Ursolic Acid Induces Apoptosis in Colorectal Cancer Cells Partially via Upregulation of MicroRNA-4500 and Inhibition of JAK2/STAT3 Phosphorylation
Abstract
:1. Introduction
2. Results
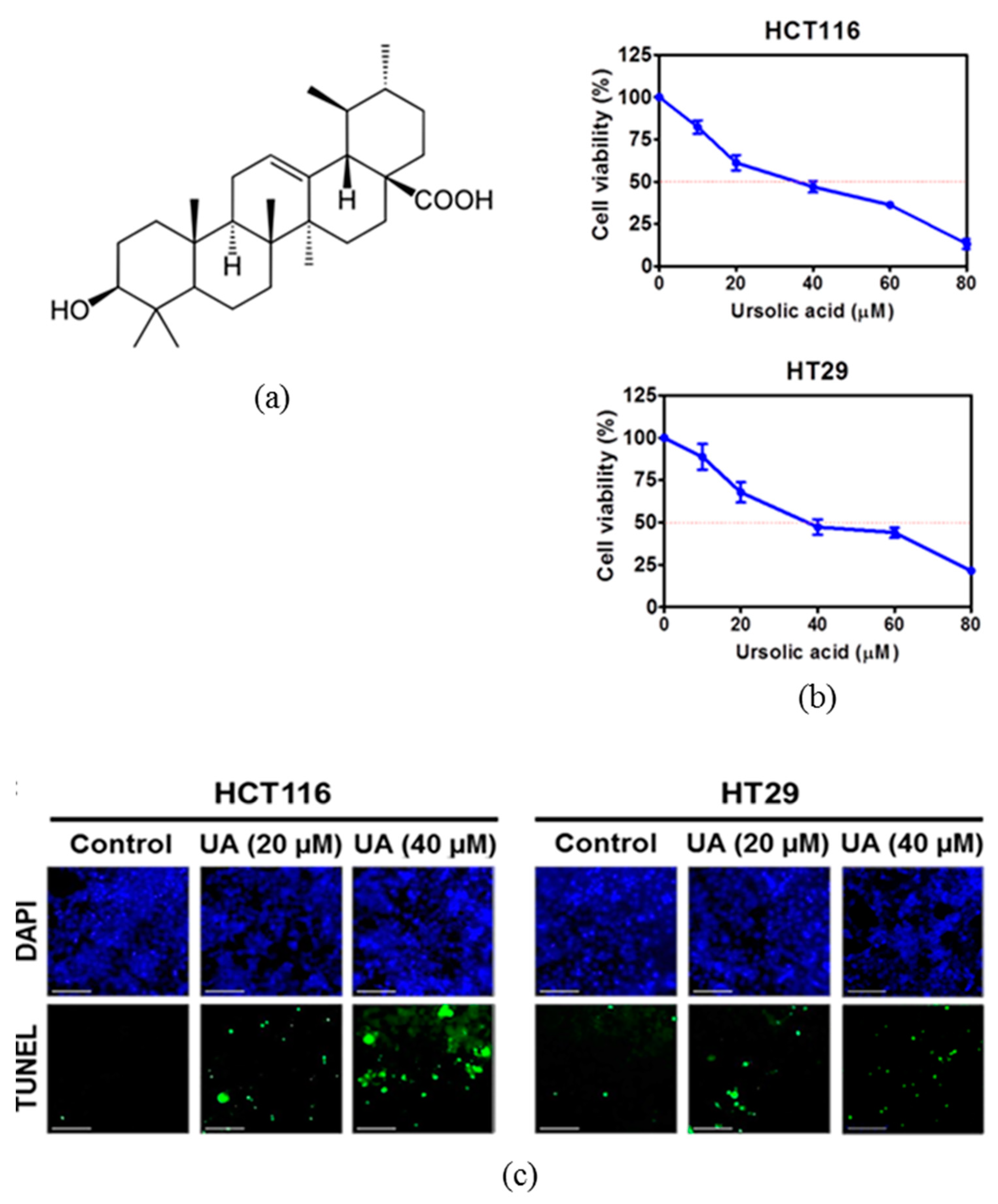
2.1. Ursolic Acid Increased Cytotoxicity and the Number of TUNEL Positive Cells in HCT116 and HT29 Cells
2.2. Ursolic Acid Increased Sub-G1 Population and Regulated Apoptotic Proteins and Attenuated the Phosphorylation of JAK2/STAT3 in HCT116 and HT29 Cells
2.3. Ursolic Acid Blocked Nuclear Translocation of STAT3 in HCT116 Cells
2.4. Inhibition of miR-4500 Suppressed Cytotoxic and Anti-Proliferative Effects of Ursolic Acid in HCT116 and HT29 Cells
2.5. Critical Role of miR-4500 in Apoptotic Effect of Ursolic Acid in HCT116 Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture
4.3. Cell Viability Assay
4.4. TUNEL Assay
4.5. Western Blotting
4.6. Sub-G1 Accumulation by Cell Cycle Analysis
4.7. Immunofluorscence
4.8. RNA Isolation and Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
4.9. RNA Isolation and Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
4.10. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PARP | Poly (ADP-ribose) polymerase |
| Caspase | Cysteine aspartyl-specific protease |
| STAT3 | Signal transducer and activator of transcription 3 |
| JAK2 | Janus kinase 2 |
| mRNA | Messenger RNA |
References
- Haller, D. Recovering the thread of life--destruction and reconstruction of the social world of patients after cancer of the colon. Pflege 1995, 8, 187–202. [Google Scholar] [PubMed]
- Zhong, Z.; Wen, Z.L.; Darnell, J.E. Stat3 and Stat4—Members of the Family of Signal Transducers and Activators of Transcription. Proc. Natl. Acad. Sci. USA 1994, 91, 4806–4810. [Google Scholar] [CrossRef] [PubMed]
- Barton, B.E.; Karras, J.G.; Murphy, T.F.; Barton, A.; Huang, H.F. Signal transducer and activator of transcription 3 (STAT3) activation in prostate cancer: Direct STAT3 inhibition induces apoptosis in prostate cancer lines. Mol. Cancer Ther. 2004, 3, 11–20. [Google Scholar] [PubMed]
- Carballo, M.; Conde, M.; El Bekay, R.; Martin-Nieto, J.; Camacho, M.J.; Monteseirin, J.; Conde, J.; Bedoya, F.J.; et al. Oxidative stress triggers STAT3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J. Biol. Chem. 1999, 274, 17580–17586. [Google Scholar] [CrossRef] [PubMed]
- Justicia, C.; Gabriel, C.; Planas, A.M. Activation of the JAK/STAT pathway following transient focal cerebral ischemia: Signaling through Jak1 and Stat3 in astrocytes. Glia 2000, 30, 253–270. [Google Scholar] [CrossRef] [Green Version]
- Kaptein, A.; Paillard, V.; Saunders, M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J. Biol. Chem. 1996, 271, 5961–5964. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E. Role of miRNA in carcinogenesis and biomarker selection: A methodological view. Expert Rev. Mol. Diagn. 2007, 7, 569–603. [Google Scholar] [CrossRef]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer—An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Markou, A.; Zavridou, M.; Lianidou, E.S. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016, 7, 19–27. [Google Scholar]
- Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [PubMed]
- Wang, J.; Li, Y.; Wang, X.; Jiang, C. Ursolic acid inhibits proliferation and induces apoptosis in human glioblastoma cell lines U251 by suppressing TGF-beta1/miR-21/PDCD4 pathway. Basic Clin. Pharmacol. Toxicol. 2012, 111, 106–112. [Google Scholar] [PubMed]
- Xiang, F.; Pan, C.; Kong, Q.; Wu, R.; Jiang, J.; Zhan, Y.; Xu, J.; Gu, X.; Kang, X. Ursolic Acid Inhibits the Proliferation of Gastric Cancer Cells by Targeting miR-133a. Oncol. Res. 2014, 22, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qian, J.; Qiang, Y.; Huang, H.; Wang, C.; Li, D.; Xu, B. Down-regulation of miR-4500 promoted non-small cell lung cancer growth. Cell Physiol. Biochem. 2014, 34, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.Y.; Tu, Y.; Deng, Y.; Guo, C.; Ning, J.; Zhu, Y.; Lv, X.; Ye, H. MiR-4500 is epigenetically downregulated in colorectal cancer and functions as a novel tumor suppressor by regulating HMGA2. Cancer Biol. Ther. 2016, 17, 1149–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.L.; Kuo, P.L.; Lin, C.C. Proliferative inhibition, cell-cycle dysregulation, and induction of apoptosis by ursolic acid in human non-small cell lung cancer A549 cells. Life Sci. 2004, 75, 2303–2316. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.Y.; Leung, H.W.; Yang, W.H.; Chen, W.H.; Lee, H.Z. Up-regulation of matrix metalloproteinase family gene involvement in ursolic acid-induced human lung non-small carcinoma cell apoptosis. Anticancer Res. 2007, 27, 145–153. [Google Scholar]
- Wang, J.; Liu, L.; Qiu, H.; Zhang, X.; Guo, W.; Chen, W.; Tian, Y.; Fu, L.; Shi, D.; Cheng, J.; et al. Ursolic acid simultaneously targets multiple signaling pathways to suppress proliferation and induce apoptosis in colon cancer cells. PLoS ONE 2013, 8, e63872. [Google Scholar] [CrossRef]
- Choi, Y.H.; Baek, J.H.; Yoo, M.A.; Chung, H.Y.; Kim, N.D.; Kim, K.W. Induction of apoptosis by ursolic acid through activation of caspases and down-regulation of c-IAPs in human prostate epithelial cells. Int. J. Oncol. 2000, 17, 565–571. [Google Scholar] [CrossRef]
- Yan, S.L.; Huang, C.Y.; Wu, S.T.; Yin, M.C. Oleanolic acid and ursolic acid induce apoptosis in four human liver cancer cell lines. Toxicol. In Vitro 2010, 24, 842–848. [Google Scholar] [CrossRef]
- Harmand, P.O.; Duval, R.; Delage, C.; Simon, A. Ursolic acid induces apoptosis through mitochondrial intrinsic pathway and caspase-3 activation in M4Beu melanoma cells. Int. J. Cancer 2005, 114, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.; Lu, Y.H.; Xie, J.H.; Wang, F.; Zou, J.N.; Yang, J.S.; Xing, Y.Y.; Xi, T. Downregulation of survivin and activation of caspase-3 through the PI3K/Akt pathway in ursolic acid-induced HepG2 cell apoptosis. AntiCancer Drugs 2009, 20, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.Z.; Xuan, Y.Y.; Zheng, S.; Dong, Q.; Zhang, S.Z. Ursolic acid inhibits proliferation and induces apoptosis of HT-29 colon cancer cells by inhibiting the EGFR/MAPK pathway. J. Zhejiang Univ. Sci. B 2009, 10, 668–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.X.; Gu, Z.L.; Yin, J.L.; Chou, W.H.; Kwok, C.Y.; Qin, Z.H.; Liang, Z.Q. Ursolic acid induces human hepatoma cell line SMMC-7721 apoptosis via p53-dependent pathway. Chinese Med. J. 2010, 123, 1915–1923. [Google Scholar]
- Xavier, C.P.R.; Lima, C.F.; Pedro, D.F.N.; Wilson, J.M.; Kristiansen, K.; Pereira-Wilson, C. Ursolic acid induces cell death and modulates autophagy through JNK pathway in apoptosis-resistant colorectal cancer cells. J. Nutr. Biochem. 2013, 24, 706–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graslkraupp, B.; Ruttkaynedecky, B.; Koudelka, H.; Bukowska, K.; Bursch, W.; Schultehermann, R. In-Situ Detection of Fragmented DNA (Tunel Assay) Fails to Discriminate among Apoptosis, Necrosis, and Autolytic Cell-Death—A Cautionary Note. Hepatology 1995, 21, 1465–1468. [Google Scholar] [CrossRef]
- Schindler, C.; Levy, D.E.; Decker, T. JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 2007, 282, 20059–20063. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; McBride, K.M.; Reich, N.C. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha 3. Proc. Natl. Acad. Sci. USA 2005, 102, 8150–8155. [Google Scholar] [CrossRef]
- Cardellina, J.H.; Gustafson, K.R.; Beutler, J.A.; Mckee, T.C.; Hallock, Y.F.; Fuller, R.W.; Boyd, M.R. National-Cancer-Institute Intramural Research on Human-Immunodeficiency-Virus Inhibitory and Antitumor Plant Natural-Products. ACS Sym. Ser. 1993, 534, 218–227. [Google Scholar]
- Zhang, B.; Wang, J.; Wang, X.; Zhu, J.; Liu, Q.; Shi, Z.; Chambers, M.C.; Zimmerman, L.J.; Shaddox, K.F.; Kim, S.; et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 513, 382–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inamura, K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers 2018, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Pagliara, V.; Saide, A.; Mitidieri, E.; d’Emmanuele di Villa Bianca, R.; Sorrentino, R.; Russo, G.; Russo, A. 5-FU targets rpL3 to induce mitochondrial apoptosis via cystathionine-beta-synthase in colon cancer cells lacking p53. Oncotarget 2016, 7, 50333–50348. [Google Scholar] [CrossRef] [PubMed]
- Tung, M.C.; Lin, P.L.; Wang, Y.C.; He, T.Y.; Lee, M.C.; Yeh, S.D.; Chen, C.Y.; Lee, H. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 2015, 6, 41692–41705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, A.; Saide, A.; Smaldone, S.; Faraonio, R.; Russo, G. Role of uL3 in Multidrug Resistance in p53-Mutated Lung Cancer Cells. Int. J. Mol. Sci. 2017, 18, 547. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, S.; Llovera, L.; Portugal, J. Chemotherapeutic targeting of cell death pathways. Anticancer Agents Med. Chem. 2012, 12, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Walsh, L.; Singh, A.; Beamer, M.; Sudini, K.; Leaman, D. Induction of Pro-Apoptotic Noxa Expression By Ursolic Acid Sensitizes Rheumatoid Arthritis Synovial Fibroblasts to Apoptosis: A Role of Mir-181a. Arthritis Rheumatol. 2014, 66, S456. [Google Scholar]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.; Shin, E.A.; Jung, J.H.; Park, J.E.; Kim, D.S.; Shim, B.S.; Kim, S.-H. Ursolic Acid Induces Apoptosis in Colorectal Cancer Cells Partially via Upregulation of MicroRNA-4500 and Inhibition of JAK2/STAT3 Phosphorylation. Int. J. Mol. Sci. 2019, 20, 114. https://doi.org/10.3390/ijms20010114
Kim K, Shin EA, Jung JH, Park JE, Kim DS, Shim BS, Kim S-H. Ursolic Acid Induces Apoptosis in Colorectal Cancer Cells Partially via Upregulation of MicroRNA-4500 and Inhibition of JAK2/STAT3 Phosphorylation. International Journal of Molecular Sciences. 2019; 20(1):114. https://doi.org/10.3390/ijms20010114
Chicago/Turabian StyleKim, Karam, Eun Ah Shin, Ji Hoon Jung, Ji Eon Park, Dong Soub Kim, Bum Sang Shim, and Sung-Hoon Kim. 2019. "Ursolic Acid Induces Apoptosis in Colorectal Cancer Cells Partially via Upregulation of MicroRNA-4500 and Inhibition of JAK2/STAT3 Phosphorylation" International Journal of Molecular Sciences 20, no. 1: 114. https://doi.org/10.3390/ijms20010114





