Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species
Abstract
:1. Introduction
2. Taxonomic Updates of the Burkholderia Sensu Lato
3. Genomic Features of the Plant-Associated Pathogenic and Beneficial Burkholderia
3.1. Genome Size
3.2. Multi-Replicon Nature
3.3. Genomic Islands and Multiple Insertion Sequences
4. Plant Pathogenic Burkholderia
4.1. Virulence Factors in Phytopathogenic Burkholderia
4.1.1. Phytotoxins
4.1.2. Secretion Systems
4.1.3. Other Virulence Factors
5. Plant-Beneficial and Symbiotic Burkholderia Species
5.1. Benevolence Factors in Plant-Beneficial and Symbiotic Burkholderia
5.1.1. Colonization of Plant Tissues and the Role of EPSs
5.1.2. Nitrogen Fixation in Diazotrophic and Legume Nodulator Burkholderia
5.1.3. Plant Growth Promotion
5.1.4. Other Benevolence Factors in Plant-Associated Burkholderia
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BCC | Burkholderia cepacia complex |
| Mbp | Million base pair |
| QS | Quorum sensing |
| AHL | Acyl homoserine lactone |
| TSS | Type secretion systems |
| EPS | Exopolysaccharide |
| ACC | Aminocyclopropane-1-carboxylate |
| IAA | Indole-3-acetic acid |
References
- O’Sullivan, L.A.; Mahenthiralingam, E. Biotechnological potential within the genus Burkholderia. Lett. Appl. Microbiol. 2005, 41, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; LiPuma, J.J.; Henry, D.; Hoste, B.; Vandemeulebroecke, K.; Gillis, M.; Speert, D.P.; Vandamme, P. Burkholderia cepacia g enomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 2001, 51, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Salles, J.F.; De Souza, F.A.; van Elsas, J.D. Molecular method to assess the diversity of Burkholderia species in environmental samples. Appl. Environ. Microbiol. 2002, 68, 1595–1603. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 2003, 5, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, W.H. Three bacterial plant pathogens: Phytomonas earyophylli sp. n., Phytomonas alliicola sp. n., and Phytomonas manihotis (Arthaud-Berthet et Sondar) Viégas. Phytopathology 1942, 32, 141–149. [Google Scholar]
- Burkholder, W.H. Sour skin, a bacterial rot of onion bulbs. Phytopathology 1950, 40, 115–117. [Google Scholar]
- Palleroni, N.J.; Holmes, B. Pseudomonas cepacia sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 1981, 31, 479–481. [Google Scholar] [CrossRef]
- Yabuuchi, E.; Kosako, Y.; Oyaizu, H.; Yano, I.; Hotta, H.; Hashimoto, Y.; Ezaki, T.; Arakawa, M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992, 36, 1251–1275. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multi-replicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef]
- Lessie, T.G.; Hendrickson, W.; Manning, B.D.; Devereux, R. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 1996, 144, 117–128. [Google Scholar] [CrossRef]
- Compant, S.; Nowak, J.; Coenye, T.; Clément, C.; Ait Barka, E. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 2008, 32, 607–626. [Google Scholar] [CrossRef]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.W.; Vandamme, P.; Morgan, S.H.; LiPuma, J.J.; Coenye, T.; Weightman, A.J.; Jones, T.H.; Mahenthiralingam, E. Development of a recA gene-based identification approach for the entire Burkholderia genus. Appl. Environ. Microbiol. 2005, 71, 3917–3927. [Google Scholar] [CrossRef]
- Ussery, D.W.; Kiil, K.; Lagesen, K.; Sicheritz-Ponten, T.; Bohlin, J.; Wassenaar, T.M. The genus Burkholderia: Analysis of 56 genomic sequences. In Microbial Pathogenomics; de Reuse, H., Bereswill, S., Eds.; Krager: Basel, Switzerland, 2009; Volume 6, pp. 140–157. [Google Scholar]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.M.; Elliott, G.N.; Bontemps, C.; Estrada-de los Santos, P.; Gross, E.; dos Reis, F.B., Jr.; Sprent, J.I.; et al. Legume-nodulating betaproteobacteria: Diversity, host range, and future prospects. Mol. Plant-Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Estrada-de los Santos, P.; Vinuesa, P.; Martínez-Aguilar, L.; Hirsch, A.M.; Caballero-Mellado, J. Phylogenetic analysis of Burkholderia species by multilocus sequence analysis. Curr. Microbiol. 2013, 67, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Angus, A.A.; Agapakis, C.M.; Fong, S.; Yerrapragada, S.; Estrada-De Los Santos, P.; Yang, P.; Song, N.; Kano, S.; Caballero-Mellado, J.; De Faria, S.M.; et al. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 2014, 9, e83779. [Google Scholar] [CrossRef] [PubMed]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef]
- Lopes-Santos, L.; Castro, D.B.A.; Ferreira-Tonin, M.; Corrêa, D.B.A.; Weir, B.S.; Park, D.; Ottoboni, L.M.M.; Neto, J.R.; Destéfano, S.A.L. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie van Leeuwenhoek 2017, 110, 727–736. [Google Scholar] [CrossRef]
- Beukes, C.W.; Palmer, M.; Manyaka, P.; Chan, W.Y.; Avontuur, J.R.; van Zyl, E.; Huntemann, M.; Clum, A.; Pillay, M.; Palaniappan, K.; et al. Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 2017, 8, 1154. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Buzard, G.S.; Ussery, D.W. Comparative Genomics in the Genus Burkholderia. In Burkholderia From Genomes to Function; Coenye, T., Mahenthiralingam, E., Eds.; Caister Academic Press: Norfolk, UK, 2014; pp. 31–50. ISBN 978-1-90823-97-3. [Google Scholar]
- Mahenthiralingam, E.; Drevinek, P. Comparative genomics of Burkholderia species. In Burkholderia: Molecular Microbiology and Genomics; Coenye, T., Vandamme, P., Eds.; Horizon Bioscience: Norfolk, UK, 2006; pp. 53–79. ISBN 978-1-904933-28-1. [Google Scholar]
- Partida-Martinez, L.P.; Monajembashi, S.; Greulich, K.O.; Hertweck, C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 2007, 17, 773–777. [Google Scholar] [CrossRef] [PubMed]
- Lackner, G.; Moebius, N.; Partida-Martinez, L.P.; Boland, S.; Hertweck, C. Evolution of an endofungal lifestyle: Deductions from the Burkholderia rhizoxinica genome. BMC Genom. 2011, 12, 210. [Google Scholar] [CrossRef] [PubMed]
- Lackner, G.; Moebius, N.; Partida-Martinez, L.; Hertweck, C. Complete genome sequence of Burkholderia rhizoxinica, an endosymbiont of Rhizopus microsporus. J. Bacteriol. 2011, 193, 783–784. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Aguilar, L.; Díaz, R.; Peña-Cabriales, J.J.; Estrada-de los Santos, P.; Dunn, M.F.; Caballero-Mellado, J. Multichromosomal genome structure and confirmation of diazotrophy in novel plant-associated Burkholderia species. Appl. Environ. Microbiol. 2008, 74, 4574–4579. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.S.; Lim, J.Y.; Park, J.; Kim, S.; Lee, H.H.; Cheong, H.; Kim, S.M.; Moon, J.S.; Hwang, I. Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genom. 2015, 16, 349. [Google Scholar] [CrossRef] [PubMed]
- Agnoli, K.; Schwager, S.; Uehlinger, S.; Vergunst, A.; Viteri, D.F.; Nguyen, D.T.; Sokol, P.A.; Carlier, A.; Eberl, L. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol. Microbiol. 2012, 83, 362–378. [Google Scholar] [CrossRef]
- Juhas, M.; Van Der Meer, J.R.; Gaillard, M.; Harding, R.M.; Hood, D.W.; Crook, D.W. Genomic islands: Tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 2009, 33, 376–393. [Google Scholar] [CrossRef]
- Lawrence, J.G.; Ochman, H. Amelioration of bacterial genomes: Rates of change and exchange. J. Mol. Evol. 1997, 44, 383–397. [Google Scholar] [CrossRef]
- Bentley, S.D.; Parkhill, J. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 2004, 38, 771–791. [Google Scholar] [CrossRef]
- Chain, P.S.; Denef, V.J.; Konstantinidis, K.T.; Vergez, L.M.; Agulló, L.; Reyes, V.L.; Hauser, L.; Córdova, M.; Gómez, L.; González, M.; et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 2006, 103, 15280–15287. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.S.; Byrne, A.; Lessie, T.G. IS406 and IS407, two gene-activating insertion sequences from Pseudomonas cepacia. Gene 1991, 105, 101–105. [Google Scholar] [CrossRef]
- Mahillon, J.; Chandler, M. Insertion sequences. Microbiol. Mol. Biol. Rev. 1998, 62, 725–774. [Google Scholar] [PubMed]
- Hübner, A.; Hendrickson, W. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2, 4, 5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J. Bacteriol. 1997, 179, 2717–2723. [Google Scholar] [CrossRef] [PubMed]
- Barsomian, G.; Lessie, T.G. Replicon fusions promoted by insertion sequences on Pseudomonas cepacia plasmid pTGL6. Mol. Gen. Genet. 1986, 204, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, T.D.; Lessie, T.G. Insertion-sequence-dependent rearrangements of Pseudomonas cepacia plasmid pTGL1. J. Bacteriol. 1987, 169, 224–230. [Google Scholar] [CrossRef]
- Nierman, W.C.; DeShazer, D.; Kim, H.S.; Tettelin, H.; Nelson, K.E.; Feldblyum, T.; Ulrich, R.L.; Ronning, C.M.; Brinkac, L.M.; Daugherty, S.C.; et al. Structural flexibility in the Burkholderia mallei genome. Proc. Natl. Acad. Sci. USA 2004, 101, 14246–14251. [Google Scholar] [CrossRef]
- Hasebe, A.; Iida, S. The novel insertion sequences IS1417, IS1418, and IS1419 from Burkholderia glumae and their strain distribution. Plasmid 2000, 44, 44–53. [Google Scholar] [CrossRef]
- Gonzalez, C.F.; Venturi, V.; Engledow, A.S. The phytopathogenic Burkholderia. Burkholderia Mol. Microbiol. Genom. 2007, 1, 153–176. [Google Scholar]
- Goto, K.; Ohata, K. New bacterial diseases of rice (brown stripe and grain rot). Ann. Phytopathol. Soc. Jpn. 1956, 21, 46–47. [Google Scholar]
- Ham, J.H.; Melanson, R.A.; Rush, M.C. Burkholderia glumae: Next major pathogen of rice? Mol. Plant Pathol. 2011, 12, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Trung, H.M.; Van, N.V.; Vien, N.V.; Lam, D.T.; Lien, M. Occurrence of rice grain rot disease in Vietnam. Int. Rice Res. Notes 1993, 18, 30. [Google Scholar]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef]
- Severini, G. Una bacteriosi dell’lxia maculate e del Gladiolus. Colvilli. Ann. Bot. (Rome) 1913, 11, 413–424. [Google Scholar]
- Hildebrand, D.C.; Palleroni, N.J.; Doudoroff, M. Synonymy of Pseudomonas gladioli Severini 1913 and Pseudomonas marginata (McCulloch 1921) Stapp 1928. Int. J. Syst. Bacteriol. 1973, 23, 433–437. [Google Scholar] [CrossRef]
- Young, J.M.; Dye, D.W.; Bradbury, J.F.; Panagopoulos, C.G.; Robbs, C.F. A proposed nomenclature and classification for plant pathogenic bacteria. N. Z. J. Agric. Res. 1978, 21, 153–177. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, S.P.; Fermor, T.R.; Stead, D.E.; Sellwood, J.E. Bacterial soft rot of Agaricus bitorquis. Plant Pathol. 1991, 40, 136–144. [Google Scholar] [CrossRef]
- Azegami, K.; Nishiyama, K.; Watanabe, Y.; Kadota, I.; Ohuchi, A.; Fukazawa, C. Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int. J. Syst. Evol. Microbiol. 1987, 37, 144–152. [Google Scholar]
- Kato, T.; Tanaka, T.; Fujita, Y. Studies on bacterial seedling blight of rice. Classification of bacteria, obtained from disease seedling of rice in Yamagata perfecture. Bull. Yamagata Agric. Exp. Stn. 1992, 26, 103–109. [Google Scholar]
- Smith, E.F. Bacteria in Relation to Plant Diseases; Carnegie Institution of Washington Publication; Carnegie Institution of Washington: Washington, DC, USA, 1911; Volume 2, pp. 1–368. [Google Scholar]
- Moffett, M.L.; Hayward, A.C.; Fahy, P.C. Five new hosts of Pseudomonas andropogonis occurring in eastern Australia: Host range and characterisation of isolates. Plant Pathol. 1986, 35, 34–43. [Google Scholar] [CrossRef]
- Cother, E.J.; Noble, D.; Peters, B.J.; Albiston, A.; Ash, G.J. A new bacterial disease of jojoba (Simmondsia chinensis) caused by Burkholderia andropogonis. Plant Pathol. 2004, 53, 129–135. [Google Scholar] [CrossRef]
- Ballard, R.W.; Palleroni, N.J.; Doudoroff, M.; Stanier, R.Y. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J. Gen. Microbiol. 1970, 60, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T. Carnation. In Horticulture in Japan; Konishi, K., Iwahori, S., Kitagawa, H., Yakuwa, T., Eds.; Asakura Publishing: Tokyo, Japan, 1994; pp. 139–144. [Google Scholar]
- Lackner, G.; Moebius, N.; Hertweck, C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2011, 5, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Partida-Martinez, L.P.; Hertweck, C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem 2007, 8, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Zhou-qi, C.; Bo, Z.; Guan-lin, X.; Bin, L.; Shi-wen, H. Research status and prospect of Burkholderia glumae, the pathogen causing bacterial panicle blight. Rice Sci. 2016, 23, 111–118. [Google Scholar] [CrossRef]
- Durbin, R.D. Bacterial phytotoxins: Mechanisms of action. Experientia 1991, 47, 776–783. [Google Scholar] [CrossRef]
- Levenberg, B.; Linton, S.N. On the biosynthesis of toxoflavin, an azapteridine antibiotic produced by Pseudomonas cocovenenans. J. Biol. Chem. 1966, 241, 846–852. [Google Scholar]
- Sato, Z.; Koiso, Y.; Iwasaki, S.; Matsuda, I.; Shirata, A. Toxins produced by Pseudomonas glumae. Jpn. J. Phytopathol. 1989, 55, 353–356. [Google Scholar] [CrossRef]
- Furuya, N.; Iiyama, K.; Shiozaki, N.; Matsuyama, N. Phytotoxin produced by Burkholderia gladioli. J. Fac. Agric. Kyushu Univ. 1997, 42, 33–37. [Google Scholar]
- Iiyama, K.; Futuya, N.; Takanami, Y.; Matsuyama, N. A role of phytotoxin in virulence of Pseudomonas glumae Kurita et Tabei. Jpn. J. Phytopathol. 1995, 61, 470–476. [Google Scholar] [CrossRef]
- Nagamatsu, T. Syntheses, Transformation, and Biological Activities of 7-Azapteridine Antibiotics: Toxoflavin, Fervenulin, Reumycin and Their Analogues. ChemInform 2001, 33, 261. [Google Scholar] [CrossRef]
- Latuasan, H.E.; Berends, W. On the origin of the toxicity of toxoflavin. Biochim. Biophys. Acta 1961, 52, 502–508. [Google Scholar] [CrossRef]
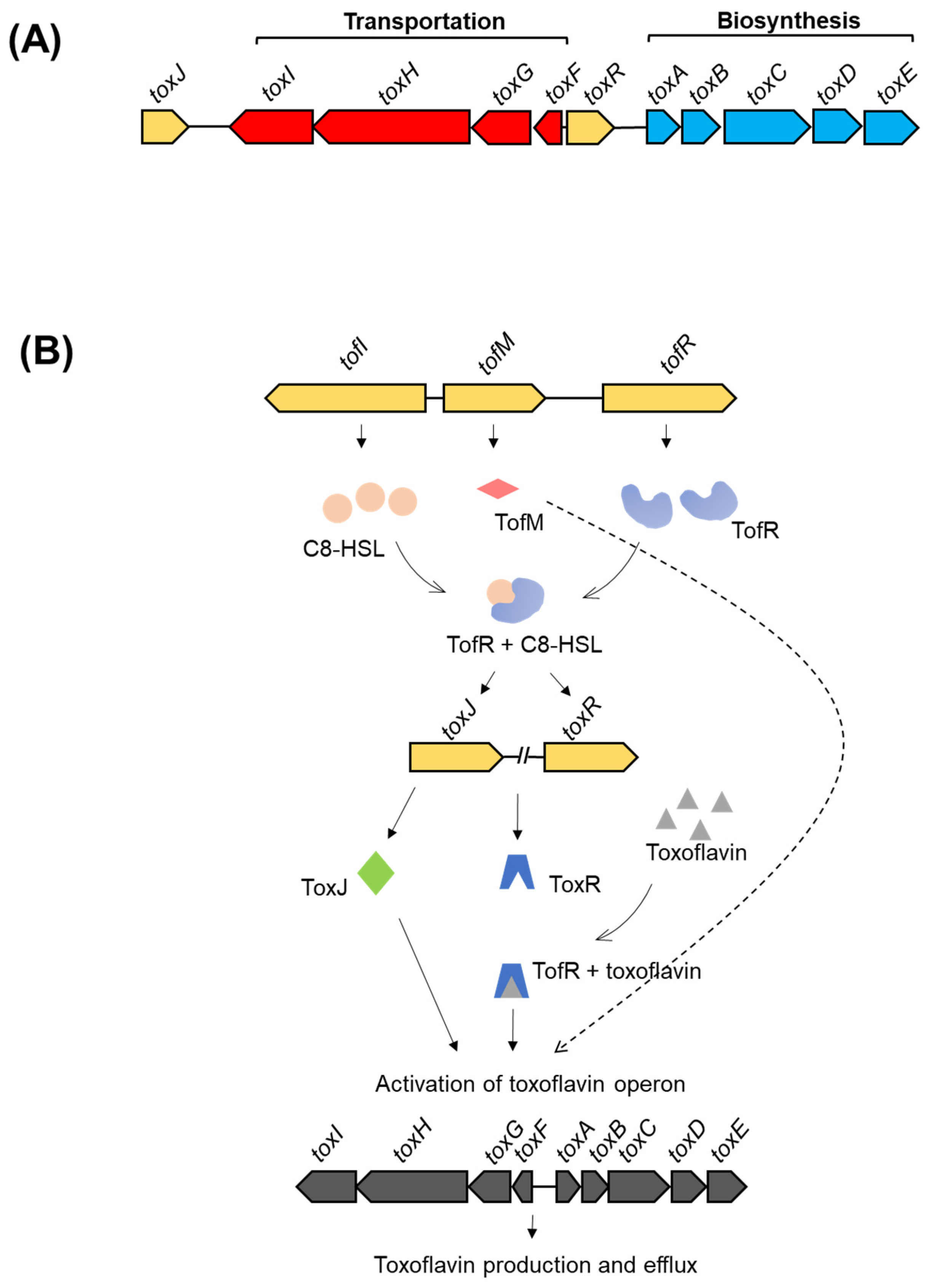
- Shingu, Y.; Yoneyama, K. Essential regulator gene toxR for toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 2004, 70, 108–114. [Google Scholar] [CrossRef]
- Suzuki, F.; Sawada, H.; Azegami, K.; Tsuchiya, K. Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 2004, 70, 97–107. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.G.; Kang, Y.; Jang, J.Y.; Jog, G.J.; Lim, J.Y.; Kim, S.; Suga, H.; Nagamatsu, T.; Hwang, I. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 2004, 54, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Barphagha, I.K.; Karki, H.S.; Ham, J.H. Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS ONE 2012, 7, e52150. [Google Scholar] [CrossRef]
- Seo, Y.S.; Lim, J.; Choi, B.S.; Kim, H.; Goo, E.; Lee, B.; Lim, J.S.; Choi, I.Y.; Moon, J.S.; Kim, J.; et al. Complete genome sequence of Burkholderia gladioli BSR3. J. Bacteriol. 2011, 193, 3149. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Kim, S.; Park, I.; Seo, Y.S. Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Mol. Plant Pathol. 2016, 17, 65–76. [Google Scholar] [CrossRef]
- Azegami, K.; Nishiyama, K.; Watanabe, Y.; Suzuki, T.; Yoshida, M.; Nose, K.; Toda, S. Tropolone as a root growth-inhibitor produced by a plant pathogenic Pseudomonas sp. causing seedling blight of rice. Jpn. J. Phytopathol. 1985, 51, 315–317. [Google Scholar] [CrossRef]
- Wakimoto, S.; Hirayae, K.; Tsuchiya, K.; Kushima, Y.; Furuya, N.; Matsuyama, N. Production of antibiotics by plant pathogenic pseudomonads. Jpn. J. Phytopathol. 1986, 52, 835–842. [Google Scholar] [CrossRef]
- Bentley, R. A fresh look at natural tropolonoids. Nat. Prod. Rep. 2008, 25, 118–138. [Google Scholar] [CrossRef]
- Solis, R.; Bertani, I.; Degrassi, G.; Devescovi, G.; Venturi, V. Involvement of quorum sensing and RpoS in rice seedling blight caused by Burkholderia plantarii. FEMS Microbiol. Lett. 2006, 259, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hashimoto, M.; Hashidoko, Y. Repression of tropolone production and induction of a Burkholderia plantarii pseudo-biofilm by carot-4-en-9, 10-diol, a cell-to-cell signaling disrupter produced by Trichoderma virens. PLoS ONE 2013, 8, e78024. [Google Scholar]
- Miwa, S.; Kihira, E.; Yoshioka, A.; Nakasone, K.; Okamoto, S.; Hatano, M.; Igarashi, M.; Eguchi, Y.; Kato, A.; Ichikawa, N.; et al. Identification of the three genes involved in controlling production of a phytotoxin tropolone in Burkholderia plantarii. J. Bacteriol. 2016, 198, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Iwasaki, S.; Kobayashi, H.; Okuda, S.; Murai, T.; Sato, Y. Rhizoxin binding to tubulin at the maytansine-binding site. Biochim. Biophys. Acta Gen. Subj. 1987, 926, 215–223. [Google Scholar] [CrossRef]
- Scherlach, K.; Busch, B.; Lackner, G.; Paszkowski, U.; Hertweck, C. Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. 2012, 124, 9753–9756. [Google Scholar] [CrossRef]
- Mitchell, R.E.; Frey, E.J. Rhizobitoxine and hydroxythreonine production by Pseudomonas andropogonis strains, and the implications to plant disease. Physiol. Mol. Plant Pathol. 1988, 32, 335–341. [Google Scholar] [CrossRef]
- Yasuta, T.; Okazaki, S.; Mitsui, H.; Yuhashi, K.I.; Ezura, H.; Minamisawa, K. DNA sequence and mutational analysis of rhizobitoxine biosynthesis genes in Bradyrhizobium elkanii. Appl. Environ. Microbiol. 2001, 67, 4999–5009. [Google Scholar] [CrossRef]
- Sugawara, M.; Okazaki, S.; Nukui, N.; Ezura, H.; Mitsui, H.; Minamisawa, K. Rhizobitoxine modulates plant–microbe interactions by ethylene inhibition. Biotechnol. Adv. 2006, 24, 382–388. [Google Scholar] [CrossRef]
- Mota, M.M.; Vieira, P. Pine Wilt Disease: A Worldwide Threat to Forest Ecosystems; Springer: Heidelberg, Germany, 2008. [Google Scholar]
- Adler, C.; Corbalán, N.S.; Seyedsayamdost, M.R.; Pomares, M.F.; de Cristóbal, R.E.; Clardy, J.; Kolter, R.; Vincent, P.A. Catecholate siderophores protect bacteria from pyochelin toxicity. PLoS ONE 2012, 7, e46754. [Google Scholar] [CrossRef]
- Le Dang, Q.; Son, S.W.; Cheon, H.M.; Choi, G.J.; Choi, Y.H.; Jang, K.S.; Lim, C.H.; Kim, J.C. Pyochelin isolated from Burkholderia arboris KRICT1 carried by pine wood nematodes exhibits phytotoxicity in pine callus. Nematology 2011, 13, 521–528. [Google Scholar] [CrossRef]
- Holland, I.B. The extraordinary diversity of bacterial protein secretion mechanisms. In Protein Secretion; Humana Press: New York, NY, USA, 2010; pp. 1–20. [Google Scholar]
- Green, E.R.; Mecsas, J. Bacterial secretion systems–an overview. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Gerlach, R.G.; Hensel, M. Protein secretion systems and adhesins: The molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 2007, 297, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Kim, J.; Kim, S.; Kim, H.; Lim, J.Y.; Kim, M.; Kwak, J.; Moon, J.S.; Hwang, I. Proteomic analysis of the proteins regulated by HrpB from the plant pathogenic bacterium Burkholderia glumae. Proteomics 2008, 8, 106–121. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Lee, H.H.; Park, I.; Seo, Y.S. Genome-Wide Analysis of Type VI System Clusters and Effectors in Burkholderia Species. Plant Pathol. J. 2018, 34, 11–22. [Google Scholar] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.R.; Van Gijsegem, F. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 2000, 54, 735–774. [Google Scholar] [CrossRef]
- Alteri, C.J.; Mobley, H.L. The versatile type VI secretion system. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Bornscheuer, U.T.; Bessler, C.; Srinivas, R.; Krishna, S.H. Optimizing lipases and related enzymes for efficient application. Trends Biotechnol. 2002, 20, 433–437. [Google Scholar] [CrossRef]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef]
- Subramoni, S.; Suárez-Moreno, Z.R.; Venturi, V. Lipases as pathogenicity factors of plant pathogens. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin, Germany, 2010; pp. 3269–3277. [Google Scholar]
- Devescovi, G.; Bigirimana, J.; Degrassi, G.; Cabrio, L.; LiPuma, J.J.; Kim, J.; Hwang, I.; Venturi, V. Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 2007, 73, 4950–4958. [Google Scholar] [CrossRef]
- Leigh, J.A.; Coplin, D.L. Exopolysaccharides in plant-bacterial interactions. Ann. Rev. Microbiol. 1992, 46, 307–346. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.; De Castro, C.; Petersen, B.O.; Duus, J.Ø.; Parrilli, M.; Holst, O. Acetyl Substitution of the O-Specific Caryan from the Lipopolysaccharide of Pseudomonas (Burkholderia) caryophylli Leads to a Block Pattern. Angew. Chem. Int. Ed. 2000, 39, 156–160. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Bustillos-Cristales, R.; Caballero-Mellado, J. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 2001, 67, 2790–2798. [Google Scholar] [CrossRef] [PubMed]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, C.; Elliott, G.N.; Simon, M.F.; Dos Reis Junior, F.B.; Gross, E.; Lawton, R.C.; Neto, N.E.; de Fatima, L.M.; De Faria, S.M.; Sprent, J.I.; et al. Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 2010, 19, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef]
- Caballero-Mellado, J.; Onofre-Lemus, J.; Estrada-de los Santos, P.; Martínez-Aguilar, L. The tomato rhizosphere, an environment rich in nitrogen-fixing Burkholderia species with capabilities of interest for agriculture and bioremediation. Appl. Environ. Microbiol. 2007, 73, 5308–5319. [Google Scholar] [CrossRef]
- Perin, L.; Martinez-Aguilar, L.; Paredes-Valdez, G.; Baldani, J.I.; Estrada-de los Santos, P.; Reis, V.M.; Caballero-Mellado, J. Burkholderia silvatlantica sp. nov., a diazotrophic bacterium associated with sugar cane and maize. Int. J. Syst. Evol. Microbiol. 2006, 56, 1931–1937. [Google Scholar] [CrossRef]
- Zhang, H.; Hanada, S.; Shigematsu, T.; Shibuya, K.; Kamagata, Y.; Kanagawa, T.; Kurane, R. Burkholderia kururiensis sp. nov., a trichloroethylene (TCE)-degrading bacterium isolated from an aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 2000, 50, 743–749. [Google Scholar] [CrossRef]
- Lynch, J.M.; Bragg, E. Microorganisms and soil aggregate stability. In Advances in Soil Science; Stewart, B.A., Ed.; Springer: New York, NY, USA, 1985; pp. 133–171. [Google Scholar]
- Jones, K.M.; Sharopova, N.; Lohar, D.P.; Zhang, J.Q.; VandenBosch, K.A.; Walker, G.C. Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide-deficient mutant. Proc. Natl. Acad. Sci. USA 2008, 105, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Vanhaverbeke, C.; Heyraud, A.; Achouak, W.; Heulin, T. Structural analysis of the exopolysaccharide from Burkholderia caribensis strain MWAP71. Carbohydr. Res. 2001, 334, 127–133. [Google Scholar] [CrossRef]
- Serrato, R.V.; Sassaki, G.L.; Cruz, L.M.; Pedrosa, F.O.; Gorin, P.A.; Iacomini, M. Culture conditions for the production of an acidic exopolysaccharide by the nitrogen-fixing bacterium Burkholderia tropica. Can. J. Microbiol. 2006, 52, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Ierano, T.; Lanzetta, R.; Molinaro, A.; Parrilli, M. The Structure of the O-Chain Polysaccharide from the Gram-Negative Endophytic Bacterium Burkholderia phytofirmans Strain PsJN. Eur. J. Org. Chem. 2008, 2008, 2303–2308. [Google Scholar] [CrossRef]
- Hallack, L.F.; Passos, D.S.; Mattos, K.A.; Agrellos, O.A.; Jones, C.; Mendonça-Previato, L.; Previato, J.O.; Todeschini, A.R. Structural elucidation of the repeat unit in highly branched acidic exopolysaccharides produced by nitrogen fixing Burkholderia. Glycobiology 2009, 20, 338–347. [Google Scholar] [CrossRef]
- Caballero-Mellado, J.; Martínez-Aguilar, L.; Paredes-Valdez, G.; Estrada-de los Santos, P. Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 2004, 54, 1165–1172. [Google Scholar] [CrossRef]
- De Zamaroczy, M.; Delorme, F.; Elmerich, C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase (nifHDK) of Azospirillum brasilense Sp7. Mol. Gen. Genet. 1989, 220, 88–94. [Google Scholar] [CrossRef]
- Ruvkun, G.B.; Ausubel, F.M. Interspecies homology of nitrogenase genes. Proc. Natl. Acad. Sci. USA 1980, 77, 191–195. [Google Scholar] [CrossRef]
- Minerdi, D.; Fani, R.; Gallo, R.; Boarino, A.; Bonfante, P. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 2001, 67, 725–732. [Google Scholar] [CrossRef]
- Foussard, M.; Garnerone, A.M.; Ni, F.; Soupène, E.; Boistard, P.; Batut, J. Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol. Microbiol. 1997, 25, 27–37. [Google Scholar] [CrossRef]
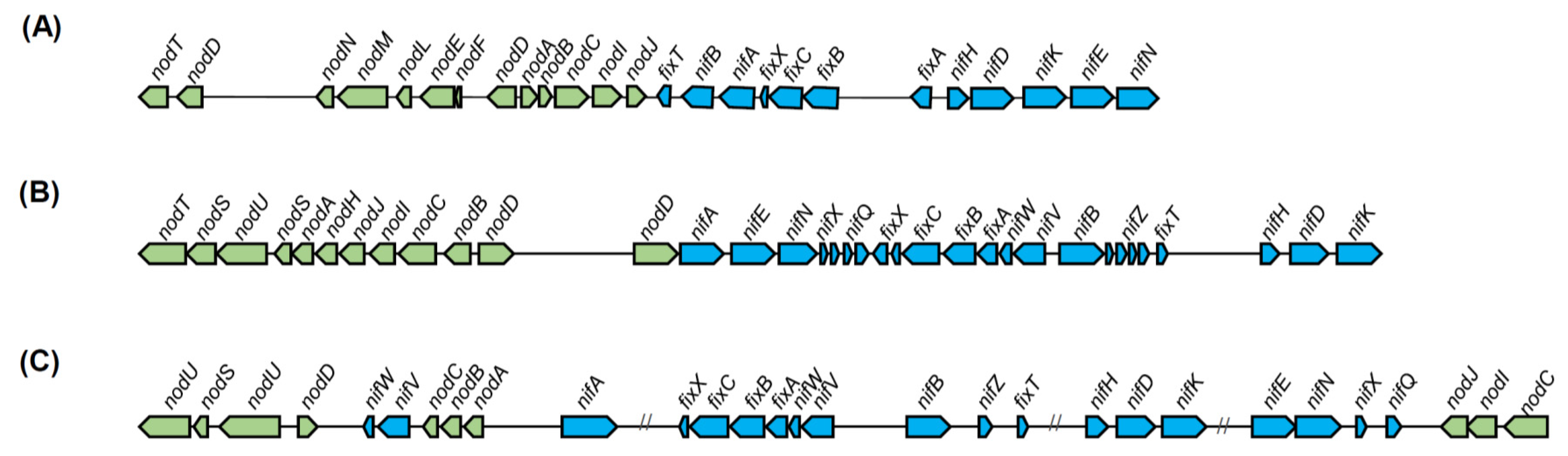
- De Meyer, S.E.; Briscoe, L.; Martínez-Hidalgo, P.; Agapakis, C.M.; Estrada-de los Santos, P.E.; Seshadri, R.; Reeve, W.; Weinstock, G.; O’Hara, G.; Howieson, J.G.; et al. Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol. Plant-Microbe Interact. 2016, 29, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Long, S.R. Rhizobium-legume nodulation: Life together in the underground. Cell 1989, 56, 203–214. [Google Scholar] [CrossRef]
- Hirsch, A.M.; Lum, M.R.; Downie, J.A. What makes the rhizobia-legume symbiosis so special? Plant Physiol. 2001, 127, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Goris, J.; Chen, W.M.; De Vos, P.; Willems, A. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 2002, 25, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Moulin, L.; Bontemps, C.; Vandamme, P.; Béna, G.; Boivin-Masson, C. Legume symbiotic nitrogen fixation by β-proteobacteria is widespread in nature. J. Bacteriol. 2003, 185, 7266–7272. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; de Faria, S.M.; Straliotto, R.; Pitard, R.M.; Simoes-Araujo, J.L.; Chou, J.H.; Chou, Y.J.; Barrios, E.; Prescott, A.R.; Elliott, G.N.; et al. Proof that Burkholderia strains form effective symbioses with legumes: A study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 2005, 71, 7461–7471. [Google Scholar] [CrossRef]
- Chen, W.M.; James, E.K.; Coenye, T.; Chou, J.H.; Barrios, E.; De Faria, S.M.; Elliott, G.N.; Sheu, S.Y.; Sprent, J.I.; Vandamme, P. Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 2006, 56, 1847–1851. [Google Scholar] [CrossRef]
- Talbi, C.; Delgado, M.J.; Girard, L.; Ramirez-Trujillo, A.; Caballero-Mellado, J.; Bedmar, E.J. Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in Moroccan soils. Appl. Environ. Microbiol. 2010, 76, 4587–4591. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for the lowering of plant ethylene concentrations by plant growth-promoting rhizobacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Onofre-Lemus, J.; Hernández-Lucas, I.; Girard, L.; Caballero-Mellado, J. ACC (1-aminocyclopropane-1-carboxylate) deaminase activity, a widespread trait in Burkholderia species, and its growth-promoting effect on tomato plants. Appl. Environ. Microbiol. 2009, 75, 6581–6590. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, Z.; Glick, B.R. The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol. Lett. 2009, 296, 131–136. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- de Oliveira-Longatti, S.M.; Marra, L.M.; Soares, B.L.; Bomfeti, C.A.; Da Silva, K.; Ferreira, P.A.A.; de Souza Moreira, F.M. Bacteria isolated from soils of the western Amazon and from rehabilitated bauxite-mining areas have potential as plant growth promoters. World J. Microbiol. Biotechnol. 2014, 30, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Selvakumar, G.; Ganeshamurthy, A.N. Draft genome sequence of phosphate-solubilizing bacterium Paraburkholderia tropica strain P-31 isolated from pomegranate (Punica granatum) rhizosphere. Genome Announc. 2016, 4, e00844-16. [Google Scholar] [CrossRef]
- Hsu, P.C.L.; O’Callaghan, M.; Condron, L.; Hurst, M.R. Use of a gnotobiotic plant assay for assessing root colonization and mineral phosphate solubilization by Paraburkholderia bryophila Ha185 in association with perennial ryegrass (Lolium perenne L.). Plant Soil 2018, 425, 43–55. [Google Scholar] [CrossRef]
- Fukaki, H.; Tasaka, M. Hormone interactions during lateral root formation. Plant Mol. Biol. 2009, 69, 437–449. [Google Scholar] [CrossRef]
- Sessitsch, A.; Coenye, T.; Sturz, A.; Vandamme, P.; Barka, E.A.; Salles, J.; Van Elsas, J.; Faure, D.; Reiter, B.; Glick, B. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef]
- Weilharter, A.; Mitter, B.; Shin, M.V.; Chain, P.S.; Nowak, J.; Sessitsch, A. Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J. Bacteriol. 2011, 193, 3383–3384. [Google Scholar] [CrossRef]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum sensing and indole-3-acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant-Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef]
- Naveed, M.; Qureshi, M.A.; Zahir, Z.A.; Hussain, M.B.; Sessitsch, A.; Mitter, B. L-Tryptophan-dependent biosynthesis of indole-3-acetic acid (IAA) improves plant growth and colonization of maize by Burkholderia phytofirmans PsJN. Ann. Microbiol. 2015, 65, 1381–1389. [Google Scholar] [CrossRef]
- Li, W.; Roberts, D.P.; Dery, P.D.; Meyer, S.L.F.; Lohrke, S.; Lumsden, R.D.; Hebbar, K.P. Broad spectrum anti-biotic activity and disease suppression by the potential biocontrol agent Burkholderia ambifaria BC-F. Crop Prot. 2002, 21, 129–135. [Google Scholar] [CrossRef]
- Vandamme, P.; Opelt, K.; Knöchel, N.; Berg, C.; Schönmann, S.; De Brandt, E.; Eberl, L.; Falsen, E.; Berg, G. Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 2007, 57, 2228–2235. [Google Scholar] [CrossRef] [PubMed]
- Farh, M.E.A.; Kim, Y.J.; Van An, H.; Sukweenadhi, J.; Singh, P.; Huq, M.A.; Yang, D.C. Burkholderia ginsengiterrae sp. nov. and Burkholderia panaciterrae sp. nov., antagonistic bacteria against root rot pathogen Cylindrocarpon destructans, isolated from ginseng soil. Arch. Microbiol. 2015, 197, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Gognies, S.; Nowak, J.; Audran, J.C.; Belarbi, A. Inhibitory effect of endophyte bacteria on Botrytis cinerea and its influence to promote the grapevine growth. Biol. Control 2002, 24, 135–142. [Google Scholar] [CrossRef]
- Barka, E.A.; Nowak, J.; Clément, C. Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl. Environ. Microbiol. 2006, 72, 7246–7252. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Carbó, M.; Gademann, K.; Eberl, L.; Carlier, A. Leaf nodule symbiosis: Function and transmission of obligate bacterial endophytes. Curr. Opin. Plant Biol. 2018, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.M. Bacterial leaf nodule symbiosis. Adv. Bot. Res. 1990, 17, 163–234. [Google Scholar]
- Sinnesael, A.; Eeckhout, S.; Janssens, S.B.; Smets, E.; Panis, B.; Leroux, O.; Verstraete, B. Detection of Burkholderia in the seeds of Psychotria punctata (Rubiaceae)–Microscopic evidence for vertical transmission in the leaf nodule symbiosis. PLoS ONE 2018, 13, e0209091. [Google Scholar] [CrossRef]
- Carlier, A.; Fehr, L.; Pinto-Carbó, M.; Schäberle, T.; Reher, R.; Dessein, S.; König, G.; Eberl, L. The genome analysis of Candidatus Burkholderia crenata reveals that secondary metabolism may be a key function of the Ardisia crenata leaf nodule symbiosis. Environ. Microbiol. 2016, 18, 2507–2522. [Google Scholar] [CrossRef]
- Sieber, S.; Carlier, A.; Neuburger, M.; Grabenweger, G.; Eberl, L.; Gademann, K. Isolation and total synthesis of kirkamide, an aminocyclitol from an obligate leaf nodule symbiont. Angew. Chem. Int. Ed. 2015, 54, 7968–7970. [Google Scholar] [CrossRef] [PubMed]
- Zolg, W.; Ottow, J.C.G. Pseudomonas glathei sp. nov., a new nitrogen-scavenging rod isolated from acid lateritic relicts in Germany. Z. Allg. Mikrobiol. 1975, 15, 287–299. [Google Scholar]
- Aizawa, T.; Ve, N.B.; Nakajima, M.; Sunairi, M. Burkholderia heleia sp. nov., a nitrogen-fixing bacterium isolated from an aquatic plant, Eleocharis dulcis, that grows in highly acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 2010, 60, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.C.; Im, W.T.; Kim, K.K.; An, D.S.; Lee, S.T. Burkholderia terrae sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Reis, V.M.; Estrada-de los Santos, P.; Tenorio-Salgado, S.; Vogel, J.; Stoffels, M.; Guyon, S.; Mavingui, P.; Baldani, V.L.D.; Schmid, M.; Baldani, J.I.; et al. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Cunha, C.; Zuleta, L.F.G.; de Almeida, L.G.P.; Ciapina, L.P.; Borges, W.L.; Pitard, R.M.; Baldani, J.I.; Straliotto, R.; de Faria, S.M.; Hungria, M.; et al. Complete genome sequence of Burkholderia phenoliruptrix BR3459a (CLA1), a heat-tolerant, nitrogen-fixing symbiont of Mimosa flocculosa. J. Bacteriol. 2012, 194, 6675–6676. [Google Scholar] [CrossRef] [PubMed]
- Elliott, G.N.; Chen, W.M.; Chou, J.H.; Wang, H.C.; Sheu, S.Y.; Perin, L.; Reis, V.M.; Moulin, L.; Simon, M.F.; Bontemps, C.; et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 2007, 173, 168–180. [Google Scholar] [CrossRef]
- Chen, W.M.; De Faria, S.M.; James, E.K.; Elliott, G.N.; Lin, K.Y.; Chou, J.H.; Sheu, S.Y.; Cnockaert, M.; Sprent, J.I.; Vandamme, P. Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 2007, 57, 1055–1059. [Google Scholar] [CrossRef]
- Nion, Y.A.; Toyota, K. Suppression of bacterial wilt and Fusarium wilt by a Burkholderia nodosa strain isolated from Kalimantan soils, Indonesia. Microbes Environ. 2008, 23, 134–141. [Google Scholar] [CrossRef]
- Martínez-Aguilar, L.; Salazar-Salazar, C.; Méndez, R.D.; Caballero-Mellado, J.; Hirsch, A.M.; Vásquez-Murrieta, M.S.; Estrada-de los Santos, P. Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Antonie van Leeuwenhoek 2013, 104, 1063–1071. [Google Scholar] [CrossRef]
- Achouak, W.; Christen, R.; Barakat, M.; Martel, M.H.; Heulin, T. Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int. J. Syst. Evol. Microbiol. 1999, 49, 787–794. [Google Scholar] [CrossRef]
- Sheu, S.Y.; Chou, J.H.; Bontemps, C.; Elliott, G.N.; Gross, E.; dos Reis Junior, F.B.; Melkonian, R.; Moulin, L.; James, E.K.; Sprent, J.I.; et al. Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 2013, 63, 435–441. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; Cnockaert, M.; Ardley, J.K.; Van Wyk, B.E.; Vandamme, P.A.; Howieson, J.G. Burkholderia dilworthii sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 2014, 64, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Chen, M.H.; Liu, W.Y.; Andrews, M.; James, E.K.; Ardley, J.K.; De Meyer, S.E.; James, T.K.; Howieson, J.G.; Coutinho, B.G.; et al. Burkholderia dipogonis sp. nov., isolated from root nodules of Dipogon lignosus in New Zealand and Western Australia. Int. J. Syst. Evol. Microbiol. 2015, 65, 4716–4723. [Google Scholar] [CrossRef] [PubMed]
- Steenkamp, E.T.; van Zyl, E.; Beukes, C.W.; Avontuur, J.R.; Chan, W.Y.; Palmer, M.; Mthombeni, L.S.; Phalane, F.L.; Sereme, T.K.; Venter, S.N. Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst. Appl. Microbiol. 2015, 38, 545–554. [Google Scholar] [CrossRef] [PubMed]
- De Meyer, S.E.; Cnockaert, M.; Ardley, J.K.; Trengove, R.D.; Garau, G.; Howieson, J.G.; Vandamme, P. Burkholderia rhynchosiae sp. nov., isolated from Rhynchosia ferulifolia root nodules. Int. J. Syst. Evol. Microbiol. 2013, 63, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; de Faria, S.M.; Chou, J.H.; James, E.K.; Elliott, G.N.; Sprent, J.I.; Bontemps, C.; Young, J.P.W.; Vandamme, P. Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 2008, 58, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Chou, J.H.; Bontemps, C.; Elliott, G.N.; Gross, E.; James, E.K.; Sprent, J.I.; Young, J.P.W.; Chen, W.M. Burkholderia symbiotica sp. nov., isolated from root nodules of Mimosa spp. native to north-east Brazil. Int. J. Syst. Evol. Microbiol. 2012, 62, 2272–2278. [Google Scholar] [CrossRef] [PubMed]
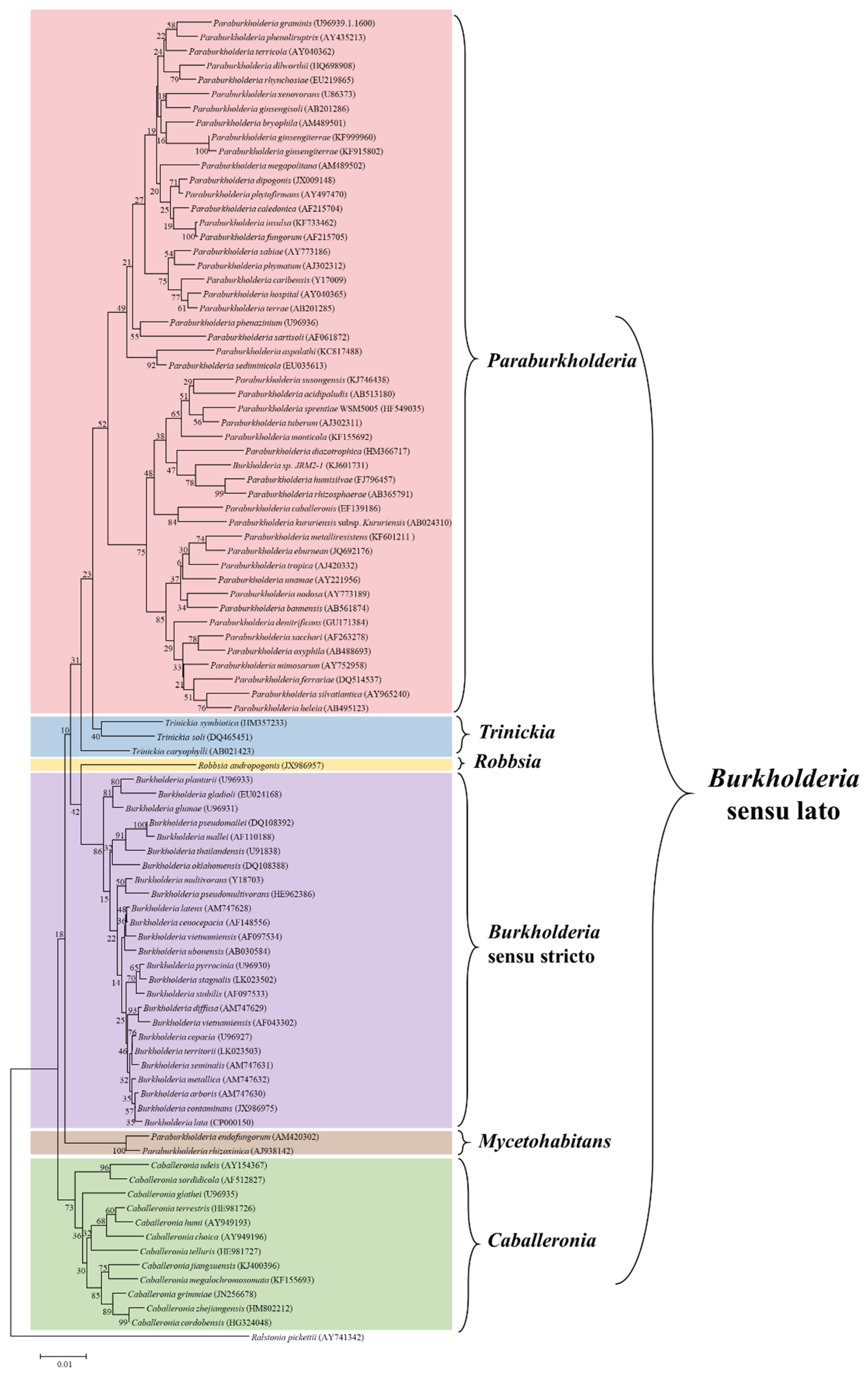

| Year | Finding | Details | Reference |
|---|---|---|---|
| 1942 | First isolation of Burkholderia | Originally named Phytomonas caryophylli; then Pseudomonas caryophylli | [5] |
| 1992 | A new Burkholderia genus was proposed | The new genus comprised seven species from the genus Pseudomonas | [8] |
| 2011 | A second genus (Caballeronia) was suggested | Based on phylogenetic analysis of multiple genes and comparative genomics; however, the evidence was not sufficient to confirm the new grouping | [15] |
| 2014 | The genus Paraburkholderia was proposed | Based on the analysis of conserved sequence in/dels | [18] |
| 2016 | Inclusion of several species in the Paraburkholderia genus and establishment of the Caballeronia genus | Eleven species were reclassified as Paraburkholderia and 14 species were transferred to the newly established Caballeronia genus | [19] |
| 2017 | Burkholderia andropogonis was separated in a newly proposed genus as Robbsia andropogonis | Based on multilocus sequence, 16S rRNA gene phylogeny, and average nucleotide identity analyses, as well as tetranucleotide signature frequency and percentage of conserved proteins | [20] |
| 2017 | Confirmation of the genetic boundaries among the 4 established groups and suggestion of a fifth division: Paraburkholderia rhizoxinica | Five groups (Burkholderia sensu stricto, Paraburkholderia, Caballeronia, Robbsia, Paraburkholderia rhizoxinica) were separated based on maximum likelihood phylogenies using the amino acid and nucleotide sequence of 106 conserved proteins | [21] |
| 2018 | Two novel genera (Mycetohabitans and Trinickia) were proposed | Based on whole-genome comparative study and phylogenetic analysis of conserved genes | [22] |
| Phytotoxin | Producing Species | Major Phytotoxic Effect | Mode of Action | References |
|---|---|---|---|---|
| Toxoflavin | B. glumae; B. gladioli | Severe damage to rice panicles and inhibition of sprout and root elongation in seedlings. | An active electron carrier between NADH and oxygen, producing reactive oxygen species | [65,67] |
| Tropolone | B. plantarii | Blight symptoms | A potential iron chelator with multiple biological roles | [51,76] |
| Rhizoxin | Mycetohabitans rhizoxinia * | Seedling blight symptoms in rice; signaling element for bacterial-fungal symbiosis | Acts on β-tubulin and blocks mitosis, inhibiting eukaryotic cell growth. | [25,80,81] |
| Rhizobitoxine | Robbsia andropogonis | Leaf chlorosis in host plants | Inhibition of methionine and ethylene biosynthetic pathways | [82,84] |
| Pyochelin | Burkholderia arboris | Necrosis in Pinus densiflora | ND | [87] |
| Category | Species | Host/Isolation | Major Virulence/Benevolence Factors | Reference |
|---|---|---|---|---|
| Phytopathogenic species | ||||
| Burkholderia gladioli | Gladiolus, Onion, Rice | Toxoflavin, Lipase, T3SS | [47,48,49,50] | |
| Burkholderia glumae | Rice and several other crops | Toxoflavin, Lipase, T3SS, T6SS, EPSs, polygalacturonases | [43,44] | |
| Burkholderia plantarii | Rice and several other crops | Tropolone, Lipase, T3SS | [42,51] | |
| Trinickia caryophylli | Carnation and onion | EPSs | [22,56,57,101] | |
| Mycetohabitans rhizoxinia | In combination with host fungus causing rice seedling rot | Rhizoxin, T3SS | [22,42,59] | |
| Robbsia andropogonis | Sorghum, velvet beans, orchids, carnation | Rhizobitoxine | [42,53,54,55,82] | |
| Plant beneficial species | ||||
| Free-living and endophytic | ||||
| Paraburkholderia phytofirmans | Cereal and other crop soils | nif, ACC deaminase, EPSs, IAA | [113,129,136,137] | |
| Paraburkholderia xenovorans | Rhizosphere | nif, ACC deaminase | [106,129] | |
| Paraburkholderia unamae | Corn, sugarcane, coffee plants, and rhizosphere | nif, ACC deaminase | [106,129] | |
| Paraburkholderia silvatlantica | Corn rhizosphere | nif, ACC deaminase | [107,129] | |
| Paraburkholderia graminis | Rhizosphere | ACC deaminase | [129] | |
| Paraburkholderia bryophila | Moss gametophytes | Siderophore, antifungal activity, phosphate solubilization | [106,134,141] | |
| Paraburkholderia kururiensis | Aquifer sample | nif, ACC deaminase, EPSs | [102,114,129] | |
| Paraburkholderia ginsengiterrae | Ginseng rhizosphere | Antifungal activity | [142] | |
| Paraburkholderia panaciterrae | Ginseng rhizosphere | Antifungal activity | [142] | |
| Caballeronia glathei | Fossil lateritic soil | nif | [150] | |
| Paraburkholderia heleia | aquatic plant from highly acidic swamps | nif | [151] | |
| Paraburkholderia megapolitana | Moss gametophytes | Siderophore, antifungal activity | [106,141] | |
| Paraburkholderia terrae | Forest soil | nif | [152] | |
| Paraburkholderia tropica | Sugarcane | nif, EPSs, phosphate solubilization | [106,112,133,153] | |
| Legume nodulators | ||||
| Paraburkholderia phenoliruptrix | Mimosa root nodules | nod, nif, ACC deaminase | [129,154] | |
| Paraburkholderia phymatum | Root nodules of mimosa and other tropical legumes | nod, nif, ACC deaminase | [123,127,129,155] | |
| Paraburkholderia tuberum | Root nodules of Papilionoid and tropical legumes | nod, nif, ACC deaminase | [123,129] | |
| Paraburkholderia mimosarum | Mimosa root nodules | nod, nif | [126] | |
| Paraburkholderia nodosa | Mimosa root nodules | nod, nif, biocontrol activity | [156,157] | |
| Paraburkholderia caballeronis | Rhizosphere of tomato | nod, nif | [158] | |
| Paraburkholderia caribensis | vertisol | nod, ACC deaminase, EPSs | [111,124,129,159] | |
| Paraburkholderia diazotrophica | Mimosa nodules | nod, nif | [160] | |
| Paraburkholderia dilworthii | Lebeckia ambigua root nodules | nod, nif | [161] | |
| Paraburkholderia dipogonis | Papilionoid legume nodules | nod, nif | [162] | |
| Paraburkholderia kirstenboschensis | Papilionoid legume nodules | nod, nif | [163] | |
| Paraburkholderia rhynchosiae | Rhynchosia ferulifolia legume | nod, nif | [164] | |
| Paraburkholderia sabiae | Mimosa nodules | nod, nif | [165] | |
| Trinickia symbiotica | Mimosa nodules | nod, nif, siderophore | [22,166] | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannaa, M.; Park, I.; Seo, Y.-S. Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species. Int. J. Mol. Sci. 2019, 20, 121. https://doi.org/10.3390/ijms20010121
Mannaa M, Park I, Seo Y-S. Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species. International Journal of Molecular Sciences. 2019; 20(1):121. https://doi.org/10.3390/ijms20010121
Chicago/Turabian StyleMannaa, Mohamed, Inmyoung Park, and Young-Su Seo. 2019. "Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species" International Journal of Molecular Sciences 20, no. 1: 121. https://doi.org/10.3390/ijms20010121
APA StyleMannaa, M., Park, I., & Seo, Y.-S. (2019). Genomic Features and Insights into the Taxonomy, Virulence, and Benevolence of Plant-Associated Burkholderia Species. International Journal of Molecular Sciences, 20(1), 121. https://doi.org/10.3390/ijms20010121







