Iron Metabolism in Cancer
Abstract
1. Introduction
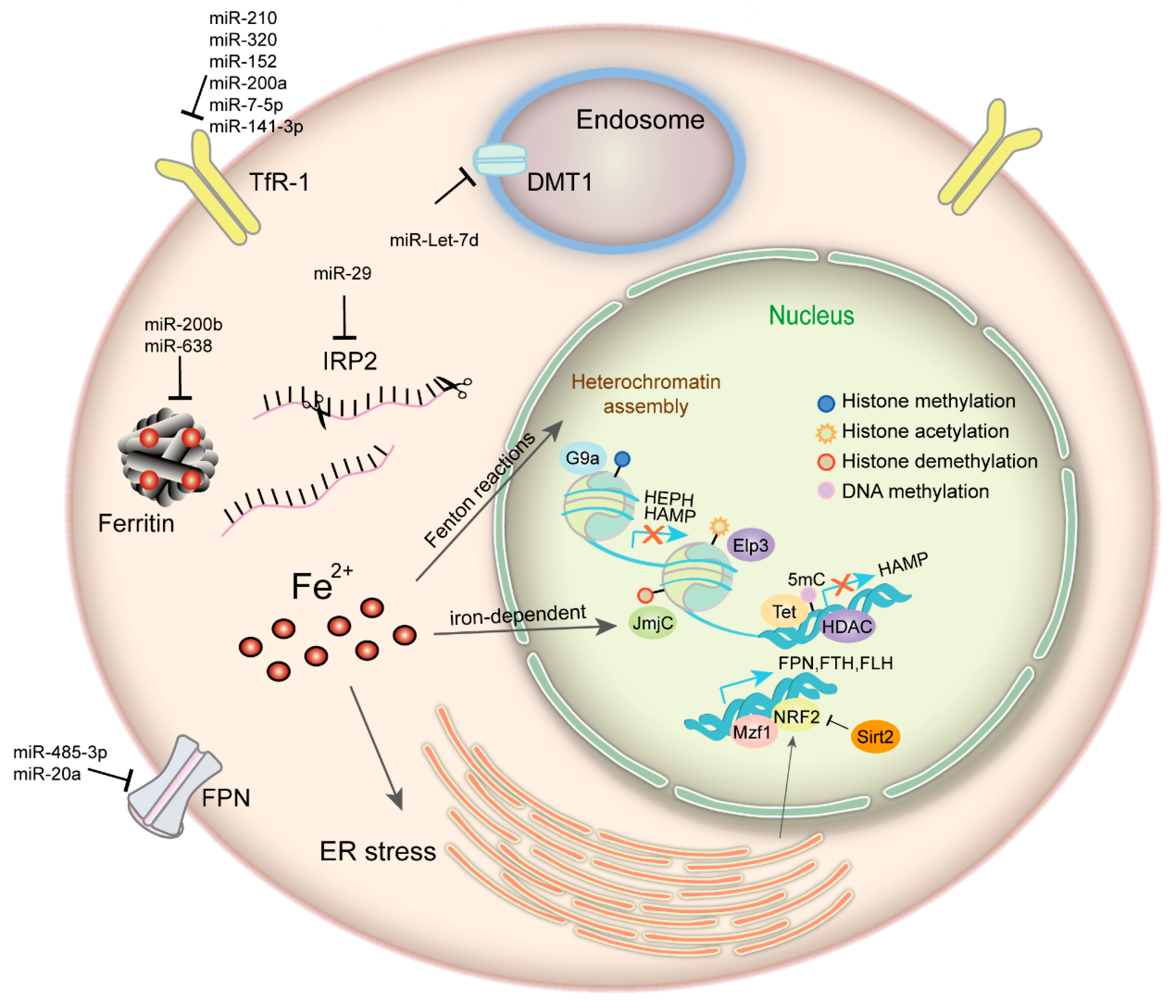
2. Regulation of Iron Homeostasis in Cancer
3. Iron and Epigenetic Regulation in Cancer
3.1. Iron Plays an Important Role in Cancer Epigenetics
3.2. Epigenetic Regulation of Iron Metabolism-Related Proteins
4. Role of Iron in Tumor Cell Biology
4.1. Iron in Cancer Cell Cycle and DNA Metabolism
4.2. Iron in Cancer Cell Demise
4.3. Iron in Tumor Metastasis and Angiogenesis
4.4. Iron in the Tumor Microenvironment
5. Iron Manipulating Strategies in Cancer
5.1. Iron is a Target for Oncotherapy
5.2. Combination Therapies and Novel Iron Modulators
6. Conclusions
Funding
Conflicts of Interest
References
- Muckenthaler, M.U.; Rivella, S.; Hentze, M.W.; Galy, B. A Red Carpet for Iron Metabolism. Cell 2017, 168, 344–361. [Google Scholar] [CrossRef]
- Recalcati, S.; Gammella, E.; Buratti, P.; Cairo, G. Molecular regulation of cellular iron balance. IUBMB Life 2017, 69, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.V.; Torti, F.M. Iron and cancer: More ore to be mined. Nat. Rev. Cancer 2013, 13, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Martinkova, P.; Brtnicky, M.; Kynicky, J.; Pohanka, M. Iron Oxide Nanoparticles: Innovative Tool in Cancer Diagnosis and Therapy. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef]
- Viktorinova, A. Iron-mediated oxidative cell death is a potential contributor to neuronal dysfunction induced by neonatal hemolytic hyperbilirubinemia. Arch. Biochem. Biophys. 2018, 654, 185–193. [Google Scholar] [CrossRef]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to tango: Regulation of Mammalian iron metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef]
- Daniels, T.R.; Delgado, T.; Helguera, G.; Penichet, M.L. The transferrin receptor part II: Targeted delivery of therapeutic agents into cancer cells. Clin. Immunol. 2006, 121, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, J.; Song, F.; Tian, M.; Shi, B.; Jiang, H.; Xu, W.; Wang, H.; Zhou, M.; Pan, X.; et al. EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett. 2016, 381, 331–340. [Google Scholar] [CrossRef]
- Bourseau-Guilmain, E.; Griveau, A.; Benoit, J.P.; Garcion, E. The importance of the stem cell marker prominin-1/CD133 in the uptake of transferrin and in iron metabolism in human colon cancer CaCO2 cells. PLoS ONE 2011, 6, e25515. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, B.; Zhang, L.; Wang, S.; Dong, D.; Lv, H.; Shang, P. Alterations in Cellular Iron Metabolism Provide More Therapeutic Opportunities for Cancer. Int. J. Mol. Sci. 2018, 19, 1545. [Google Scholar] [CrossRef]
- Crielaard, B.J.; Lammers, T.; Rivella, S. Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 2017, 16, 400–423. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Jiang, J.N.; Fang, X.D.; Ji, F.J. STEAP1 Regulates Tumorigenesis and Chemoresistance During Peritoneal Metastasis of Gastric Cancer. Front. Physiol. 2018, 9, 1132. [Google Scholar] [CrossRef]
- Burnell, S.E.A.; Spencer-Harty, S.; Howarth, S.; Bodger, O.; Kynaston, H.; Morgan, C.; Doak, S.H. STEAP2 Knockdown Reduces the Invasive Potential of Prostate Cancer Cells. Sci. Rep. 2018, 8, 6252. [Google Scholar] [CrossRef] [PubMed]
- Whiteland, H.; Spencer-Harty, S.; Morgan, C.; Kynaston, H.; Thomas, D.H.; Bose, P.; Fenn, N.; Lewis, P.; Jenkins, S.; Doak, S.H. A role for STEAP2 in prostate cancer progression. Clin. Exp. Metastasis 2014, 31, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Xu, R.; Wang, S.; Yang, N.; Ni, S.; Zhang, Q.; Xu, Y.; Zhang, X.; Zhang, C.; Wei, Y.; et al. Six-Transmembrane Epithelial Antigen of Prostate 3 Predicts Poor Prognosis and Promotes Glioblastoma Growth and Invasion. Neoplasia 2018, 20, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Bredell, B.X.; Anderson, E.R.; Martin, A.; Mays, C.; Nagao-Kitamoto, H.; Huang, S.; Gyorffy, B.; Greenson, J.K.; Hardiman, K.; et al. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9608–E9617. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP proteins: From structure to applications in cancer therapy. Mol. Cancer Res. 2012, 10, 573–587. [Google Scholar] [CrossRef]
- Barroca-Ferreira, J.; Pais, J.P.; Santos, M.M.; Goncalves, A.M.; Gomes, I.M.; Sousa, I.; Rocha, S.M.; Passarinha, L.A.; Maia, C.J. Targeting STEAP1 Protein in Human Cancer: Current Trends and Future Challenges. Curr. Cancer Drug Targets 2018, 18, 222–230. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, F. Iron homeostasis and tumorigenesis: Molecular mechanisms and therapeutic opportunities. Protein Cell 2015, 6, 88–100. [Google Scholar] [CrossRef]
- Brookes, M.J.; Hughes, S.; Turner, F.E.; Reynolds, G.; Sharma, N.; Ismail, T.; Berx, G.; McKie, A.T.; Hotchin, N.; Anderson, G.J.; et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut 2006, 55, 1449–1460. [Google Scholar] [CrossRef]
- Wolff, N.A.; Garrick, M.D.; Zhao, L.; Garrick, L.M.; Ghio, A.J.; Thevenod, F. A role for divalent metal transporter (DMT1) in mitochondrial uptake of iron and manganese. Sci. Rep. 2018, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Ramakrishnan, S.K.; Weisz, K.; Triner, D.; Xie, L.; Attili, D.; Pant, A.; Gyorffy, B.; Zhan, M.; Carter-Su, C.; et al. Iron Uptake via DMT1 Integrates Cell Cycle with JAK-STAT3 Signaling to Promote Colorectal Tumorigenesis. Cell Metab. 2016, 24, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Hann, H.W.; Ye, Z.; Yin, C.; Wang, Y.; Fang, W.; Wan, S.; Wang, C.; Tao, K. Ferritin level prospectively predicts hepatocarcinogenesis in patients with chronic hepatitis B virus infection. Oncol. Lett. 2018, 16, 3499–3508. [Google Scholar] [CrossRef] [PubMed]
- Song, A.; Eo, W.; Kim, S.; Shim, B.; Lee, S. Significance of serum ferritin as a prognostic factor in advanced hepatobiliary cancer patients treated with Korean medicine: A retrospective cohort study. BMC Complement. Altern. Med. 2018, 18, 176. [Google Scholar] [CrossRef] [PubMed]
- Lipper, C.H.; Karmi, O.; Sohn, Y.S.; Darash-Yahana, M.; Lammert, H.; Song, L.; Liu, A.; Mittler, R.; Nechushtai, R.; Onuchic, J.N.; et al. Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, 272–277. [Google Scholar] [CrossRef]
- Mittler, R.; Darash-Yahana, M.; Sohn, Y.S.; Bai, F.; Song, L.; Cabantchik, I.Z.; Jennings, P.A.; Onuchic, J.N.; Nechushtai, R. NEET Proteins: A New Link Between Iron Metabolism, Reactive Oxygen Species, and Cancer. Antioxid. Redox Signal. 2018. [Google Scholar] [CrossRef]
- Gu, Z.; Wang, H.; Xia, J.; Yang, Y.; Jin, Z.; Xu, H.; Shi, J.; De Domenico, I.; Tricot, G.; Zhan, F. Decreased ferroportin promotes myeloma cell growth and osteoclast differentiation. Cancer Res. 2015, 75, 2211–2221. [Google Scholar] [CrossRef]
- Xue, D.; Zhou, C.X.; Shi, Y.B.; Lu, H.; He, X.Z. Decreased expression of ferroportin in prostate cancer. Oncol. Lett. 2015, 10, 913–916. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, S.; Chen, Y.; Zhang, D.; Yuan, L.; Cong, H.; Liu, S. An important role of the hepcidin-ferroportin signaling in affecting tumor growth and metastasis. Acta Biochim. Biophys. Sin. 2015, 47, 703–715. [Google Scholar] [CrossRef]
- Moussa, R.S.; Park, K.C.; Kovacevic, Z.; Richardson, D.R. Ironing out the role of the cyclin-dependent kinase inhibitor, p21 in cancer: Novel iron chelating agents to target p21 expression and activity. Free Radic. Boil. Med. 2018. [Google Scholar] [CrossRef]
- Shan, Z.; Wei, Z.; Shaikh, Z.A. Suppression of ferroportin expression by cadmium stimulates proliferation, EMT, and migration in triple-negative breast cancer cells. Toxicol. Appl. Pharmacol. 2018, 356, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, C.N.; Ruwe, T.A.; Shawki, A.; Xin, V.; Vieth, K.R.; Valore, E.V.; Qiao, B.; Ganz, T.; Nemeth, E.; Mackenzie, B.; et al. Calcium is an essential cofactor for metal efflux by the ferroportin transporter family. Nat. Commun. 2018, 9, 3075. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.; Galy, B.; Schwanhaeusser, B.; Blake, J.; Bahr-Ivacevic, T.; Benes, V.; Selbach, M.; Muckenthaler, M.U.; Hentze, M.W. Iron regulatory protein-1 and -2: Transcriptome-wide definition of binding mRNAs and shaping of the cellular proteome by iron regulatory proteins. Blood 2011, 118, e168–e179. [Google Scholar] [CrossRef] [PubMed]
- Kwok, J.C.; Richardson, D.R. The iron metabolism of neoplastic cells: Alterations that facilitate proliferation? Crit. Rev. Oncol. Hematol. 2002, 42, 65–78. [Google Scholar] [CrossRef]
- Horniblow, R.D.; Bedford, M.; Hollingworth, R.; Evans, S.; Sutton, E.; Lal, N.; Beggs, A.; Iqbal, T.H.; Tselepis, C. BRAF mutations are associated with increased iron regulatory protein-2 expression in colorectal tumorigenesis. Cancer Sci. 2017, 108, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Holmes-Hampton, G.P.; Ghosh, M.C.; Rouault, T.A. Methods for Studying Iron Regulatory Protein 1: An Important Protein in Human Iron Metabolism. Methods Enzym. 2018, 599, 139–155. [Google Scholar]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 2014, 5, 176. [Google Scholar] [CrossRef] [PubMed]
- Pantopoulos, K. Iron metabolism and the IRE/IRP regulatory system: An update. Ann. N. Y. Acad. Sci. 2004, 1012, 1–13. [Google Scholar] [CrossRef]
- Du, F.; Qian, Z.M.; Gong, Q.; Zhu, Z.J.; Lu, L.; Ke, Y. The iron regulatory hormone hepcidin inhibits expression of iron release as well as iron uptake proteins in J774 cells. J. Nutr. Biochem. 2012, 23, 1694–1700. [Google Scholar] [CrossRef]
- Yang, J.; Xu, T.; Gomez, D.R.; Yuan, X.; Nguyen, Q.N.; Jeter, M.; Song, Y.; Komaki, R.; Hu, Y.; Hahn, S.M.; et al. Nomograms incorporating genetic variants in BMP/Smad4/Hamp pathway to predict disease outcomes after definitive radiotherapy for non-small cell lung cancer. Cancer Med. 2018, 7, 2247–2255. [Google Scholar] [CrossRef]
- Vela, D.; Vela-Gaxha, Z. Differential regulation of hepcidin in cancer and non-cancer tissues and its clinical implications. Exp. Mol. Med. 2018, 50, e436. [Google Scholar] [CrossRef]
- Huang, X. Iron overload and its association with cancer risk in humans: Evidence for iron as a carcinogenic metal. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2003, 533, 153–171. [Google Scholar] [CrossRef]
- Cairo, G.; Recalcati, S. Iron-regulatory proteins: Molecular biology and pathophysiological implications. Expert Rev. Mol. Med. 2007, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kabat, G.C.; Rohan, T.E. Does excess iron play a role in breast carcinogenesis? An unresolved hypothesis. Cancer Causes Control 2007, 18, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.D.; Coffman, L.G.; Chou, J.W.; Black, M.A.; Bergh, J.; D’Agostino, R., Jr.; Torti, S.V.; Torti, F.M. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011, 71, 6728–6737. [Google Scholar] [CrossRef] [PubMed]
- Galaris, D.; Skiada, V.; Barbouti, A. Redox signaling and cancer: The role of “labile” iron. Cancer Lett. 2008, 266, 21–29. [Google Scholar] [CrossRef]
- Tong, W.H.; Maio, N.; Zhang, D.L.; Palmieri, E.M.; Ollivierre, H.; Ghosh, M.C.; McVicar, D.W.; Rouault, T.A. TLR-activated repression of Fe-S cluster biogenesis drives a metabolic shift and alters histone and tubulin acetylation. Blood Adv. 2018, 2, 1146–1156. [Google Scholar] [CrossRef]
- Buzas, D.M.; Nakamura, M.; Kinoshita, T. Epigenetic role for the conserved Fe-S cluster biogenesis protein AtDRE2 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 13565–13570. [Google Scholar] [CrossRef]
- Sturm, A.; Ivics, Z.; Vellai, T. The mechanism of ageing: Primary role of transposable elements in genome disintegration. Cell. Mol. Life Sci. 2015, 72, 1839–1847. [Google Scholar] [CrossRef]
- Sfera, A.; Bullock, K.; Price, A.; Inderias, L.; Osorio, C. Ferrosenescence: The iron age of neurodegeneration? Mech. Ageing Dev. 2018, 174, 63–75. [Google Scholar] [CrossRef]
- Cao, L.L.; Liu, H.; Yue, Z.; Liu, L.; Pei, L.; Gu, J.; Wang, H.; Jia, M. Iron chelation inhibits cancer cell growth and modulates global histone methylation status in colorectal cancer. Biometals 2018, 31, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Rao, A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014, 30, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Cascella, B.; Mirica, L.M. Kinetic analysis of iron-dependent histone demethylases: Alpha-ketoglutarate substrate inhibition and potential relevance to the regulation of histone demethylation in cancer cells. Biochemistry 2012, 51, 8699–8701. [Google Scholar] [CrossRef] [PubMed]
- Clifton, I.J.; McDonough, M.A.; Ehrismann, D.; Kershaw, N.J.; Granatino, N.; Schofield, C.J. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J. Inorg. Biochem. 2006, 100, 644–669. [Google Scholar] [CrossRef]
- Wilson, S.; Fan, L.; Sahgal, N.; Qi, J.; Filipp, F.V. The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget 2017, 8, 30328–30343. [Google Scholar] [CrossRef] [PubMed]
- Kuo, K.T.; Huang, W.C.; Bamodu, O.A.; Lee, W.H.; Wang, C.H.; Hsiao, M.; Wang, L.S.; Yeh, C.T. Histone demethylase JARID1B/KDM5B promotes aggressiveness of non-small cell lung cancer and serves as a good prognostic predictor. Clin. Epigenet. 2018, 10, 107. [Google Scholar] [CrossRef]
- Harmeyer, K.M.; Facompre, N.D.; Herlyn, M.; Basu, D. JARID1 Histone Demethylases: Emerging Targets in Cancer. Trends Cancer 2017, 3, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Mao, Y.; Du, G.; He, C.; Han, S. Overexpression of JARID1B is associated with poor prognosis and chemotherapy resistance in epithelial ovarian cancer. Tumour Boil. 2015, 36, 2465–2472. [Google Scholar] [CrossRef]
- Ponnaluri, V.K.; Maciejewski, J.P.; Mukherji, M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem. Biophys. Res. Commun. 2013, 436, 115–120. [Google Scholar] [CrossRef]
- Tamanaha, E.; Guan, S.; Marks, K.; Saleh, L. Distributive Processing by the Iron(II)/alpha-Ketoglutarate-Dependent Catalytic Domains of the TET Enzymes Is Consistent with Epigenetic Roles for Oxidized 5-Methylcytosine Bases. J. Am. Chem. Soc. 2016, 138, 9345–9348. [Google Scholar] [CrossRef]
- Zhao, B.; Yang, Y.; Wang, X.; Chong, Z.; Yin, R.; Song, S.H.; Zhao, C.; Li, C.; Huang, H.; Sun, B.F.; et al. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res. 2014, 42, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; DesMarais, T.L.; Tong, Z.; Yao, Y.; Costa, M. Oxidative stress alters global histone modification and DNA methylation. Free Radic. Boil. Med. 2015, 82, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Udali, S.; Guarini, P.; Ruzzenente, A.; Ferrarini, A.; Guglielmi, A.; Lotto, V.; Tononi, P.; Pattini, P.; Moruzzi, S.; Campagnaro, T.; et al. DNA methylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin. Epigenet. 2015, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wu, Q.; Monga, J.; Xie, E.; Wang, H.; Wang, S.; Zhang, H.; Wang, Z.Y.; Zhou, T.; Shi, Y.; et al. HDAC1 Governs Iron Homeostasis Independent of Histone Deacetylation in Iron-Overload Murine Models. Antioxid. Redox Signal. 2018, 28, 1224–1237. [Google Scholar] [CrossRef] [PubMed]
- Pasricha, S.R.; Lim, P.J.; Duarte, T.L.; Casu, C.; Oosterhuis, D.; Mleczko-Sanecka, K.; Suciu, M.; Da Silva, A.R.; Al-Hourani, K.; Arezes, J.; et al. Hepcidin is regulated by promoter-associated histone acetylation and HDAC3. Nat. Commun. 2017, 8, 403. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, J.; Feng, J.; Wang, J. E4BP4 promotes thyroid cancer proliferation by modulating iron homeostasis through repression of hepcidin. Cell Death Dis. 2018, 9, 987. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Wu, Q.; Guo, X.; Zhang, Z.; Shen, Y.; Wang, F. MBD5 regulates iron metabolism via methylation-independent genomic targeting of Fth1 through KAT2A in mice. Br. J. Haematol. 2014, 166, 279–291. [Google Scholar] [CrossRef]
- Ohba, C.; Nabatame, S.; Iijima, Y.; Nishiyama, K.; Tsurusaki, Y.; Nakashima, M.; Miyake, N.; Tanaka, F.; Ozono, K.; Saitsu, H.; et al. De novo WDR45 mutation in a patient showing clinically Rett syndrome with childhood iron deposition in brain. J. Hum. Genet. 2014, 59, 292–295. [Google Scholar] [CrossRef]
- Wang, Y.F.; Zhang, J.; Su, Y.; Shen, Y.Y.; Jiang, D.X.; Hou, Y.Y.; Geng, M.Y.; Ding, J.; Chen, Y. G9a regulates breast cancer growth by modulating iron homeostasis through the repression of ferroxidase hephaestin. Nat. Commun. 2017, 8, 274. [Google Scholar] [CrossRef]
- Jeong, S.M.; Lee, J.; Finley, L.W.; Schmidt, P.J.; Fleming, M.D.; Haigis, M.C. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene 2015, 34, 2115–2124. [Google Scholar] [CrossRef]
- Guaraldo, M.; Santambrogio, P.; Rovelli, E.; Di Savino, A.; Saglio, G.; Cittaro, D.; Roetto, A.; Levi, S. Characterization of human mitochondrial ferritin promoter: Identification of transcription factors and evidences of epigenetic control. Sci. Rep. 2016, 6, 33432. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, S.; Wang, X.; Guo, W.; Wang, L.; Zhang, D.; Yuan, L.; Zhang, Z.; Xu, Y.; Liu, S. Disordered signaling governing ferroportin transcription favors breast cancer growth. Cell Signal. 2015, 27, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Fujiwara, T.; Ota, U.; Hatta, S.; Ichikawa, S.; Kobayashi, M.; Okitsu, Y.; Fukuhara, N.; Onishi, Y.; Ishizuka, M.; et al. Dynamics of absorption, metabolism, and excretion of 5-aminolevulinic acid in human intestinal Caco-2 cells. Biochem. Biophys. Rep. 2017, 11, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Park, S.H.; Chang, H.C.; Shapiro, J.S.; Vassilopoulos, A.; Sawicki, K.T.; Chen, C.; Shang, M.; Burridge, P.W.; Epting, C.L.; et al. Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J. Clin. Investig. 2017, 127, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Kato, H.; Hada, H.; Itoh-Nakadai, A.; Fujiwara, T.; Muto, A.; Inoguchi, Y.; Ichiyanagi, K.; Hojo, W.; Tomosugi, N.; et al. Iron-heme-Bach1 axis is involved in erythroblast adaptation to iron deficiency. Haematologica 2017, 102, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Schaar, D.G.; Medina, D.J.; Moore, D.F.; Strair, R.K.; Ting, Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp. Hematol. 2009, 37, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, M.; Bogdan, A.R.; Hashimoto, K.; Tsuji, Y. Regulation of transferrin receptor-1 mRNA by the interplay between IRE-binding proteins and miR-7/miR-141 in the 3’-IRE stem-loops. RNA 2018, 24, 468–479. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Kosaka, N.; Ochiya, T.; Kato, T. Micromanaging Iron Homeostasis: Hypoxia-inducible micro-RNA-210 suppresses iron homeostasis-related proteins. J. Boil. Chem. 2012, 287, 34110–34119. [Google Scholar] [CrossRef]
- Shpyleva, S.; Pogribna, M.; Cozart, C.; Bryant, M.S.; Muskhelishvili, L.; Tryndyak, V.P.; Ross, S.A.; Beland, F.A.; Pogribny, I.P. Interstrain differences in the progression of nonalcoholic steatohepatitis to fibrosis in mice are associated with altered hepatic iron metabolism. J. Nutr. Biochem. 2014, 25, 1235–1242. [Google Scholar] [CrossRef]
- Kindrat, I.; Tryndyak, V.; de Conti, A.; Shpyleva, S.; Mudalige, T.K.; Kobets, T.; Erstenyuk, A.M.; Beland, F.A.; Pogribny, I.P. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget 2016, 7, 1276–1287. [Google Scholar] [CrossRef]
- Andolfo, I.; De Falco, L.; Asci, R.; Russo, R.; Colucci, S.; Gorrese, M.; Zollo, M.; Iolascon, A. Regulation of divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA Let-7d in erythroid cells. Haematologica 2010, 95, 1244–1252. [Google Scholar] [CrossRef]
- Chan, J.J.; Kwok, Z.H.; Chew, X.H.; Zhang, B.; Liu, C.; Soong, T.W.; Yang, H.; Tay, Y. A FTH1 gene:pseudogene:microRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 2018, 46, 1998–2011. [Google Scholar] [CrossRef] [PubMed]
- Shpyleva, S.I.; Tryndyak, V.P.; Kovalchuk, O.; Starlard-Davenport, A.; Chekhun, V.F.; Beland, F.A.; Pogribny, I.P. Role of ferritin alterations in human breast cancer cells. Breast Cancer Res. Treat. 2011, 126, 63–71. [Google Scholar] [CrossRef] [PubMed]
- White, K.; Lu, Y.; Annis, S.; Hale, A.E.; Chau, B.N.; Dahlman, J.E.; Hemann, C.; Opotowsky, A.R.; Vargas, S.O.; Rosas, I.; et al. Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension. EMBO Mol. Med. 2015, 7, 695–713. [Google Scholar] [CrossRef] [PubMed]
- Castoldi, M.; Vujic Spasic, M.; Altamura, S.; Elmen, J.; Lindow, M.; Kiss, J.; Stolte, J.; Sparla, R.; D’Alessandro, L.A.; Klingmuller, U.; et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Investig. 2011, 121, 1386–1396. [Google Scholar] [CrossRef]
- Liao, Y.; Du, X.; Lonnerdal, B. miR-214 regulates lactoferrin expression and pro-apoptotic function in mammary epithelial cells. J. Nutr. 2010, 140, 1552–1556. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Lonnerdal, B. miR-584 mediates post-transcriptional expression of lactoferrin receptor in Caco-2 cells and in mouse small intestine during the perinatal period. Int. J. Biochem. Cell Boil. 2010, 42, 1363–1369. [Google Scholar] [CrossRef]
- Babu, K.R.; Muckenthaler, M.U. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. J. Mol. Med. 2016, 94, 347–359. [Google Scholar] [CrossRef]
- Sangokoya, C.; Doss, J.F.; Chi, J.T. Iron-responsive miR-485-3p regulates cellular iron homeostasis by targeting ferroportin. PLoS Genet. 2013, 9, e1003408. [Google Scholar] [CrossRef]
- Thevenod, F. Iron and Its Role in Cancer Defense: A Double-Edged Sword. Met. Ions Life Sci. 2018, 18. [Google Scholar] [CrossRef]
- Le, N.T.; Richardson, D.R. The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biochim. Biophys. Acta 2002, 1603, 31–46. [Google Scholar] [CrossRef]
- Yu, Y.; Kovacevic, Z.; Richardson, D.R. Tuning cell cycle regulation with an iron key. Cell Cycle 2007, 6, 1982–1994. [Google Scholar] [CrossRef] [PubMed]
- Steegmann-Olmedillas, J.L. The role of iron in tumour cell proliferation. Clin. Transl. Oncol. 2011, 13, 71–76. [Google Scholar] [CrossRef]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Novel chelators for cancer treatment: Where are we now? Antioxid. Redox Signal. 2013, 18, 973–1006. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Chou, C.H.; Shun, C.T.; Mao, T.L.; Wen, W.F.; Chen, C.D.; Chen, S.U.; Yang, Y.S.; Ho, H.N. Iron suppresses ovarian granulosa cell proliferation and arrests cell cycle through regulating p38 mitogen-activated protein kinase/p53/p21 pathway. Biol. Reprod. 2017, 97, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Manz, D.H.; Torti, S.V.; Torti, F.M. Iron-responsive element-binding protein 2 plays an essential role in regulating prostate cancer cell growth. Oncotarget 2017, 8, 82231–82243. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Song, J.; Yung, B.C.; Zhou, Z.; Wu, A.; Chen, X. Emerging Strategies of Cancer Therapy Based on Ferroptosis. Adv. Mater. 2018, 30, e1704007. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Lu, B.; Chen, X.B.; Ying, M.D.; He, Q.J.; Cao, J.; Yang, B. The Role of Ferroptosis in Cancer Development and Treatment Response. Front. Pharmacol. 2017, 8, 992. [Google Scholar] [CrossRef]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Boil. Med. 2018. [Google Scholar] [CrossRef]
- Alvarez, S.W.; Sviderskiy, V.O.; Terzi, E.M.; Papagiannakopoulos, T.; Moreira, A.L.; Adams, S.; Sabatini, D.M.; Birsoy, K.; Possemato, R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 2017, 551, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Kroemer, G.; Tang, D. The tumor suppressor protein p53 and the ferroptosis network. Free Radic. Boil. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Lee, D.H.; Lee, Y.S.; Jo, M.J.; Jeong, Y.A.; Kwon, W.T.; Choudry, H.A.; Bartlett, D.L.; Lee, Y.J. Molecular crosstalk between ferroptosis and apoptosis: Emerging role of ER stress-induced p53-independent PUMA expression. Oncotarget 2017, 8, 115164–115178. [Google Scholar] [CrossRef] [PubMed]
- El Hout, M.; Dos Santos, L.; Hamai, A.; Mehrpour, M. A promising new approach to cancer therapy: Targeting iron metabolism in cancer stem cells. Semin. Cancer Biol. 2018, 53, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Borgstrom, B.; Stegmayr, J.; Abassi, Y.; Kruszyk, M.; Leffler, H.; Persson, L.; Albinsson, S.; Massoumi, R.; Scheblykin, I.G.; et al. The Molecular Basis for Inhibition of Stemlike Cancer Cells by Salinomycin. ACS Cent. Sci. 2018, 4, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Chen, Z.; Wu, D.; Chen, L. Ferritinophagy/ferroptosis: Iron-related newcomers in human diseases. J. Cell. Physiol. 2018, 233, 9179–9190. [Google Scholar] [CrossRef]
- Ryu, M.S.; Duck, K.A.; Philpott, C.C. Ferritin iron regulators, PCBP1 and NCOA4, respond to cellular iron status in developing red cells. Blood Cells Mol. Dis. 2018, 69, 75–81. [Google Scholar] [CrossRef]
- Huang, T.; Sun, Y.; Li, Y.; Wang, T.; Fu, Y.; Li, C.; Li, C. Growth Inhibition of a Novel Iron Chelator, DpdtC, against Hepatoma Carcinoma Cell Lines Partly Attributed to Ferritinophagy-Mediated Lysosomal ROS Generation. Oxid. Med. Cell. Longev. 2018, 2018, 4928703. [Google Scholar] [CrossRef]
- Wylie, A.; Jones, A.E.; D’Brot, A.; Lu, W.J.; Kurtz, P.; Moran, J.V.; Rakheja, D.; Chen, K.S.; Hammer, R.E.; Comerford, S.A.; et al. p53 genes function to restrain mobile elements. Genes Dev. 2016, 30, 64–77. [Google Scholar] [CrossRef]
- Kaomongkolgit, R.; Cheepsunthorn, P.; Pavasant, P.; Sanchavanakit, N. Iron increases MMP-9 expression through activation of AP-1 via ERK/Akt pathway in human head and neck squamous carcinoma cells. Oral Oncol. 2008, 44, 587–594. [Google Scholar] [CrossRef]
- Shan, L. Bimodal lentiviral vector encoding myc-tagged human ferritin heavy chain and green fluorescent protein (GFP). In Molecular Imaging and Contrast Agent Database (MICAD); National Center for Biotechnology Information (US): Bethesda, MD, USA, 2004. [Google Scholar]
- Lin, R.; Huang, J.; Wang, L.; Li, Y.; Lipowska, M.; Wu, H.; Yang, J.; Mao, H. Bevacizumab and near infrared probe conjugated iron oxide nanoparticles for vascular endothelial growth factor targeted MR and optical imaging. Biomater. Sci. 2018, 6, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, K.; Khare, V.; Evstatiev, R.; Kulnigg-Dabsch, S.; Jambrich, M.; Strobl, H.; Gasche, C. Increased expression of HIF2alpha during iron deficiency-associated megakaryocytic differentiation. J. Thromb. Haemost. 2015, 13, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.V.; Sahni, S.; Kovacevic, Z.; Richardson, D.R. Interplay of the iron-regulated metastasis suppressor NDRG1 with epidermal growth factor receptor (EGFR) and oncogenic signaling. J. Boil. Chem. 2017, 292, 12772–12782. [Google Scholar] [CrossRef]
- Sahni, S.; Krishan, S.; Richardson, D.R. NDRG1 as a molecular target to inhibit the epithelial-mesenchymal transition: The case for developing inhibitors of metastasis. Future Med. Chem. 2014, 6, 1241–1244. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, C.Y.; Xia, R.H.; Han, J.; Sun, B.; Sun, S.Y.; Li, J. The MYB/miR-130a/NDRG2 axis modulates tumor proliferation and metastatic potential in salivary adenoid cystic carcinoma. Cell Death Dis. 2018, 9, 917. [Google Scholar] [CrossRef]
- Yang, C.L.; Zheng, X.L.; Ye, K.; Ge, H.; Sun, Y.N.; Lu, Y.F.; Fan, Q.X. NDRG2 suppresses proliferation, migration, invasion and epithelial-mesenchymal transition of esophageal cancer cells through regulating the AKT/XIAP signaling pathway. Int. J. Biochem. Cell Boil. 2018, 99, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Lei, L.; Hong, L.L.; Ling, Z.Q. Potential role of NDRG2 in reprogramming cancer metabolism and epithelial-to-mesenchymal transition. Histol. Histopathol. 2018, 33, 655–663. [Google Scholar]
- Guo, H.F.; Tsai, C.L.; Terajima, M.; Tan, X.; Banerjee, P.; Miller, M.D.; Liu, X.; Yu, J.; Byemerwa, J.; Alvarado, S.; et al. Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe(2+)-binding. Nat. Commun. 2018, 9, 512. [Google Scholar] [CrossRef]
- Zhang, A.; Wang, B.; Yang, M.; Shi, H.; Gan, W. beta2-microglobulin induces epithelial-mesenchymal transition in human renal proximal tubule epithelial cells in vitro. BMC Nephrol. 2015, 16, 60. [Google Scholar] [CrossRef]
- Song, E.; Ramos, S.V.; Huang, X.; Liu, Y.; Botta, A.; Sung, H.K.; Turnbull, P.C.; Wheeler, M.B.; Berger, T.; Wilson, D.J.; et al. Holo-lipocalin-2-derived siderophores increase mitochondrial ROS and impair oxidative phosphorylation in rat cardiomyocytes. Proc. Natl. Acad. Sci. USA 2018, 115, 1576–1581. [Google Scholar] [CrossRef]
- Ganz, T.; Nemeth, E. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 2015, 15, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.P.; Elliott, R.L. Decreased Iron in Cancer Cells and Their Microenvironment Improves Cytolysis of Breast Cancer Cells by Natural Killer Cells. Anticancer. Res. 2017, 37, 2297–2305. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.M.; He, X.; Liang, T.; Wu, K.C.; Yan, Y.C.; Lu, L.N.; Yang, G.; Luo, Q.Q.; Yung, W.H.; Ke, Y. Lipopolysaccharides upregulate hepcidin in neuron via microglia and the IL-6/STAT3 signaling pathway. Mol. Neurobiol. 2014, 50, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Bj, R.; Dw, V.P.; Lm, G.; Ac, L.; Ra, G.R.; Ahn, C.; Kl, F.; Jf, H. Plasma ferritin concentration is positively associated with in vivo fatty acid mobilization and insulin resistance in obese women. Exp. Physiol. 2018, 103, 1443–1447. [Google Scholar]
- Marques, O.; Porto, G.; Rema, A.; Faria, F.; Cruz Paula, A.; Gomez-Lazaro, M.; Silva, P.; Martins da Silva, B.; Lopes, C. Local iron homeostasis in the breast ductal carcinoma microenvironment. BMC Cancer 2016, 16, 187. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.N.; Lei, Y.H.; Chang, Y.Z. The regulation of iron metabolism in the mononuclear phagocyte system. Expert Rev. Hematol. 2013, 6, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Arezes, J.; Costa, M.; Vieira, I.; Dias, V.; Kong, X.L.; Fernandes, R.; Vos, M.; Carlsson, A.; Rikers, Y.; Porto, G.; et al. Non-transferrin-bound iron (NTBI) uptake by T lymphocytes: Evidence for the selective acquisition of oligomeric ferric citrate species. PLoS ONE 2013, 8, e79870. [Google Scholar] [CrossRef]
- Daher, R.; Karim, Z. Iron metabolism: State of the art. Transfus Clin. Boil. 2017, 24, 115–119. [Google Scholar] [CrossRef]
- Knutson, M.D. Non-transferrin-bound iron transporters. Free Radic. Boil. Med. 2018. [Google Scholar] [CrossRef]
- Sayadi, A.; Nguyen, A.T.; Bard, F.A.; Bard-Chapeau, E.A. Zip14 expression induced by lipopolysaccharides in macrophages attenuates inflammatory response. Inflamm. Res. 2013, 62, 133–143. [Google Scholar] [CrossRef]
- Pyle, C.J.; Azad, A.K.; Papp, A.C.; Sadee, W.; Knoell, D.L.; Schlesinger, L.S. Elemental Ingredients in the Macrophage Cocktail: Role of ZIP8 in Host Response to Mycobacterium tuberculosis. Int. J. Mol. Sci. 2017, 18, 2375. [Google Scholar] [CrossRef] [PubMed]
- Hvidberg, V.; Maniecki, M.B.; Jacobsen, C.; Hojrup, P.; Moller, H.J.; Moestrup, S.K. Identification of the receptor scavenging hemopexin-heme complexes. Blood 2005, 106, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.J.; Andersen, C.B.; Moestrup, S.K. CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J. Boil. Chem. 2013, 288, 18834–18841. [Google Scholar] [CrossRef]
- White, C.; Yuan, X.; Schmidt, P.J.; Bresciani, E.; Samuel, T.K.; Campagna, D.; Hall, C.; Bishop, K.; Calicchio, M.L.; Lapierre, A.; et al. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 2013, 17, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.; Mertens, C.; Brune, B. Macrophage iron homeostasis and polarization in the context of cancer. Immunobiology 2015, 220, 295–304. [Google Scholar] [CrossRef]
- Agoro, R.; Taleb, M.; Quesniaux, V.F.J.; Mura, C. Cell iron status influences macrophage polarization. PLoS ONE 2018, 13, e0196921. [Google Scholar] [CrossRef]
- Zhou, Y.; Que, K.T.; Zhang, Z.; Yi, Z.J.; Zhao, P.X.; You, Y.; Gong, J.P.; Liu, Z.J. Iron overloaded polarizes macrophage to proinflammation phenotype through ROS/acetyl-p53 pathway. Cancer Med. 2018, 7, 4012–4022. [Google Scholar] [CrossRef]
- Costa da Silva, M.; Breckwoldt, M.O.; Vinchi, F.; Correia, M.P.; Stojanovic, A.; Thielmann, C.M.; Meister, M.; Muley, T.; Warth, A.; Platten, M.; et al. Iron Induces Anti-tumor Activity in Tumor-Associated Macrophages. Front. Immunol. 2017, 8, 1479. [Google Scholar] [CrossRef] [PubMed]
- Tang, X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013, 332, 3–10. [Google Scholar] [CrossRef]
- Dignass, A.; Farrag, K.; Stein, J. Limitations of Serum Ferritin in Diagnosing Iron Deficiency in Inflammatory Conditions. Int. J. Chronic Dis. 2018, 2018, 9394060. [Google Scholar] [CrossRef]
- Meng, F.; Zhen, S.; Song, B. HBV-specific CD4+ cytotoxic T cells in hepatocellular carcinoma are less cytolytic toward tumor cells and suppress CD8+ T cell-mediated antitumor immunity. APMIS 2017, 125, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Q.; De Marchi, T.; Timmermans, A.M.; Beekhof, R.; Trapman-Jansen, A.M.; Foekens, R.; Look, M.P.; van Deurzen, C.H.; Span, P.N.; Sweep, F.C.; et al. Ferritin heavy chain in triple negative breast cancer: A favorable prognostic marker that relates to a cluster of differentiation 8 positive (CD8+) effector T-cell response. Mol. Cell. Proteom. 2014, 13, 1814–1827. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Paragas, N.; Ned, R.M.; Qiu, A.; Viltard, M.; Leete, T.; Drexler, I.R.; Chen, X.; Sanna-Cherchi, S.; Mohammed, F.; et al. Scara5 is a ferritin receptor mediating non-transferrin iron delivery. Dev. Cell 2009, 16, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; He, K.; Liu, P.; Xu, L.X. Iron participated in breast cancer chemoresistance by reinforcing IL-6 paracrine loop. Biochem. Biophys. Res. Commun. 2016, 475, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Legrand, D. Overview of Lactoferrin as a Natural Immune Modulator. J. Pediatr. 2016, 173, S10–S15. [Google Scholar] [CrossRef]
- Wisgrill, L.; Wessely, I.; Spittler, A.; Forster-Waldl, E.; Berger, A.; Sadeghi, K. Human lactoferrin attenuates the proinflammatory response of neonatal monocyte-derived macrophages. Clin. Exp. Immunol. 2018, 192, 315–324. [Google Scholar] [CrossRef]
- Borkham-Kamphorst, E.; van de Leur, E.; Zimmermann, H.W.; Karlmark, K.R.; Tihaa, L.; Haas, U.; Tacke, F.; Berger, T.; Mak, T.W.; Weiskirchen, R. Protective effects of lipocalin-2 (LCN2) in acute liver injury suggest a novel function in liver homeostasis. Biochim. Biophys. Acta 2013, 1832, 660–673. [Google Scholar] [CrossRef]
- Karlsen, J.R.; Borregaard, N.; Cowland, J.B. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J. Boil. Chem. 2010, 285, 14088–14100. [Google Scholar] [CrossRef]
- Johansen, C.; Bertelsen, T.; Ljungberg, C.; Mose, M.; Iversen, L. Characterization of TNF-alpha- and IL-17A-Mediated Synergistic Induction of DEFB4 Gene Expression in Human Keratinocytes through IkappaBzeta. J. Investig. Dermatol. 2016, 136, 1608–1616. [Google Scholar] [CrossRef]
- Moschen, A.R.; Gerner, R.R.; Wang, J.; Klepsch, V.; Adolph, T.E.; Reider, S.J.; Hackl, H.; Pfister, A.; Schilling, J.; Moser, P.L.; et al. Lipocalin 2 Protects from Inflammation and Tumorigenesis Associated with Gut Microbiota Alterations. Cell Host Microbe 2016, 19, 455–469. [Google Scholar] [CrossRef]
- Koh, S.A.; Lee, K.H. HGF mediated upregulation of lipocalin 2 regulates MMP9 through nuclear factor-kappaB activation. Oncol. Rep. 2015, 34, 2179–2187. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; He, K.; Li, J.; Cheng, M.; Song, H.; Liu, J.; Liu, P. Tumor associated macrophages deliver iron to tumor cells via Lcn2. Int. J. Physiol. Pathophysiol. Pharmacol. 2018, 10, 105–114. [Google Scholar] [PubMed]
- Lisowska-Myjak, B.; Skarzynska, E.; Wilczynska, P.; Jakimiuk, A. Correlation between the concentrations of lactoferrin and neutrophil gelatinase-associated lipocalin in meconium. Biometals 2018, 31, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.C.; Chaze, T.; Coic, Y.M.; Injarabian, L.; Jonsson, F.; Lombion, N.; Selimoglu-Buet, D.; Souphron, J.; Ridley, C.; Vonaesch, P.; et al. MUB40 Binds to Lactoferrin and Stands as a Specific Neutrophil Marker. Cell Chem. Boil. 2018, 25, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, F.; Pisa, L.; Mellone, S.; Cillo, M.; Lepore, A.; Leonardi, A. NGAL promotes recruitment of tumor infiltrating leukocytes. Oncotarget 2018, 9, 30761–30772. [Google Scholar] [CrossRef] [PubMed]
- Candido, S.; Abrams, S.L.; Steelman, L.S.; Lertpiriyapong, K.; Fitzgerald, T.L.; Martelli, A.M.; Cocco, L.; Montalto, G.; Cervello, M.; Polesel, J.; et al. Roles of NGAL and MMP-9 in the tumor microenvironment and sensitivity to targeted therapy. Biochim. Biophys. Acta 2016, 1863, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Fillebeen, C.; Wilkinson, N.; Charlebois, E.; Katsarou, A.; Wagner, J.; Pantopoulos, K. Hepcidin-mediated hypoferremic response to acute inflammation requires a threshold of Bmp6/Hjv/Smad signaling. Blood 2018, 132, 1829–1841. [Google Scholar] [CrossRef]
- Arosio, P. New signaling pathways for hepcidin regulation. Blood 2014, 123, 1433–1434. [Google Scholar] [CrossRef] [PubMed]
- Zuo, E.; Lu, Y.; Yan, M.; Pan, X.; Cheng, X. Increased expression of hepcidin and associated upregulation of JAK/STAT3 signaling in human gastric cancer. Oncol. Lett. 2018, 15, 2236–2244. [Google Scholar] [CrossRef] [PubMed]
- Blanchette-Farra, N.; Kita, D.; Konstorum, A.; Tesfay, L.; Lemler, D.; Hegde, P.; Claffey, K.P.; Torti, F.M.; Torti, S.V. Contribution of three-dimensional architecture and tumor-associated fibroblasts to hepcidin regulation in breast cancer. Oncogene 2018, 37, 4013–4032. [Google Scholar] [CrossRef] [PubMed]
- Colucci, S.; Pagani, A.; Pettinato, M.; Artuso, I.; Nai, A.; Camaschella, C.; Silvestri, L. The immunophilin FKBP12 inhibits hepcidin expression by binding the BMP type I receptor ALK2 in hepatocytes. Blood 2017, 130, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Neshastehriz, A.; Khosravi, Z.; Ghaznavi, H.; Shakeri-Zadeh, A. Gold-coated iron oxide nanoparticles trigger apoptosis in the process of thermo-radiotherapy of U87-MG human glioma cells. Radiat. Environ. Biophys. 2018, 57, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.V.; Manz, D.H.; Paul, B.T.; Blanchette-Farra, N.; Torti, F.M. Iron and Cancer. Annu. Rev. Nutr. 2018, 38, 97–125. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, Y.; Lai, H.; Li, X.; Chen, T. Iron(II)-Polypyridyl Complexes Inhibit the Growth of Glioblastoma Tumor and Enhance TRAIL-Induced Cell Apoptosis. Chem. Asian J. 2018, 13, 2730–2738. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, F.; Wei, Z.; Li, X.; Zhao, H.; Lv, H.; Ge, R.; Ma, H.; Zhang, H.; Yang, B.; et al. Magnetic delivery of Fe3O4@polydopamine nanoparticle-loaded natural killer cells suggest a promising anticancer treatment. Biomater. Sci. 2018, 6, 2714–2725. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhao, H.Y.; Du, J.; Wang, F. Anti-tumor activities of four chelating agents against human neuroblastoma cells. In Vivo 2005, 19, 233–236. [Google Scholar] [PubMed]
- Li, P.; Zheng, X.; Shou, K.; Niu, Y.; Jian, C.; Zhao, Y.; Yi, W.; Hu, X.; Yu, A. The iron chelator Dp44mT suppresses osteosarcoma's proliferation, invasion and migration: in vitro and in vivo. Am. J. Transl. Res. 2016, 8, 5370–5385. [Google Scholar] [PubMed]
- Lui, G.Y.; Kovacevic, Z.; Richardson, V.; Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Targeting cancer by binding iron: Dissecting cellular signaling pathways. Oncotarget 2015, 6, 18748–18779. [Google Scholar] [CrossRef] [PubMed]
- Breccia, M.; Alimena, G. Efficacy and safety of deferasirox in myelodysplastic syndromes. Ann. Hematol. 2013, 92, 863–870. [Google Scholar] [CrossRef]
- Simoes, R.V.; Veeraperumal, S.; Serganova, I.S.; Kruchevsky, N.; Varshavsky, J.; Blasberg, R.G.; Ackerstaff, E.; Koutcher, J.A. Inhibition of prostate cancer proliferation by Deferiprone. NMR Biomed. 2017, 30. [Google Scholar] [CrossRef] [PubMed]
- Knickle, A.; Fernando, W.; Greenshields, A.L.; Rupasinghe, H.P.V.; Hoskin, D.W. Myricetin-induced apoptosis of triple-negative breast cancer cells is mediated by the iron-dependent generation of reactive oxygen species from hydrogen peroxide. Food Chem. Toxicol. 2018, 118, 154–167. [Google Scholar] [CrossRef]
- Bajbouj, K.; Shafarin, J.; Hamad, M. High-Dose Deferoxamine Treatment Disrupts Intracellular Iron Homeostasis, Reduces Growth, and Induces Apoptosis in Metastatic and Nonmetastatic Breast Cancer Cell Lines. Technol. Cancer Res. Treat. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Mertens, C.; Akam, E.A.; Rehwald, C.; Brune, B.; Tomat, E.; Jung, M. Intracellular Iron Chelation Modulates the Macrophage Iron Phenotype with Consequences on Tumor Progression. PLoS ONE 2016, 11, e0166164. [Google Scholar] [CrossRef] [PubMed]
- Busti, F.; Marchi, G.; Ugolini, S.; Castagna, A.; Girelli, D. Anemia and Iron Deficiency in Cancer Patients: Role of Iron Replacement Therapy. Pharmaceuticals 2018, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.H.; Huang, P.H.; Hsu, Y.J.; Peng, Y.J.; Lee, C.H.; Wang, J.C.; Chen, J.W.; Lin, S.J. Inhibition of hypoxia inducible factor-1alpha attenuates abdominal aortic aneurysm progression through the down-regulation of matrix metalloproteinases. Sci. Rep. 2016, 6, 28612. [Google Scholar] [CrossRef] [PubMed]
- Kontoghiorghes, G.J. Ethical issues and risk/benefit assessment of iron chelation therapy: Advances with deferiprone/deferoxamine combinations and concerns about the safety, efficacy and costs of deferasirox. Hemoglobin 2008, 32, 1–15. [Google Scholar] [CrossRef]
- Di Nicola, M.; Barteselli, G.; Dell’Arti, L.; Ratiglia, R.; Viola, F. Functional and Structural Abnormalities in Deferoxamine Retinopathy: A Review of the Literature. BioMed Res. Int. 2015, 2015, 249617. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.L.; Hatef, A.; Imran ul-Haq, M.; Nair, N.; Unniappan, S.; Kizhakkedathu, J.N. Clinically approved iron chelators influence zebrafish mortality, hatching morphology and cardiac function. PLoS ONE 2014, 9, e109880. [Google Scholar] [CrossRef]
- Tury, S.; Assayag, F.; Bonin, F.; Chateau-Joubert, S.; Servely, J.L.; Vacher, S.; Becette, V.; Caly, M.; Rapinat, A.; Gentien, D.; et al. The iron chelator deferasirox synergises with chemotherapy to treat triple-negative breast cancers. J. Pathol. 2018, 246, 103–114. [Google Scholar] [CrossRef]
- Piro, E.; Lentini, M.; Levato, L.; Russo, A.; Molica, S. Sustained Erythroid Response in a Patient with Myelofibrosis Receiving Concomitant Treatment with Ruxolitinib and Deferasirox. Chemotherapy 2018, 63, 107–110. [Google Scholar] [CrossRef]
- Chang, Y.C.; Lo, W.J.; Huang, Y.T.; Lin, C.L.; Feng, C.C.; Lin, H.T.; Cheng, H.C.; Yeh, S.P. Deferasirox has strong anti-leukemia activity but may antagonize theanti-leukemia effect of doxorubicin. Leuk. Lymphoma 2017, 58, 1–12. [Google Scholar] [CrossRef]
- Ikeda, R.; Vermeulen, L.C.; Jiang, Z.; Lau, E.; Kolesar, J.M. Gemcitabine and paclitaxel suppress the production of vascular endothelial growth factor induced by deferoxamine in human non-small cell lung cancer A549 cells. Exp. Ther. Med. 2010, 1, 853–857. [Google Scholar] [CrossRef]
- Shinoda, S.; Kaino, S.; Amano, S.; Harima, H.; Matsumoto, T.; Fujisawa, K.; Takami, T.; Yamamoto, N.; Yamasaki, T.; Sakaida, I. Deferasirox, an oral iron chelator, with gemcitabine synergistically inhibits pancreatic cancer cell growth in vitro and in vivo. Oncotarget 2018, 9, 28434–28444. [Google Scholar] [CrossRef] [PubMed]
- Eyvazzadeh, N.; Shakeri-Zadeh, A.; Fekrazad, R.; Amini, E.; Ghaznavi, H.; Kamran Kamrava, S. Gold-coated magnetic nanoparticle as a nanotheranostic agent for magnetic resonance imaging and photothermal therapy of cancer. Lasers Med. Sci. 2017, 32, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wang, X.; Zhou, S.; Zhang, Y. IONP-doped nanoparticles for highly effective NIR-controlled drug release and combination tumor therapy. Int. J. Nanomed. 2017, 12, 3751–3766. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhou, Z.; Mao, H.; Yang, L. Magnetic nanoparticles for precision oncology: Theranostic magnetic iron oxide nanoparticles for image-guided and targeted cancer therapy. Nanomedicine 2017, 12, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, X.; Cheng, Y.; Guo, X.; Yuan, W. Iron Oxide Nanoparticles-Based Vaccine Delivery for Cancer Treatment. Mol. Pharm. 2018, 15, 1791–1799. [Google Scholar] [CrossRef]
- Alarifi, S.; Ali, D.; Alkahtani, S.; Alhader, M.S. Iron oxide nanoparticles induce oxidative stress, DNA damage, and caspase activation in the human breast cancer cell line. Boil. Trace Element Res. 2014, 159, 416–424. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Y.; Peng, H.; Fang, X.; Zhang, X.; Chen, Q.; Wang, X.; Yang, W.; Sha, X. Blocking Autophagic Flux Enhances Iron Oxide Nanoparticle Photothermal Therapeutic Efficiency in Cancer Treatment. ACS Appl. Mater. Interfaces 2018, 10, 27701–27711. [Google Scholar] [CrossRef]
- Tsai, M.F.; Hsu, C.; Yeh, C.S.; Hsiao, Y.J.; Su, C.H.; Wang, L.F. Tuning the Distance of Rattle-Shaped IONP@Shell-in-Shell Nanoparticles for Magnetically-Targeted Photothermal Therapy in the Second Near-Infrared Window. ACS Appl. Mater. Interfaces 2018, 10, 1508–1519. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Duan, J.; Wang, J.; Liu, Q.; Shang, R.; Yang, X.; Lu, P.; Xia, C.; Wang, L.; Dou, K. Superparamagnetic iron oxide nanoparticles modified with polyethylenimine and galactose for siRNA targeted delivery in hepatocellular carcinoma therapy. Int. J. Nanomed. 2018, 13, 1851–1865. [Google Scholar] [CrossRef] [PubMed]
- Truffi, M.; Colombo, M.; Sorrentino, L.; Pandolfi, L.; Mazzucchelli, S.; Pappalardo, F.; Pacini, C.; Allevi, R.; Bonizzi, A.; Corsi, F.; et al. Multivalent exposure of trastuzumab on iron oxide nanoparticles improves antitumor potential and reduces resistance in HER2-positive breast cancer cells. Sci. Rep. 2018, 8, 6563. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhanl, D.; Du, Y. The immunotherapeutic effect of Fe3O4 nanoparticles as adjuvants on mice H22 live cancer. J. Nanosci. Nanotechnol. 2010, 10, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Wang, J.; Chen, B.A.; Ding, J.H.; Gao, C.; Cheng, J.; Zhao, G.; Bao, W.; Gao, F.; Xia, G.H.; et al. Influence of magnetic Fe3O4 nanoparticle on functions of lymphocytes and macrophages in mice. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2010, 18, 136–139. [Google Scholar] [PubMed]
- Jeong, J.; Kwon, E.K.; Cheong, T.C.; Park, H.; Cho, N.H.; Kim, W. Synthesis of multifunctional Fe(3)O(4)-CdSe/ZnS nanoclusters coated with lipid A toward dendritic cell-based immunotherapy. ACS Appl. Mater. Interfaces 2014, 6, 5297–5307. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, Z.; Gu, N.; Wang, J. Effects of DMSA-coated Fe3O4 magnetic nanoparticles on global gene expression of mouse macrophage RAW264.7 cells. Toxicol. Lett. 2011, 205, 130–139. [Google Scholar] [CrossRef]
- Jaidev, L.R.; Chellappan, D.R.; Bhavsar, D.V.; Ranganathan, R.; Sivanantham, B.; Subramanian, A.; Sharma, U.; Jagannathan, N.R.; Krishnan, U.M.; Sethuraman, S. Multi-functional nanoparticles as theranostic agents for the treatment & imaging of pancreatic cancer. Acta Biomater. 2017, 49, 422–433. [Google Scholar] [PubMed]
- Zheng, X.C.; Ren, W.; Zhang, S.; Zhong, T.; Duan, X.C.; Yin, Y.F.; Xu, M.Q.; Hao, Y.L.; Li, Z.T.; Li, H.; et al. The theranostic efficiency of tumor-specific, pH-responsive, peptide-modified, liposome-containing paclitaxel and superparamagnetic iron oxide nanoparticles. Int. J. Nanomed. 2018, 13, 1495–1504. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yu, L.; Ding, J.; Chen, Y. Iron Metabolism in Cancer. Int. J. Mol. Sci. 2019, 20, 95. https://doi.org/10.3390/ijms20010095
Wang Y, Yu L, Ding J, Chen Y. Iron Metabolism in Cancer. International Journal of Molecular Sciences. 2019; 20(1):95. https://doi.org/10.3390/ijms20010095
Chicago/Turabian StyleWang, Yafang, Lei Yu, Jian Ding, and Yi Chen. 2019. "Iron Metabolism in Cancer" International Journal of Molecular Sciences 20, no. 1: 95. https://doi.org/10.3390/ijms20010095
APA StyleWang, Y., Yu, L., Ding, J., & Chen, Y. (2019). Iron Metabolism in Cancer. International Journal of Molecular Sciences, 20(1), 95. https://doi.org/10.3390/ijms20010095





