Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber
Abstract
:1. Introduction
2. Results
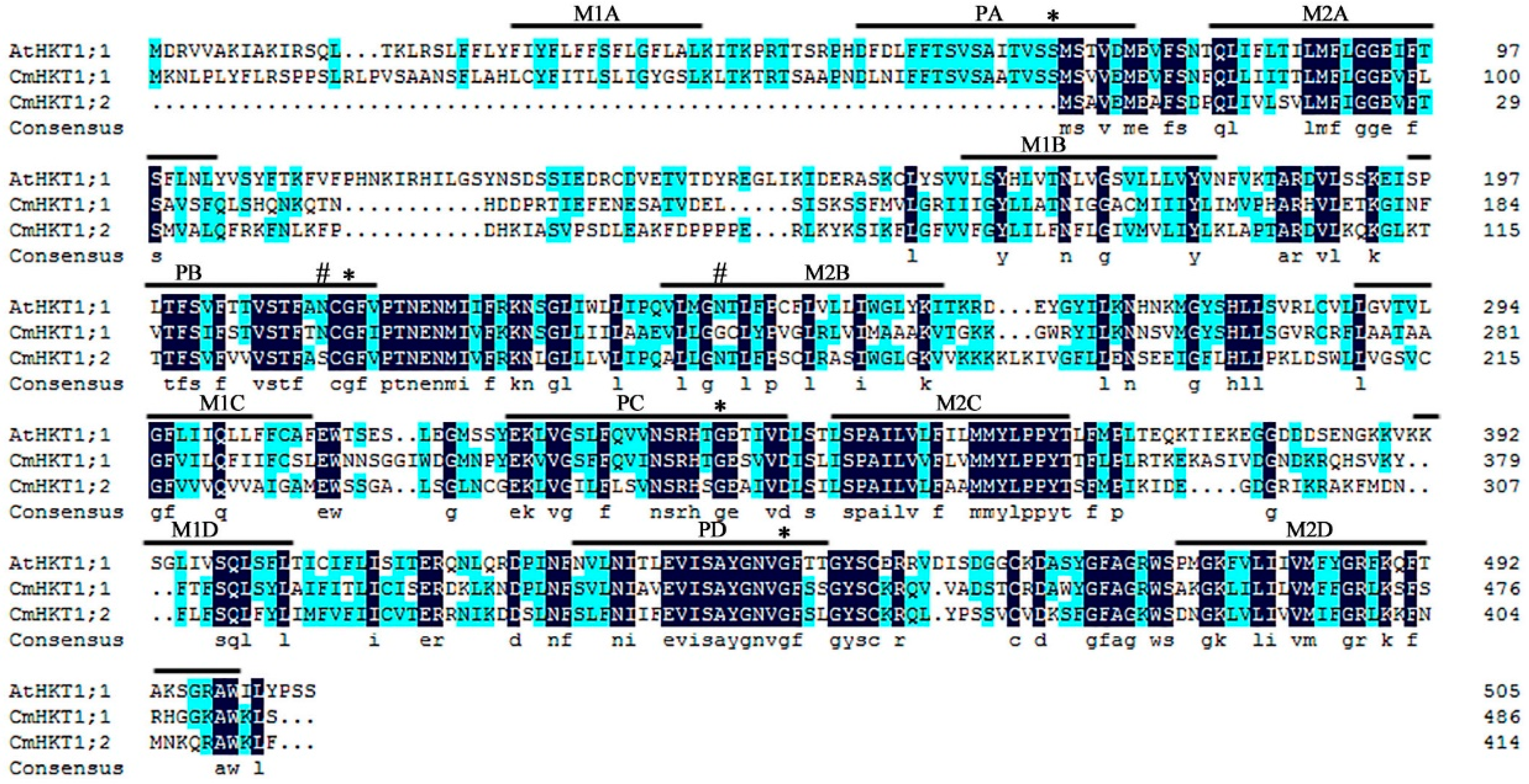
2.1. Identification and Characterization of Pumpkin CmHKT Genes
2.2. NaCl Stress-Induced CmHKT1;1 Upregulated in Pumpkin
2.3. CmHKT1;1 Is Expressed at the Plasma Membrane of Stelar Cells in Pumpkin Roots
2.4. CmHKT1;1 Prefers Na+ Transport over K+ Transport
2.5. Ectopic Expression of CmHKT1;1 in Cucumber Confers Salt Tolerance in Transgenic Plants
2.6. CmHKT1;1 Transgenic Cucumber as Rootstock Improve the Growth of Wild-Type Cucumber Scion under Salt Stress
3. Discussion
3.1. CmHKT1;1 Is a Pumpkin Na+-Selective Class I HKT
3.2. CmHKT1;1 Enhances Salinity Tolerance in Transgenic Cucumber
3.3. CmHKT1;1 Is Critical for Regulation of Shoot Na+ Accumulation in Grafted Cucumber through Limiting Na+ Transport from the Roots to the Shoots
4. Materials and Methods
4.1. Plant Material, Growth Conditions, and Stress Treatments
4.2. Transient Gene Expression in Tobacco
4.3. Cation Uptake Experiments in Yeast Cells
4.4. Cucumber Transformation
4.5. RT-PCR and Quantitative Real-Time PCR
4.6. Determination of Plant Dry Weight, Na+, and K+ Concentrations
4.7. Determination of Chlorophyll Content and Electrolyte Leakage
4.8. In Situ Hybridization
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Uozumi, N.; Kim, E.J.; Rubio, F.; Yamaguchi, T.; Muto, S.; Tsuboi, A.; Bakker, E.P.; Nakamura, T.; Schroeder, J.I. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000, 122, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.J.; Liu, Y.; Kang, D.; Fan, K.J.; Wang, C.Y.; Wang, G.Y.; Liu, Y.J. Two alternative splicing variants of maize HKT1;1 confer salt tolerance in transgenic tobacco plants. Plant Cell Tissue Organ Cult. 2015, 123, 569–578. [Google Scholar] [CrossRef]
- Mäser, P.; Eckelman, B.; Vaidyanathan, R.; Horie, T.; Fairbairn, D.J.; Kubo, M.; Yamagami, M.; Yamaguchi, K.; Nishimura, M.; Uozumi, N.; et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS. Lett. 2002, 531, 157–161. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Xu, B.; Athman, A.; Conn, S.J.; Jordans, C.; Byrt, C.S.; Hare, R.A.; Tyerman, S.D.; Tester, M.; et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012, 30, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef] [PubMed]
- Davenport, R.J.; Munoz-Mayor, A.; Jha, D.; Essah, P.A.; Rus, A.; Tester, M. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 2007, 30, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Møller, I.S.; Gilliham, M.; Jha, D.; Mayo, G.M.; Roy, S.J.; Coates, J.C.; Haseloff, J.; Tester, M. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 2009, 21, 2163–2178. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; Konomi, M.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 44, 928–938. [Google Scholar]
- Berthomieu, P.; Conéjéro, G.; Nublat, A.; Brackenbury, W.J.; Lambert, C.; Savio, C.; Uozumi, N.; Oiki, S.; Yamada, K.; Cellier, F.; et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003, 22, 2004–2014. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Xu, B.; Krishnan, M.; Lightfoot, D.J.; Athman, A.; Jacobs, A.K.; Watson-Haigh, N.S.; Plett, D.; Munns, R.; Tester, M. The Na+ transporter, TaHKT1; 5-D, limits shoot Na+ accumulation in bread wheat. Plant J. 2014, 80, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cao, Y.B.; Wang, Z.Q.P.; Wang, Z.Q.; Shi, J.P.; Liang, X.Y.; Song, W.B.; Chen, Q.J.; Lai, J.S.; Jiang, C.F. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.; Safwat, G.; Gilliham, M.; Møller, I.S.; Roy, S.; Shirley, N.; Jacobs, A.; Johnson, A.; Tester, M. Improved salinity tolerance of rice through cell type-specific expression of AtHTK1;1. PLoS ONE 2010, 5, e12571. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, X.; Gu, H.; Wu, B.; Zhang, H.; Yuan, X.; Cui, X. GmHKT1;4, a novel soybean gene regulating Na+/K+ ratio in roots enhances salt tolerance in transgenic plants. Plant Growth Regul. 2014, 73, 299–308. [Google Scholar] [CrossRef]
- Jones, J.R.W.; Pike, L.M.; Yourman, L.F. Salinity influences cucumber growth and yield. J. Am. Soc. Hortic. Sci. 1989, 114, 547–551. [Google Scholar]
- Trajkova, F.; Papadantonakis, N. Comparative effects of NaCl and CaCl2 salinity on cucumber grown in a closed hydroponic system. HortScience 2006, 41, 437–441. [Google Scholar]
- Huang, Y.; Tang, R.; Cao, Q.L.; Bie, Z.L. Improving the fruit yield and quality of cucumber by grafting onto the salt tolerant rootstock under NaCl stress. Sci. Hortic. 2009, 122, 26–31. [Google Scholar] [CrossRef]
- Colla, G.; Rouphael, Y.; Rea, E.; Cardarelli, M. Grafting cucumber plants enhance tolerance to sodium chloride and sulfate salinization. Sci. Hortic. 2012, 135, 177–185. [Google Scholar] [CrossRef]
- Shu, S.; Guo, S.R.; Sun, J.; Yuan, L.Y. Effects of salt stress on the structure and function of the photosynthetic apparatus in Cucumis sativus and its protection by exogenous putrescine. Physiol. Plant 2012, 146, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Kere, G.M.; Guo, Q.; Shen, J.; Xu, J.; Chen, J. Heritability and gene effects for salinity tolerance in cucumber (Cucumis sativus L.) estimated by generation mean analysis. Sci. Hortic. 2013, 159, 122–127. [Google Scholar] [CrossRef]
- Zhu, J.; Bie, Z.L.; Huang, Y.; Han, X.Y. Effect of grafting on the growth and ion concentrations of cucumber seedlings under NaCl stress. Soil Sci. Plant Nutr. 2008, 54, 895–902. [Google Scholar] [CrossRef]
- Huang, Y.; Bie, Z.L.; Liu, P.Y.; Niu, M.L.; Zhen, A.; Liu, Z.X.; Lei, B.; Gu, D.J.; Lu, C.; Wang, B.T. Reciprocal grafting between cucumber and pumpkin demonstrates the roles of the rootstock in the determination of cucumber salt tolerance and sodium accumulation. Sci. Hortic. 2013, 149, 47–54. [Google Scholar] [CrossRef]
- Niu, M.; Xie, J.; Sun, J.; Huang, Y.; Kong, Q.; Nawaz, M.A.; Bie, Z. A shoot based Na+ tolerance mechanism observed in pumpkin—an important consideration for screening salt tolerant rootstocks. Sci. Hortic. 2017, 218, 38–47. [Google Scholar] [CrossRef]
- Lei, B.; Huang, Y.; Sun, J.Y.; Xie, J.J.; Niu, M.L.; Liu, Z.X.; Fan, M.L.; Bie, Z.L. Scanning ion-selective electrode technique and X-ray microanalysis provide direct evidence of contrasting Na+ transport ability from root to shoot in salt-sensitive cucumber and salt-tolerant pumpkin under NaCl stress. Physiol. Plant. 2014, 152, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Durell, S.R.; Guy, H.R. Structural models of the KtrB, TrkH, and Trk1,2 symporters based on the structure of the KcsA K+ channel. Biophys. J. 1999, 77, 789–807. [Google Scholar] [CrossRef]
- Kato, Y.; Sakaguchi, M.; Mori, Y.; Saito, K.; Nakamura, T.; Bakker, E.P.; Sato, Y.; Goshima, S.; Uozumi, N. Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. USA 2001, 98, 6488–6493. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Park, H.C.; Ali, A.; Oh, D.H.; Aman, R.; Kropornicka, A.; Hong, H.; Choi, W.; Chung, W.S.; Kim, W.Y.; et al. TsHKT1;2, a HKT1 homolog from the extremophile Arabidopsis relative Thellungiella salsuginea, shows K+ specificity in the presence of NaCl. Plant Physiol. 2012, 158, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef] [PubMed]
- Rus, A.; Baxter, I.; Muthukumar, B.; Gustin, J.; Lahner, B.; Yakubova, E.; Salt, D.E. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006, 2, e210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Navarro, A.; Rubio, F. High-affinity potassium and sodium transport systems in plants. J. Exp. Bot. 2006, 57, 1149–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, F.; Horie, T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010, 33, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Corratgé-Faillie, C.; Jabnoune, M.; Zimmermann, S.; Very, A.A.; Fizames, C.; Sentenac, H. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cell. Mol. Life Sci. 2010, 67, 2511–2532. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.W.; Yao, X.; Luo, W.; Jha, D.; Tester, M.; Horie, T.; Schroeder, J.I. AtHKT1;1 mediates Nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS ONE 2011, 6, e24725. [Google Scholar] [CrossRef] [PubMed]
- Byrt, C.S.; Platten, J.D.; Spielmeyer, W.; James, R.A.; Lagudah, E.S.; Dennis, E.S.; Tester, M.; Munns, R. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007, 143, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Jing, W.; Xiao, L.; Jin, Y.; Shen, L.; Zhang, W. The rice high-affinity potassium transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 2015, 168, 1076–1090. [Google Scholar] [CrossRef] [PubMed]
- Jabnoune, M.; Espeout, S.; Mieulet, D.; Fizames, C.; Verdeil, J.L.; Conejero, G.; Rodriguez-Navarro, A.; Sentenac, H.; Guiderdoni, E.; Abdelly, C.; et al. Diversity in expression patterns and functional properties in the rice HKT transporter family. Plant Physiol. 2009, 150, 1955–1971. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Ren, Z.J.; Liu, Z.Q.; Feng, X.; Guo, R.Q.; Li, B.G.; Li, L.G.; Jing, H.C. SbHKT1; 4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+/K+ balance under Na+ stress. J. Integr. Plant Biol. 2014, 56, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Jaime-Pérez, N.; Pineda, B.; Garcia-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olias, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Yoshida, K.; Nakayama, H.; Yamada, K.; Oiki, S.; shinmyo, A. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 2001, 27, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Mian, A.; Oomen, R.J.F.J.; Isayenkov, S.; Sentenac, H.; Maathuis, F.J.M.; Very, A.A. Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 2011, 68, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Henderson, S.W.; Dunlevy, J.D.; Wu, Y.; Blackmore, D.H.; Walker, R.R.; Edwards, E.J.; Gilliham, M.; Walker, A.R. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks. New Phytol. 2018, 217, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Jiang, J.; Chung, J.S.; Wang, B.; Chen, J.; Xin, Z.; Shi, H. Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 2011, 52, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Almeida, P.M.F.; de Boer, G.J.; de Boer, A.H. Assessment of natural variation in the first pore domain of the tomato HKT1;2 transporter and characterization of mutated versions of SiHKT1;2 expressed in Xenopus laevis oocytes and via complementation of the salt sensitive athkt1;1 mutant. Front. Plant Sci. 2014, 5, 600. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, T.; Furuhashi, M.; Sakaoka, S.; Morikami, A.; Tsukagoshi, H. Ectopic expression of Mesembryanthemum crystallinum sodium transporter McHKT2 provides salt stress tolerance in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2017, 81, 2139–2144. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Han, N.; Ding, T.; Zhou, F.; Wang, B. SsHKT1;1 is a potassium transporter of the C3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 2014, 41, 790. [Google Scholar] [CrossRef]
- Almeida, P.; Katschnig, D.; de Boer, A.H. HKT transporters-state of the art. Int. J. Mol. Sci. 2013, 14, 20359–20385. [Google Scholar] [CrossRef] [PubMed]
- Estañ, M.T.; Martinez-Rodriguez, M.M.; Perez-Alfocea, F.; Flowers, T.J.; Bolarin, M.C. Grafting raises the salt tolerance of tomato through limiting the transport of sodium and chloride to the shoot. J. Exp. Bot. 2005, 56, 703–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouphael, Y.; Cardarelli, M.; Rea, E.; Colla, G. Improving melon and cucumber photosynthetic activity, mineral composition, and growth performance under salinity stress by grafting onto Cucurbita hybrid rootstocks. Photosynthetica 2012, 50, 180–188. [Google Scholar] [CrossRef]
- Yetisir, H.; Uygur, V. Responses of grafted watermelon onto different gourd species to salinity stress. J. Plant Nutr. 2010, 33, 315–327. [Google Scholar] [CrossRef]
- Bañuls, J.; Primo-Millo, E. Effects of salinity on some citrus scion-rootstock combinations. Ann. Bot. 1995, 76, 97–102. [Google Scholar] [CrossRef]
- Santa-Cruz, A.; Martinez-Rodriguez, M.M.; Perez-Alfocea, F.; Romero-Aranda, R.; Bolarin, M.C. The rootstock effect on the tomato salinity response depends on the genotype. Plant Sci. 2002, 162, 825–831. [Google Scholar] [CrossRef]
- Chen, G.X.; Fu, X.P.; Lips, S.H.; Sagi, M. Control of plant growth resides in the shoot, and not in the root, in reciprocal grafts of flacca and wild-type tomato (Lysopersicon esculentum), in the presence and absence of salinity stress. Plant Soil 2003, 256, 205–215. [Google Scholar] [CrossRef]
- Hassell, R.L.; Memmott, F. Grafting methods for watermelon production. HortScience 2008, 43, 1677–1679. [Google Scholar]
- Nawaz, M.A.; Jiao, Y.; Chen, C.; Shireen, F.; Zheng, Z.; Imtiaz, M.; Bie, Z.; Huang, Y. Melatonin pretreatment improves vanadium stress tolerance of watermelon seedlings by reducing vanadium concentration in the leaves and regulating melatonin biosynthesis and antioxidant-related gene expression. J. Plant Physiol. 2018, 220, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Sheludko, Y.V.; Sindarovska, Y.R.; Gerasymenko, I.M.; Bannikova, M.A.; Kuchuk, N.V. Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bioeng. 2007, 96, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.K.; Cai, X.; Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007, 51, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Garciadeblás, B.; Rodríguez-Navarro, A. The sal1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 1996, 8, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guan, C.; Wang, S.-M. Coordination of AtHKT1;1 and AtSOS1 facilitates Na+ and K+ homeostasis in Arabidopsis thaliana under salt stress. J. Plant Biol. 2014, 57, 282–290. [Google Scholar] [CrossRef]
- Haro, R.; Rodriguez-Navarro, A. Functional analysis of the M2D helix of the TRK1 potassium transporter of Saccharomyces cerevisiae. Biochim. Biophys. Acta 2003, 1613, 1–6. [Google Scholar] [CrossRef]
- Rodriguez-Navarro, A.; Ramos, J. Dual system for potassium transport in Sacchromyces cerevisiae. J. Bacteriol. 1984, 159, 940–945. [Google Scholar] [PubMed]
- Haro, R.; Banuelos, M.A.; Senn, M.A.E.; Barrero-Gil, J.; Rodriguez-Navarro, A. HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol. 2005, 139, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Asins, M.J.; Villalta, I.; Aly, M.M.; Olias, R.; De Morales, P.A.; Huertas, R.; Li, J.; Jaime-Perez, N.; Haro, R.; Raga, V.; et al. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environ. 2013, 36, 1171–1191. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Wang, Z.; Yao, F.; Gao, L.; Ma, S.; Sui, X.; Zhang, Z. Down-regulating CsHT1, a cucumber pollen-specific hexose transporter, inhibits pollen germination, tube growth, and seed development. Plant Physiol. 2015, 168, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.L.; Meng, F.Z.; Wang, H.Y.; Wei, Y.X.; Li, R.F.; Wang, Z.Y.; Hu, L.P.; Wang, S.H.; Zhang, Z.X. Molecular cloning, characteristics and low temperature response of raffinose synthase gene in Cucumis sativus L. J. Plant Physiol. 2012, 169, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.W.; Lee, U.; Vierling, E. Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol. 2003, 132, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Kouchi, H.; Hata, S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 1993, 238, 106–119. [Google Scholar] [PubMed]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Cao, H.; Cheng, J.; He, X.; Sohail, H.; Niu, M.; Huang, Y.; Bie, Z. Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber. Int. J. Mol. Sci. 2018, 19, 2648. https://doi.org/10.3390/ijms19092648
Sun J, Cao H, Cheng J, He X, Sohail H, Niu M, Huang Y, Bie Z. Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber. International Journal of Molecular Sciences. 2018; 19(9):2648. https://doi.org/10.3390/ijms19092648
Chicago/Turabian StyleSun, Jingyu, Haishun Cao, Jintao Cheng, Xiaomeng He, Hamza Sohail, Mengliang Niu, Yuan Huang, and Zhilong Bie. 2018. "Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber" International Journal of Molecular Sciences 19, no. 9: 2648. https://doi.org/10.3390/ijms19092648
APA StyleSun, J., Cao, H., Cheng, J., He, X., Sohail, H., Niu, M., Huang, Y., & Bie, Z. (2018). Pumpkin CmHKT1;1 Controls Shoot Na+ Accumulation via Limiting Na+ Transport from Rootstock to Scion in Grafted Cucumber. International Journal of Molecular Sciences, 19(9), 2648. https://doi.org/10.3390/ijms19092648




