://doi.org/10.3390/ijms19082335