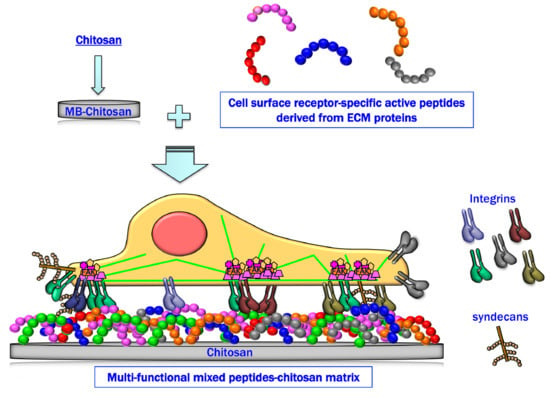
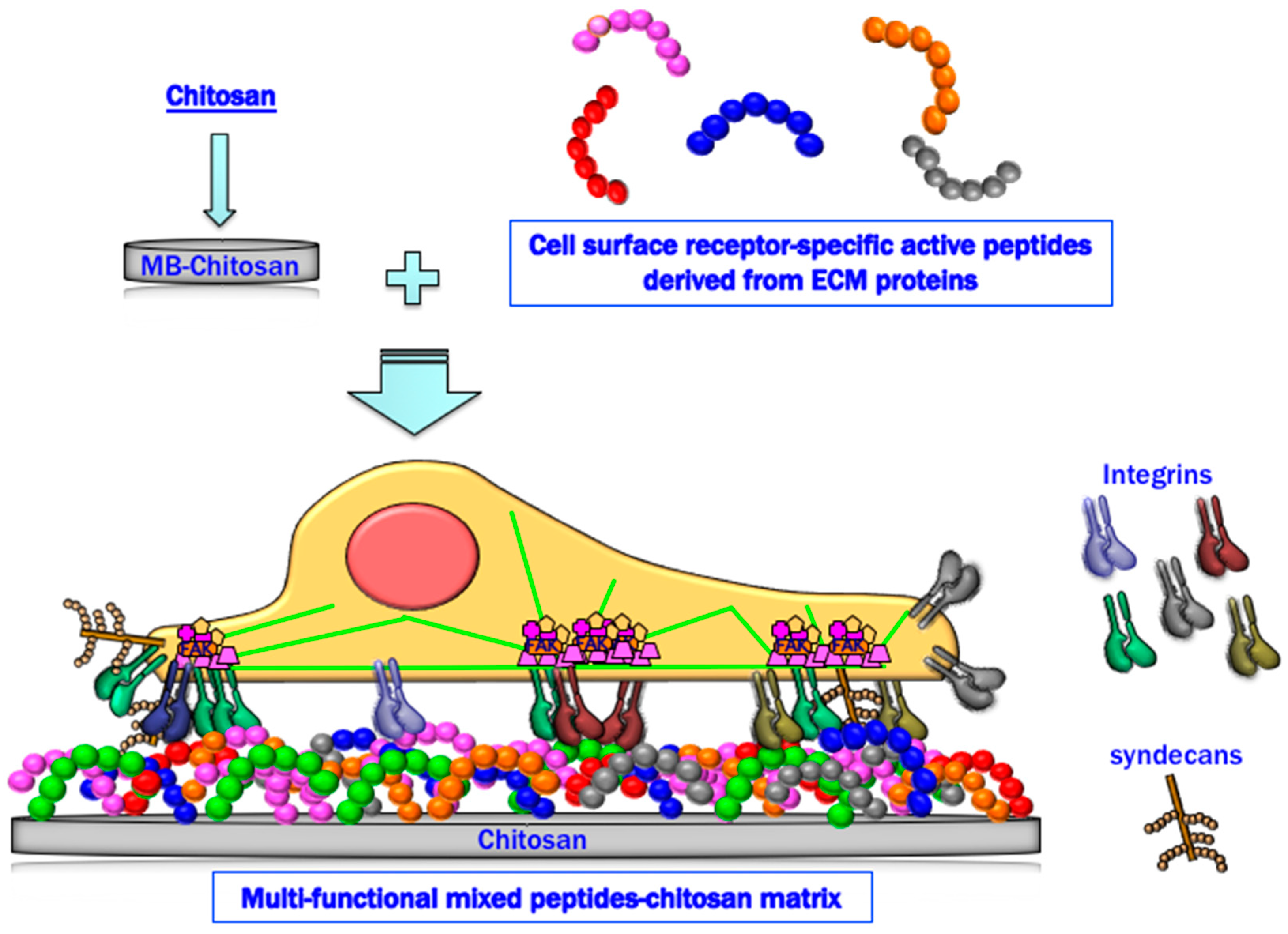
Mixed Peptide-Conjugated Chitosan Matrices as Multi-Receptor Targeted Cell-Adhesive Scaffolds
Abstract
:1. Introduction
2. Preparation of Peptide–Chitosan Matrices
3. Properties of Peptide–Chitosan Matrices
4. Cell Attachment Activities of Mixed Peptide–Chitosan Matrices Depending on Integrins and Syndecans
5. Cell Attachment Activities of Mixed Peptides–Chitosan Matrices Depending on Different Integrin Subtypes
6. Cell Attachment of Mixed Peptide–Chitosan Matrices Interacting with Multiple Receptors
7. Neurite Outgrowth Activities of Mixed Peptide–Chitosan Matrices
8. Summary and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pina, S.; Oliveira, J.M.; Reis, R.L. Natural-based nanocomposites for bone tissue engineering and regenerative medicine: A review. Adv. Mater. 2015, 27, 1143–1169. [Google Scholar] [CrossRef] [PubMed]
- Camponeschi, F.; Atrei, A.; Rocchigiani, G.; Mencuccini, L.; Uva, M.; Barbucci, R. New formulations of polysaccharide-based hydrogels for drug release and tissue engineering. Gels 2015, 1, 3–23. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, T.; Jiang, C. Biomacromolecules as carriers in drug delivery and tissue engineering. Acta Pharm. Sin. B 2018, 8, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Croisier, F.; Jérôme, C. Chitosan-based biomaterials for tissue engineering. Eur. Polym. J. 2013, 49, 780–792. [Google Scholar] [CrossRef]
- Anitha, A.; Sowmya, S.; Kumar, P.T.S.; Deepthi, S.; Chennazhi, K.P.; Ehrlich, H.; Tsurkan, M.; Jayakumar, R. Chitin and chitosan in selected biomedical applications. Prog. Polym. Sci. 2014, 39, 1644–1667. [Google Scholar] [CrossRef]
- Zargar, V.; Asghari, M.; Dashti, A. A review on chitin and chitosan polymers: Structure, chemistry, solubility, derivatives, and applications. ChemBioEng Rev. 2015, 2, 204–226. [Google Scholar] [CrossRef]
- Balagangadharan, K.; Dhivya, S.; Selvamurugan, N. Chitosan based nanofibers in bone tissue engineering. Int. J. Biol. Macromol. 2017, 104, 1372–1382. [Google Scholar] [CrossRef] [PubMed]
- Salehi, E.; Daraei, P.; Arabi Shamsabadi, A. A review on chitosan-based adsorptive membranes. Carbohydr. Polym. 2016, 152, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Ikeda, K.; Yamamoto, K.; Ishizaki, H.; Yoshizawa, Y.; Yanagiguchi, K.; Yamada, S.; Hayashi, Y. Fabrication and characteristics of chitosan sponge as a tissue engineering scaffold. BioMed Res. Int. 2014, 2014, 786892. [Google Scholar] [CrossRef] [PubMed]
- Rampino, A.; Borgogna, M.; Blasi, P.; Bellich, B.; Cesaro, A. Chitosan nanoparticles: Preparation, size evolution and stability. Int. J. Pharm. 2013, 455, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Maharjan, S.; Cho, K.H.; Cui, L.; Park, I.K.; Choi, Y.J.; Cho, C.S. Chitosan-based particulate systems for the delivery of mucosal vaccines against infectious diseases. Int. J. Biol. Macromol. 2018, 110, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Saravanakumar, G.; Kim, K.; Kwon, I.C. Targeted delivery of low molecular drugs using chitosan and its derivatives. Adv. Drug Deliv. Rev. 2010, 62, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Pella, M.C.G.; Lima-Tenorio, M.K.; Tenorio-Neto, E.T.; Guilherme, M.R.; Muniz, E.C.; Rubira, A.F. Chitosan-based hydrogels: From preparation to biomedical applications. Carbohydr. Polym. 2018, 196, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, F.; Oveisi, Z.; Samani, S.M.; Amoozgar, Z. Chitosan based hydrogels: Characteristics and pharmaceutical applications. Res. Pharm. Sci. 2015, 10, 1–16. [Google Scholar] [PubMed]
- Muzzarelli, R.A. Chitins and chitosans as immunoadjuvants and non-allergenic drug carriers. Mar. Drugs 2010, 8, 292–312. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Chung, Y.C.; Wang, I.J.; Young, T.H. Control of cell attachment on pH-responsive chitosan surface by precise adjustment of medium pH. Biomaterials 2012, 33, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Lu, X.; Hong, Y.; Xi, T.; Zhang, D. The molecular mechanism of mediation of adsorbed serum proteins to endothelial cells adhesion and growth on biomaterials. Biomaterials 2013, 34, 5747–5758. [Google Scholar] [CrossRef] [PubMed]
- Kanta, J. Collagen matrix as a tool in studying fibroblastic cell behavior. Cell Adhes. Migr. 2015, 9, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Zeltz, C.; Gullberg, D. The integrin-collagen connection—A glue for tissue repair? J. Cell Sci. 2016, 129, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Zollinger, A.J.; Smith, M.L. Fibronectin, the extracellular glue. Matrix Biol. 2017, 60–61, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y. Laminin: Loss-of-function studies. Cell. Mol. Life Sci. 2017, 74, 1095–1115. [Google Scholar] [CrossRef] [PubMed]
- Rahmany, M.B.; Van Dyke, M. Biomimetic approaches to modulate cellular adhesion in biomaterials: A review. Acta Biomater. 2013, 9, 5431–5437. [Google Scholar] [CrossRef] [PubMed]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Sreejalekshmi, K.G.; Nair, P.D. Biomimeticity in tissue engineering scaffolds through synthetic peptide modifications-altering chemistry for enhanced biological response. J. Biomed. Mater. Res. A 2011, 96, 477–491. [Google Scholar] [CrossRef] [PubMed]
- Rubert Perez, C.M.; Stephanopoulos, N.; Sur, S.; Lee, S.S.; Newcomb, C.; Stupp, S.I. The powerful functions of peptide-based bioactive matrices for regenerative medicine. Ann. Biomed. Eng. 2015, 43, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.I.; Rahman, S.; Queen, A.; Ahamad, S.; Ali, S.; Kim, J.; Hassan, M.I. Implications of molecular diversity of chitin and its derivatives. Appl. Microbiol. Biotechnol. 2017, 101, 3513–3536. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ma, L.; Mao, Z.; Gao, C. Chitosan-based biomaterials for tissue repair and regeneration. Adv. Polym. Sci. 2011, 244, 81–127. [Google Scholar] [CrossRef]
- Bellis, S.L. Advantages of RGD peptides for directing cell association with biomaterials. Biomaterials 2011, 32, 4205–4210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Singha, K.; Son, S.; Kim, J.; Namgung, R.; Yun, C.O.; Kim, W.J. A review of RGD-functionalized nonviral gene delivery vectors for cancer therapy. Cancer Gene Ther. 2012, 19, 741–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansson, A.; Hashom, N.; Falson, F.; Rousselle, P.; Jordan, O.; Borchard, G. In vitro evaluation of an RGD-functionalized chitosan derivative for enhanced cell adhesion. Carbohydr. Polym. 2012, 90, 1494–1500. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.; Jones, J.P.; Lei, P.; Andreadis, S.T.; Baker, O.J. Laminin-111 peptides conjugated to fibrin hydrogels promote formation of lumen containing parotid gland cell clusters. Biomacromolecules 2016, 17, 2293–2301. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.C.; Yoo, S.Y.; Kim, H.; Lee, J. Chitosan-based multifunctional platforms for local delivery of therapeutics. Mar. Drugs 2017, 15, 60. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, P.; Grove, T.; Edgar, K.J.; Goldstein, A.S. Surface grafting of chitosan shell, polycaprolactone core fiber meshes to confer bioactivity. J. Bioact. Compat. Pol. 2015, 30, 258–274. [Google Scholar] [CrossRef]
- Costa, F.M.; Maia, S.R.; Gomes, P.A.; Martins, M.C. Dhvar5 antimicrobial peptide (AMP) chemoselective covalent immobilization results on higher antiadherence effect than simple physical adsorption. Biomaterials 2015, 52, 531–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, K.; Ruan, Q.; Liberman, D.; White, S.N.; Moradian-Oldak, J. Repairing human tooth enamel with leucine-rich amelogenin peptide–chitosan hydrogel. J. Mater. Res. 2016, 31, 556–563. [Google Scholar] [CrossRef]
- Mochizuki, M.; Kadoya, Y.; Wakabayashi, Y.; Kato, K.; Okazaki, I.; Yamada, M.; Sato, T.; Sakairi, N.; Nishi, N.; Nomizu, M. Laminin-1 peptide-conjugated chitosan membranes as a novel approach for cell engineering. FASEB J. 2003, 17, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Hozumi, K.; Nomizu, M. Construction and activity of a synthetic basement membrane with active laminin peptides and polysaccharides. Chemistry 2011, 17, 10500–10508. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Kumai, J.; Yamada, Y.; Nomizu, M. Active peptide-conjugated chitosan matrices as an artificial basement membrane. Polymers 2015, 7, 281–297. [Google Scholar] [CrossRef]
- Colognato, H.; Yurchenco, P.D. Form and function: The laminin family of heterotrimers. Dev. Dyn. 2000, 218, 213–234. [Google Scholar] [CrossRef] [Green Version]
- Miner, J.H.; Yurchenco, P.D. Laminin functions in tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2004, 20, 255–284. [Google Scholar] [CrossRef] [PubMed]
- Iorio, V.; Troughton, L.D.; Hamill, K.J. Laminins: Roles and utility in wound repair. Adv. Wound Care 2015, 4, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.H.; Li, C.; Mudd, J.L.; Go, G.; Sutherland, A.E. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development 2004, 131, 2247–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, T.; Giltay, R.; Talts, U.; Timpl, R.; Talts, J.F. Expression and distribution of laminin alpha1 and alpha2 chains in embryonic and adult mouse tissues: An immunochemical approach. Exp. Cell Res. 2002, 275, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Patton, B.L. Laminins of the neuromuscular system. Microsc. Res. Tech. 2000, 51, 247–261. [Google Scholar] [CrossRef]
- Fleischmajer, R.; Kuroda, K.; Utani, A.; Douglas MacDonald, E.; Perlish, J.S.; Arikawa-Hirasawa, E.; Sekiguchi, K.; Sanzen, N.; Timpl, R.; Yamada, Y. Differential expression of laminin alpha chains during proliferative and differentiation stages in a model for skin morphogenesis. Matrix Biol. 2000, 19, 637–647. [Google Scholar] [CrossRef]
- Hamill, K.J.; McLean, W.H. The alpha-3 polypeptide chain of laminin 5: Insight into wound healing responses from the study of genodermatoses. Clin. Exp. Dermatol. 2005, 30, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, R.; Horn, N.; Selg, M.; Wendler, O.; Pausch, F.; Sorokin, L.M. Expression and function of laminins in the embryonic and mature vasculature. Physiol. Rev. 2005, 85, 979–1000. [Google Scholar] [CrossRef] [PubMed]
- Petajaniemi, N.; Korhonen, M.; Kortesmaa, J.; Tryggvason, K.; Sekiguchi, K.; Fujiwara, H.; Sorokin, L.; Thornell, L.E.; Wondimu, Z.; Assefa, D.; et al. Localization of laminin alpha4-chain in developing and adult human tissues. J. Histochem. Cytochem. 2002, 50, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, Y.; Miner, J.H. Molecular dissection of laminin alpha 5 in vivo reveals separable domain-specific roles in embryonic development and kidney function. Dev. Biol. 2006, 296, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.H.; Cunningham, J.; Sanes, J.R. Roles for laminin in embryogenesis: Exencephaly, syndactyly, and placentopathy in mice lacking the laminin alpha5 chain. J. Cell Biol. 1998, 143, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Durbeej, M. Laminins. Cell Tissue Res. 2010, 339, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, M.D.; Merzouki, A.; Lavertu, M.; Thibault, M.; Jean, M.; Darras, V. Chitosans for delivery of nucleic acids. Adv. Drug Deliv. Rev. 2013, 65, 1234–1270. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.H.; Wang, D.M.; Hsieh, H.J.; Liu, H.C.; Hsien, T.Y.; Lai, J.Y.; Hou, L.T. Preparation and characterization of RGD-immobilized chitosan scaffolds. Biomaterials 2005, 26, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Tigli, R.S.; Gumusderelioglu, M. Evaluation of RGD- or EGF-immobilized chitosan scaffolds for chondrogenic activity. Int. J. Biol. Macromol. 2008, 43, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.; Lee, Y.C. Thiolation of chitosan. Attachment of proteins via thioether formation. Biomacromolecules 2005, 6, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Nomizu, M. Cell adhesion activity of peptides conjugated to polysaccharides. Curr. Protoc. Cell Biol. 2018, 80, e53. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Suzuki, N.; Nielsen, P.K.; Nomizu, M.; Yamada, Y. Laminin alpha1 chain LG4 module promotes cell attachment through syndecans and cell spreading through integrin alpha2beta1. J. Biol. Chem. 2006, 281, 32929–32940. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Otagiri, D.; Yamada, Y.; Sasaki, A.; Fujimori, C.; Wakai, Y.; Uchida, T.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Cell surface receptor-specific scaffold requirements for adhesion to laminin-derived peptide-chitosan membranes. Biomaterials 2010, 31, 3237–3243. [Google Scholar] [CrossRef] [PubMed]
- Masuko, T.; Iwasaki, N.; Yamane, S.; Funakoshi, T.; Majima, T.; Minami, A.; Ohsuga, N.; Ohta, T.; Nishimura, S. Chitosan-RGDSGGC conjugate as a scaffold material for musculoskeletal tissue engineering. Biomaterials 2005, 26, 5339–5347. [Google Scholar] [CrossRef] [PubMed]
- Lv, T.; Liang, W.; Li, L.; Cui, X.; Wei, X.; Pan, H.; Li, B. Novel calcitonin gene-related peptide/chitosan-strontium-calcium phosphate cement: Enhanced proliferation of human umbilical vein endothelial cells in vitro. J. Biomed. Mater. Res. B 2018. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, M.; Wang, X.; Chen, Y.; Yan, Y.; Zhang, L.; Zhang, L. Peptide-modified chitosan hydrogels promote skin wound healing by enhancing wound angiogenesis and inhibiting inflammation. Am. J. Transl. Res. 2017, 9, 2352–2362. [Google Scholar] [PubMed]
- Ikemoto, S.; Mochizuki, M.; Yamada, M.; Takeda, A.; Uchinuma, E.; Yamashina, S.; Nomizu, M.; Kadoya, Y. Laminin peptide-conjugated chitosan membrane: Application for keratinocyte delivery in wounded skin. J. Biomed. Mater. Res. A 2006, 79, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Fujimori, C.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Suppression of cell adhesion through specific integrin crosstalk on mixed peptide-polysaccharide matrices. Biomaterials 2015, 37, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Nakamura, K.; Hori, H.; Miyagi, M.; Nagao, R.; Takasaki, K.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Mixed fibronectin-derived peptides conjugated to a chitosan matrix effectively promotes biological activities through integrins, alpha4beta1, alpha5beta1, alphavbeta3, and syndecan. Biores. Open Access 2016, 5, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Sasaki, A.; Yamada, Y.; Otagiri, D.; Kobayashi, K.; Fujimori, C.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Reconstitution of laminin-111 biological activity using multiple peptide coupled to chitosan scaffolds. Biomaterials 2012, 33, 4241–4250. [Google Scholar] [CrossRef] [PubMed]
- Otagiri, D.; Yamada, Y.; Hozumi, K.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Cell attachment and spreading activity of mixed laminin peptide-chitosan membranes. Biopolymers 2013, 100, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Kobayashi, K.; Katagiri, F.; Kikkawa, Y.; Kadoya, Y.; Nomizu, M. Syndecan- and integrin-binding peptides synergistically accelerate cell adhesion. FEBS Lett. 2010, 584, 3381–3385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hozumi, K.; Yamagata, N.; Otagiri, D.; Fujimori, C.; Kikkawa, Y.; Kadoya, Y.; Nomizu, M. Mixed peptide-chitosan membranes to mimic the biological activities of a multifunctional laminin alpha1 chain LG4 module. Biomaterials 2009, 30, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Urushibata, S.; Hozumi, K.; Ishikawa, M.; Katagiri, F.; Kikkawa, Y.; Nomizu, M. Identification of biologically active sequences in the laminin alpha2 chain G domain. Arch. Biochem. Biophys. 2010, 497, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Yasa, I.C.; Gunduz, N.; Kilinc, M.; Guler, M.O.; Tekinay, A.B. Basal lamina mimetic nanofibrous peptide networks for skeletal myogenesis. Sci. Rep. 2015, 5, 16460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheele, S.; Falk, M.; Franzen, A.; Ellin, F.; Ferletta, M.; Lonai, P.; Andersson, B.; Timpl, R.; Forsberg, E.; Ekblom, P. Laminin alpha1 globular domains 4-5 induce fetal development but are not vital for embryonic basement membrane assembly. Proc. Natl. Acad. Sci. USA 2005, 102, 1502–1506. [Google Scholar] [CrossRef] [PubMed]
- Streuli, C.H.; Akhtar, N. Signal co-operation between integrins and other receptor systems. Biochem. J. 2009, 418, 491–506. [Google Scholar] [CrossRef] [PubMed]
- Tschumperlin, D.J. Matrix, mesenchyme, and mechanotransduction. Ann. Am. Thorac. Soc. 2015, 12, S24–S29. [Google Scholar] [CrossRef] [PubMed]
- Legate, K.R.; Wickstrom, S.A.; Fassler, R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009, 23, 397–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvard, D.; Pouwels, J.; De Franceschi, N.; Ivaska, J. Integrin inactivators: Balancing cellular functions in vitro and in vivo. Nat. Rev. Mol. Cell Biol. 2013, 14, 430–442. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.M.; Bhattacharya, R.; deHart, G.W.; Jones, J.C. Transdominant regulation of integrin function: Mechanisms of crosstalk. Cell. Signal. 2010, 22, 578–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roper, J.A.; Williamson, R.C.; Bass, M.D. Syndecan and integrin interactomes: Large complexes in small spaces. Curr. Opin. Struct. Biol. 2012, 22, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Delaney, M.K.; Du, X. Inside-out, outside-in, and inside-outside-in: G protein signaling in integrin-mediated cell adhesion, spreading, and retraction. Curr. Opin. Cell Biol. 2012, 24, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Lukjanenko, L.; Jung, M.J.; Hegde, N.; Perruisseau-Carrier, C.; Migliavacca, E.; Rozo, M.; Karaz, S.; Jacot, G.; Schmidt, M.; Li, L.; et al. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nat. Med. 2016, 22, 897–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheung, R.C.; Ng, T.B.; Wong, J.H.; Chan, W.Y. Chitosan: An update on potential biomedical and pharmaceutical applications. Mar. Drugs 2015, 13, 5156–5186. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, L.; Stossel, M.; Ronchi, G.; Geuna, S.; Yin, Y.; Mommert, S.; Martensson, L.; Metzen, J.; Grothe, C.; Dahlin, L.B.; et al. Regeneration of long-distance peripheral nerve defects after delayed reconstruction in healthy and diabetic rats is supported by immunomodulatory chitosan nerve guides. BMC Neurosci. 2017, 18, 53. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Matsumoto, I.; Suzuki, M.; Kaneko, M.; Nitta, K.; Seguchi, R.; Ooi, A.; Takemura, H. Chitosan tubes can restore the function of resected phrenic nerves. Interact. Cardiovasc. Thorac. Surg. 2015, 21, 8–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapira, Y.; Tolmasov, M.; Nissan, M.; Reider, E.; Koren, A.; Biron, T.; Bitan, Y.; Livnat, M.; Ronchi, G.; Geuna, S.; et al. Comparison of results between chitosan hollow tube and autologous nerve graft in reconstruction of peripheral nerve defect: An experimental study. Microsurgery 2016, 36, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, F.; Cobianchi, S.; Heimann, C.; Phillips, J.B.; Udina, E.; Navarro, X. Stabilization, rolling, and addition of other extracellular matrix proteins to collagen hydrogels improve regeneration in chitosan guides for long peripheral nerve gaps in rats. Neurosurgery 2017, 80, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Arslan, E.; Garip, I.C.; Gulseren, G.; Tekinay, A.B.; Guler, M.O. Bioactive supramolecular peptide nanofibers for regenerative medicine. Adv. Healthc. Mater. 2014, 3, 1357–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Peptides | Sequence a | Original Proteins | Receptors | References |
|---|---|---|---|---|
| 531 | CGG-GEFYFDLRLKGDKY | COL4-α1 | Integrin α3β1 | [63] |
| FIB1 | CGG-YAVTGRGDSPAS | FN | Integrin αvβ3 Integrin α5β1 | [64] |
| ePRARI-C | CGG-QPPRARITGYII | FN | Integrin α4β1 syndecans | [64] |
| A99 | CGG-AGTFALRGDNPQG | LM-α1 | Integrin αvβ3 | [58,65,66] |
| A112 | CGG-VLIKGGRARKHV | LM-α1 | N.D. b | [65] |
| AG73 | CGG-RKRLQVQSIRT | LM-α1 | syndecans | [57,58,65,66,67,68] |
| EF1 | CGG-DYATLQLQEGRLHFXFDLG | LM-α1 | Integrin α2β1 | [57] |
| EF1zz | CGG-ATLQLQEGRLHFXFDLGKGR X:Nle, a modified EF1 | LM-α1 | Integrin α2β1 | [63,65,67,68] |
| A2G10 | CGG-SYWYRIEASRTG | LM-α2 | Integrin α6β1 | [66,69] |
| B31 | CGG-TNLRIKFVKLHT | LM-β2 | syndecans | [65] |
| C16 | CGG-KAFDITYVRLKF | LM-γ1 | Integrin β1 syndecans | [65,66] |
| C68 | CGG-TSIKIRGTYSER | LM-γ1 | N.D. b | [65] |
| Group | Biological Activities on HDFs a | PC12 Cell N.O. c | Peptides | ||||
|---|---|---|---|---|---|---|---|
| Attachment | Spreading b | Inhibitory Effect | Receptors | ||||
| EDTA | Heparin | ||||||
| 1 | + | S | + | − | Integrin | + | A99a |
| 2 | + | S | + | − | Integrin | − | EF1zz |
| 3 | + | s | + | + | Integrin/ syndecan | + | A13, AG32, AG103, C16, C57, C64 |
| 4 | + | s | + | + | Integrin/ syndecan | − | A3, A55, A65, A119, A167, A174, AG10, AG28, AG56, B30, B133, B160, C59, C68 |
| 5 | + | w | − | + | Syndecan | + | A206, AG73, B20, B31 |
| 6 | − | − | − | − | − | + | A25, A112, A194 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hozumi, K.; Nomizu, M. Mixed Peptide-Conjugated Chitosan Matrices as Multi-Receptor Targeted Cell-Adhesive Scaffolds. Int. J. Mol. Sci. 2018, 19, 2713. https://doi.org/10.3390/ijms19092713
Hozumi K, Nomizu M. Mixed Peptide-Conjugated Chitosan Matrices as Multi-Receptor Targeted Cell-Adhesive Scaffolds. International Journal of Molecular Sciences. 2018; 19(9):2713. https://doi.org/10.3390/ijms19092713
Chicago/Turabian StyleHozumi, Kentaro, and Motoyoshi Nomizu. 2018. "Mixed Peptide-Conjugated Chitosan Matrices as Multi-Receptor Targeted Cell-Adhesive Scaffolds" International Journal of Molecular Sciences 19, no. 9: 2713. https://doi.org/10.3390/ijms19092713
APA StyleHozumi, K., & Nomizu, M. (2018). Mixed Peptide-Conjugated Chitosan Matrices as Multi-Receptor Targeted Cell-Adhesive Scaffolds. International Journal of Molecular Sciences, 19(9), 2713. https://doi.org/10.3390/ijms19092713