Furosine, a Maillard Reaction Product, Triggers Necroptosis in Hepatocytes by Regulating the RIPK1/RIPK3/MLKL Pathway
Abstract
1. Introduction
2. Results
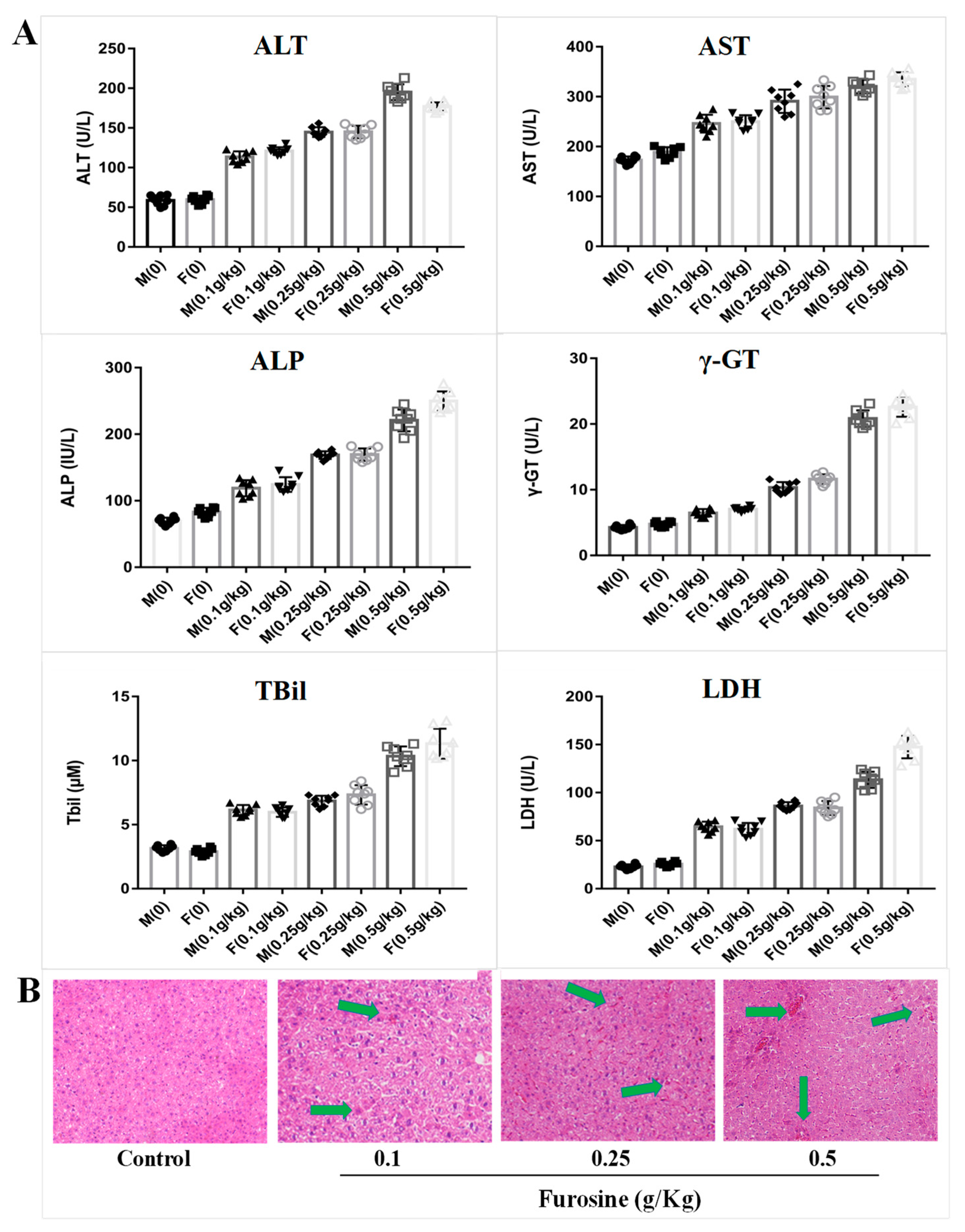
2.1. Furosine Increases the Expressios of Liver Biochemical Indicators
2.2. Furosine Exerts and Adverse Effect on the Liver
2.3. Transcriptomics Analysis of Furosine Metabolites in Liver Tissue, Serum, and Red Blood Cells
2.4. Screening of Specific Metabolites in Liver Tissue, Serum, and RBC
2.5. Furosine Enhanced the Expression of PLA2-3, RIPK-1, RIPK-3, P-MLKL, TNF-α, and IL-1β in Liver Tissue and Primary Hepatocytes
3. Discussion
4. Materials and Methods
4.1. Mice Chronic Toxicity Model
4.2. Histopathologial Test
4.3. Biochemical Analysis
4.4. Sample Preparation for Metabonomics Analysis
4.5. Quantitative Real-Time PCR (q-PCR) Analysis
4.6. Primary Hepatocyte Separation and Culture
4.7. Cell Viability Detection
4.8. Cell siRNA Treatment
4.9. Protein Sample Preparation and Western Blot Detection
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MRP | Maillard reaction product |
| LPC (18:0) | lysophosphatidylcholine 18:0 |
| PLA2-3 | phospholipase A2 gamma |
| RIPK1 | receptor-interacting serine/threonine-protein kinase 1 |
| RIPK3 | receptor-interacting serine/threonine-protein kinase 3 |
| MLKL | mixed lineage kinase domain-like protein |
References
- Huang, M.; Zhang, X.; Karangwa, E. Comparation sensory characteristic, non-volatile compounds, volatile compounds and antioxidant activity of MRPs by novel gradient temperature-elevating and traditional isothermal methods. J. Food Sci. Technol. 2015, 52, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Finot, P.A.; Bricout, J.; Viani, R.; Mauron, J. Identification of a new lysine derivative obtained upon acid hydrolysis of heated milk. Experientia 1968, 24, 1097–1099. [Google Scholar] [CrossRef]
- Villamiel, M.; Mddel, C.; Corzo, N.; Olano, A. Presence of furosine in honeys. J. Sci. Food Agric. 2001, 81, 790–793. [Google Scholar] [CrossRef]
- Rajchl, A.; Čížková, H.; Voldřich, M.; Jirušková, M.; Ševčík, R. Evaluation of shelf life and heat treatment of tomato products. Czech J. Food Sci. 2009, 27, 130–133. [Google Scholar] [CrossRef]
- Seiquer, I.; Díaz-Alguacil, J.; Delgado-Andrade, C.; López-Frías, M.; Muñoz, H.A.; Galdó, G.; María, P.N. Diets rich in Maillard reaction products affect protein digestibility in adolescent males aged 11-14 y. Am. J. Clin. Nutr. 2006, 83, 1082–1088. [Google Scholar] [CrossRef] [PubMed]
- Troise, A.D.; Fiore, A.; Wiltafsky, M.; Fogliano, V. Quantification of N-epsilon-2-Furoylmethyl-l-lysine furosine, Nepsilon-Carboxymethyl-l-lysine CML, N-epsilon-Carboxyethyl-l-lysine CEL and total lysine through stable isotope dilution assay and tandem mass spectrometry. Food Chem. 2015, 188, 357–364. [Google Scholar] [CrossRef]
- Saeed, Y.; Wang, J.Q.; Zheng, N. Furosine induces DNA damage and cell death in selected human cell lines: A strong toxicant to kidney Hek-293 cells. Food Sci. Biotechnol. 2017, 26, 1093–1101. [Google Scholar] [CrossRef]
- Li, H.Y.; Xing, L.; Wang, J.Q.; Zheng, N. Toxicology studies of furosine in vitro/in vivo and exploration of the related mechanism. Toxicol. Lett. 2018, 291, 101–111. [Google Scholar] [CrossRef]
- Li, X.R.; Luo, Y.J.; Wang, L.J.; Li, Y.H.; Shi, Y.B.; Yin, C.; Xue, M. Acute and subacute toxicity of ethanol extracts from Salvia przewalskii Maxim in rodents. J. Ethnopharmacol. 2010, 131, 110–115. [Google Scholar] [CrossRef]
- Wasan, K.M.; Najafi, S.; Wong, J.; Kwong, M.; Pritchard, P.H. Assessing plasma lipid levels, body weight, and hepatic and renal toxicity following chronic oral administration of a water soluble phytostanol compound, FM-VP4, to gerbils. J. Pharm. Pharm. Sci. 2001, 4, 228–234. [Google Scholar]
- Burgart, L.J. Cholangitis in viral disease. Mayo Clin. Proc. 1998, 73, 479–482. [Google Scholar] [CrossRef]
- Tarazov, P.G. High serum level of alkaline phosphatase is not a contraindication for embolization of the hepatic artery in liver cancer. Am. J. Roentgenol. 1991, 157, 887. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Liu, S.; Yi, Y.; Ni, X.; Wang, J.; Huang, J.; Fu, Y.; Cao, Y.; Zhou, J.; Fan, J.; et al. Serum gamma-glutamyl transferase levels affect the prognosis of patients with intrahepatic cholangiocarcinoma who receive postoperative adjuvant transcatheter arterial chemoembolization: A propensity score matching study. Int. J. Surg. 2017, 37, 24–28. [Google Scholar] [CrossRef]
- Everhart, J.E.; Wright, E.C. Association of γ-glutamyl transferase GGT activity with treatment and clinical outcomes in chronic hepatitis C HCV. Hepatology 2013, 57, 1725–1733. [Google Scholar] [CrossRef]
- Mathurin, P.; Hollebecque, A.; Arnalsteen, L.; Buob, D.; Leteurtre, E.; Caiazzo, R.; Pigeyre, M.; Verkindt, H.; Dharancy, S.; Louvet, A.; et al. Prospective study of the long-term effects of bariatric surgery on liver injury in patients without advanced disease. Gastroenterology 2009, 137, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Blomme, B.; Van, S.C.; Vanhuysse, J.; Colle, I.; Callewaert, N.; Van, V.H. Impact of elevation of total bilirubin level and etiology of the liver disease on serum N-glycosylation patterns in mice and humans. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, 615–624. [Google Scholar] [CrossRef][Green Version]
- Moon, J.S. Role of Bilirubin in Diabetic Vascular Complications: Can Bilirubin Predict More than Just Liver Disease? Diabetes Metab. 2015, 39, 384–386. [Google Scholar] [CrossRef]
- Arnoldi, A.; Resta, D.; Brambilla, F.; Boschin, G.; D’Agostina, A.; Sirtori, E.; O’Kane, F. Parameters for the evaluation of the thermal damage and nutraceutical potential of lupin-based ingredients and food products. Mol. Nutr. Food Res. 2007, 51, 431–436. [Google Scholar] [CrossRef]
- Kotoh, K.; Enjoji, M.; Kato, M.; Kohjima, M.; Nakamuta, M.; Takayanagi, R. A new parameter using serum lactate dehydrogenase and alanine aminotransferase level is useful for predicting the prognosis of patients at an early stage of acute liver injury: A retrospective study. Comp. Hepatol. 2008, 7, 1–8. [Google Scholar] [CrossRef]
- Vlassara, H. Advanced Glycation in Health and Disease: Role of the Modern Environment. Ann. N. Y. Acad. Sci. 2005, 1043, 452–460. [Google Scholar] [CrossRef]
- George, J.; Chandrakasan, G. Lactate Dehydrogenase Isoenzymes in Dimethyl nitrosamine-Induced Hepatic Fibrosis in Rats. J. Clin. Biochemnutr. 2010, 22, 51–62. [Google Scholar]
- Li, M.; Zeng, M.; He, Z.; Zheng, Z.; Qin, F.; Tao, G.; Zhang, S.; Chen, J. Increased accumulation of protein-Bound Nε-Carboxymethyl lysine in tissues of healthy rats after chronic oral Nε-Carboxymethyl lysine. J. Agric. Food. Chem. 2015, 63, 1658–1663. [Google Scholar] [CrossRef]
- Shyur, L.F.; Yang, N.S. Metabolomics for phytomedicine research and drug development. Curr. Opin. Chem. Biol. 2008, 12, 66–71. [Google Scholar] [CrossRef]
- Lao, Y.M.; Jiang, J.G.; Yan, L. Application of metabonomic analytical techniques in the modernization and toxicology research of traditional Chinese medicine. Br. J. Pharmacol. 2009, 157, 1128–1141. [Google Scholar] [CrossRef]
- Mercier, K.A.; Aljazrawe, M.; Poon, R.; Acuff, Z.; Alman, B. A Metabolomics Pilot Study on Desmoid Tumors and Novel Drug Candidates. Sci. Rep. 2018, 8, 584. [Google Scholar] [CrossRef]
- Gookin, J.L.; Mathews, K.G.; Cullen, J.; Seiler, G. Qualitative metabolomics profiling of serum and bile from dogs with gallbladder mucocele formation. PLoS ONE 2018, 13, e0191076. [Google Scholar] [CrossRef]
- Scholz, H.; Eder, C. Lysophosphatidylcholine activates caspase-1 in microglia via a novel pathway involving two inflammasomes. J. Neuroimmunol. 2017, 310, 107–110. [Google Scholar] [CrossRef]
- Yoder, M.; Zhuge, Y.; Yuan, Y.; Holian, O.; Kuo, S.; Van, B.R.; Thomas, L.L.; Lum, H. Bioactive lysophosphatidylcholine 16:0 and 18:0 are elevated in lungs of asthmatic subjects. Allergy Asthma Immun. 2014, 6, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Newton, K.; Lamkanfi, M.; Mariathasan, S.; Dixit, V.M.; Monack, D.M. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 2010, 207, 1745–1755. [Google Scholar] [CrossRef]
- Drzazga, A.; Sowińska, A.; Koziołkiewicz, M. Lysophosphatidylcholine and lysophosphatidylinosiol--novel promissing signaling molecules and their possible therapeutic activity. Acta Pol. Pharm. 2014, 71, 887–899. [Google Scholar]
- Cui, Y.; Liu, X.; Wang, M.; Liu, L.; Sun, X.; Ma, L.; Xie, W.; Wang, C.; Tang, S.; Wang, D.; et al. Lysophosphatidylcholine and Amide as Metabolites for Detecting Alzheimer Disease Using Ultrahigh-Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry–Based Metabonomics. J. Neuropath. Exp. Neurol. 2014, 73, 954–963. [Google Scholar] [CrossRef]
- Liao, W.T.; Liu, B.; Chen, J.; Cui, J.H.; Gao, Y.X.; Liu, F.Y.; Xu, G.; Sun, B.D.; Zhang, E.L.; Yuan, Z.B.; et al. Metabolite Modulation in Human Plasma in the Early Phase of Acclimatization to Hypobaric Hypoxia. Sci. Rep. 2016, 6, 22589. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, Y.; Lian, M.; Fan, Z.; Tian, Y.; Wang, Y.; Kang, H.; Liu, S.; Liu, S.; Li, T.; et al. Metabolic profiling of hepatitis B virus-related hepatocellular carcinoma with diverse differentiation grades. Oncol. Lett. 2017, 13, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Eder, C. Mechanisms of interleukin-1β release. Immunobiology 2009, 214, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Farooqui, A.A.; Horrocks, L.A. Phospholipase A2-generated lipid mediators in the brain: The good, the bad, and the ugly. Neurosci. A Rev. J. Bring. Neurobiol. Neurol. Psychiatry 2006, 12, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Freeman, L.; Guo, H.; David, C.N.; Brickey, W.J.; Jha, S.; Ting, J.P. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J. Exp. Med. 2017, 214, 1351–1370. [Google Scholar] [CrossRef] [PubMed]
- Garlanda, C.; Dinarello, C.; Mantovani, A. The Interleukin-1 Family: Back to the Future. Immunity 2013, 39, 1003–1018. [Google Scholar] [CrossRef]
- Jamilloux, Y.; Pierini, R.; Querenet, M.; Juruj, C.; Fauchais, A.L.; Jauberteau, M.O.; Jarraud, S.; Lina, G.; Etienne, J.; Roy, C.R.; et al. Inflammasome activation restricts Legionella pneumophila replication in primary microglial cells through flagellin detection. Glia 2013, 61, 539–549. [Google Scholar] [CrossRef]
- Inose, Y.; Kato, Y.; Kitagawa, K.; Uchiyama, S.; Shibata, N. Activated microglia in ischemic stroke penumbra upregulate MCP-1 and CCR2 expression in response to lysophosphatidylcholine derived from adjacent neurons and astrocytes. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2015, 35, 209–223. [Google Scholar] [CrossRef]
- Lammerding, L.; Slowik, A.; Johann, S.; Beyer, C.; Zendedel, A. Post-Stroke Inflammasome Expression and Regulation in the Peri-Infarct Area by Gonadal Steroids after Transient Focal Ischemia in the Rat Brain. J. Neuroendocrinol. 2015, 103, 460–475. [Google Scholar] [CrossRef]
- Man, S.M.; Hopkins, L.J.; Nugent, E.; Cox, S.; Glück, I.M.; Tourlomousis, P.; Wright, J.A.; Cicuta, P.; Monie, T.P.; Bryant, C.E. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA 2014, 111, 7403–7408. [Google Scholar] [CrossRef] [PubMed]
- Martens, L.K.; Kirschner, K.M.; Warnecke, C.; Scholz, H. Hypoxia-inducible factor-1 HIF-1 is a transcriptional activator of the TrkB neurotrophin receptor gene. J. Biol. Chem. 2007, 282, 14379–14388. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Li, G.R. TRPC1/TRPC3 channels mediate lysophosphatidylcholine-induced apoptosis in cultured human coronary artery smooth muscles cells. Oncotarget 2016, 7, 50937–50951. [Google Scholar] [CrossRef]
- Burke, J.E.; Dennis, E.A. Phospholipase A Biochemistry. Cardiovasc. Drug Ther. 2009, 23, 49–59. [Google Scholar]
- Atsumi, G.; Murakami, M.K.; Hadano, A.; Tajima, M.; Kudo, I. Distinct roles of two intracellular phospholipase A2s in fatty acid release in the cell death pathway. Proteolytic fragment of type IVA cytosolic phospholipase A2alpha inhibits stimulus-induced arachidonate release, whereas that of type VI Ca2+-indep. J. Biol. Chem. 2000, 275, 18248–18258. [Google Scholar] [CrossRef]
- Clark, J.D.; Lin, L.L.; Kriz, R.W.; Ramesha, C.S.; Sultzman, L.A.; Lin, A.Y.; Milona, N.; Knopf, J.L. A novel arachidonic acid-selective cytosolic PLA2 contains a Ca2+-dependent translocation domain with homology to PKC and GAP. Cell 1991, 65, 1043–1051. [Google Scholar] [CrossRef]
- Hattori, K.; Adachi, H.; Matsuzawa, A.; Yamamoto, K.; Tsujimoto, M.; Aoki, J.; Hattori, M.; Arai, H.; Inoue, K. cDNA cloning and expression of intracellular platelet-activating factor PAF acetylhydrolase II. Its homology with plasma PAF acetylhydrolase. J. Biol. Chem. 1996, 271, 33032–33038. [Google Scholar] [CrossRef]
- Laufs, U.; Marra, D.; Node, K.; Liao, J.K. 3-Hydroxy-3-methylglutaryl-CoA Reductase Inhibitors Attenuate Vascular Smooth Muscle Proliferation by Preventing Rho GTPase-induced Down-regulation of p27 Kip. J. Biol. Chem. 1999, 274, 21926–21931. [Google Scholar] [CrossRef]
- Larsson, P.K.; Claesson, H.E.; Kennedy, B.P. Multiple splice variants of the human calcium-independent phospholipase A2 and their effect on enzyme activity. J. Biol. Chem. 1998, 273, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.M.; Michikawa, M.; Kim, S.U.; Nagai, A. Lysophosphatidylcholine increases the neurotoxicity of Alzheimer’s amyloid β1-42 peptide: Role of oligomer formation. J. Neurosci. 2015, 292, 159–169. [Google Scholar] [CrossRef]
- Tischfield, J.A. A reassessment of the low molecular weight phospholipase A2 gene family in mammals. J. Biol. Chem. 1997, 272, 17247–17250. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.L.; Babensee, J.E. Procoagulant phenotype of endothelial cells after coculture with biomaterial-treated blood cells. J. Biomed. Mater Res. A 2005, 72, 269–278. [Google Scholar] [CrossRef]
- Valentin, E.; Ghomashchi, F.; Gelb, M.H.; Lazdunski, M.; Lambeau, G. On the diversity of secreted phospholipases ACloning, tissue distribution, and functional expression of two novel mouse group II enzymes. J. Biol. Chem. 1999, 274, 31195–31202. [Google Scholar] [CrossRef]
- Tjoelker, L.W.; Wilder, C.; Eberhardt, C.; Stafforini, D.M.; Dietsch, G.; Schimpf, B.; Hooper, S.; Le, T.H.; Cousens, L.S.; Zimmerman, G.A.; et al. Anti-inflammatory properties of a platelet-activating factor acetylhydrolase. Nature 1995, 374, 549–553. [Google Scholar] [CrossRef]
- Sundaram, J.R.; Chan, E.S.; Poore, C.P.; Pareek, T.K.; Cheong, W.F.; Shui, G.; Tang, N.; Low, C.M.; Wenk, M.R.; Kesavapany, S. Cdk5/p25-induced cytosolic PLA2-mediated lysophosphatidylcholine production regulates neuroinflammation and triggers neurodegeneration. J. Neurosci. 2012, 32, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Muthalif, M.M.; Ljuca, F.; Roaten, J.B.; Pentapaty, N.; Uddin, M.R.; Malik, K.U. Ca2+/calmodulin-dependent protein kinase II and cytosolic phospholipase A2 contribute to mitogenic signaling in myeloblastic leukemia U-937 cells. J. Pharmacol. Exp. Ther. 2001, 298, 272–278. [Google Scholar] [PubMed]
- Christofferson, D.E.; Yuan, J. Necroptosis as an alternative form of programmed cell death. Curr. Opin. Cell Biol. 2010, 22, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, P.; Galluzzi, L.; Vanden, B.T.; Kroemer, G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010, 11, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Challa, S.; Moquin, D.; Genga, R.; Ray, T.D.; Guildford, M.; Chan, F.K. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009, 137, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Degterev, A.; Hitomi, J.; Germscheid, M.; Chen, I.L.; Korkina, O.; Teng, X.; Abbott, D.; Cuny, G.D.; Yuan, C.; Wagner, G.; et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008, 4, 313–321. [Google Scholar] [CrossRef]
- He, S.; Wang, L.; Miao, L.; Wang, T.; Du, F.; Zhao, L.; Wang, X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009, 137, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Shao, J.; Lin, J.; Zhang, N.; Lu, B.J.; Lin, S.C.; Dong, M.Q.; Han, J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009, 325, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Shang, L.; Li, N.; Wang, S.; Wang, M.; Huang, Y.; Chen, D.; Huang, J.; Xiong, K. Mixed lineage kinase domain-like protein induces RGC-5 necroptosis following elevated hydrostatic pressure. Acta Biochim. Biophys. Sin. 2017, 49, 879–889. [Google Scholar] [CrossRef]
- Cai, Z.; Jitkaew, S.; Zhao, J.; Chiang, H.C.; Choksi, S.; Liu, J.; Ward, Y.; Wu, L.G.; Liu, Z.G. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 2014, 16, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, W.; Ren, J.; Huang, D.; He, W.T.; Song, Y.; Yang, C.; Li, W.; Zheng, X.R.; Chen, P.; et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014, 24, 105–121. [Google Scholar] [CrossRef]
- Hildebrand, J.M.; Tanzer, M.C.; Lucet, I.S.; Young, S.N.; Spall, S.K.; Sharma, P.; Pierotti, C.; Garnier, J.M.; Dobson, R.C.; Webb, A.I.; et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. USA 2014, 111, 15072–15077. [Google Scholar] [CrossRef]
- Mulay, S.R.; Desai, J.; Kumar, S.V.; Eberhard, J.N.; Thomasova, D.; Romoli, S.; Grigorescu, M.; Kulkarni, O.P.; Popper, B.; Vielhauer, V.; et al. Cytotoxicity of crystals involves RIPK3-MLKL-mediated necroptosis. Nat. Commun. 2016, 7, 10274. [Google Scholar] [CrossRef]
- Rodriguez, D.A.; Weinlich, R.; Brown, S.; Guy, C.; Fitzgerald, P.; Dillon, C.P.; Oberst, A.; Quarato, G.; Low, J.; Cripps, J.G.; et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2015, 23, 76–88. [Google Scholar] [CrossRef]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef]
- Wang, H.; Sun, L.; Su, L.; Rizo, J.; Liu, L.; Wang, L.F.; Wang, F.S.; Wang, X. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP. Mol. Cell 2014, 54, 133–146. [Google Scholar] [CrossRef]
- Pierdomenico, M.; Negroni, A.; Stronati, L.; Vitali, R.; Prete, E.; Bertin, J.; Gough, P.J.; Aloi, M.; Cucchiara, S. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am. J. Gastroenterol. 2014, 1092, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Bao, Y.H.; Wang, Y.; Jiang, J.Y. The role of necroptosis in neurosurgical diseases. Braz. J. Med. Biol. Res. 2015, 484, 292–298. [Google Scholar] [CrossRef]
- Weinlich, R.; Oberst, A.; Beere, H.M.; Green, D.R. Necroptosis in development, inflammation and disease. Nat. Rev. Mol. Cell Biol. 2016, 182, 127–136. [Google Scholar] [CrossRef]
- Günther, C.; Martini, E.; Wittkopf, N.; Amann, K.; Weigmann, B.; Neumann, H.; Waldner, M.J.; Hedrick, S.M.; Tenzer, S.; Neurath, M.F.; et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 2011, 4777, 335–339. [Google Scholar] [CrossRef] [PubMed]
- Seifert, L.; Werba, G.; Tiwari, S.; Ly, N.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.; et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016, 532, 245–249. [Google Scholar] [CrossRef]
- Yang, X.; Chao, X.; Wang, Z.T.; Ding, W.X. The end of RIPK1-RIPK3-MLKL-mediated necroptosis in acetaminophen-induced hepatotoxicity? Hepatology 2016, 64, 311–312. [Google Scholar] [CrossRef]
- Ito, Y.; Ofengeim, D.; Najafov, A.; Das, S.; Saberi, S.; Li, Y.; Hitomi, J.; Zhu, H.; Chen, H.; Mayo, L.; et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 2016, 353, 603–608. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Primer Sequences (5′→3′) | |
|---|---|---|
| Forward Primer | Reverse Primer | |
| PLA2-3 | CCGCAGAGAAGAAGGAGCAGTT | CACGCACAGGAGGCAGAACA |
| RIPK-1 | TGAAGCCCACAGCGATTCTT | GCCATCTTTCTCCCCCAAGA |
| RIPK-3 | ACATGCATGGTCATGCACACACAT | TTGAGACATCTCTTTTTGGAG |
| MLKL | CCCGAGTTGTTGCAGGAGAT | TCTCCAAGATTCCATCCGCAG |
| TNF-α | GTCCCCAAAGGGATGAGAAGTT | GTTTGCTACGACGTGGGCTACA |
| IL-1β | TGTGAAATGCCACCTTTTGA | GCTCAAAGGTTTGGAAGCAG |
| GAPDH | CAATGAATAGGGCTACAGCA | AGGGAGATGCTCAGTGTTGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Wang, Y.; Yang, H.; Zhang, Y.; Xing, L.; Wang, J.; Zheng, N. Furosine, a Maillard Reaction Product, Triggers Necroptosis in Hepatocytes by Regulating the RIPK1/RIPK3/MLKL Pathway. Int. J. Mol. Sci. 2019, 20, 2388. https://doi.org/10.3390/ijms20102388
Li H, Wang Y, Yang H, Zhang Y, Xing L, Wang J, Zheng N. Furosine, a Maillard Reaction Product, Triggers Necroptosis in Hepatocytes by Regulating the RIPK1/RIPK3/MLKL Pathway. International Journal of Molecular Sciences. 2019; 20(10):2388. https://doi.org/10.3390/ijms20102388
Chicago/Turabian StyleLi, Huiying, Yizhen Wang, Huaigu Yang, Yangdong Zhang, Lei Xing, Jiaqi Wang, and Nan Zheng. 2019. "Furosine, a Maillard Reaction Product, Triggers Necroptosis in Hepatocytes by Regulating the RIPK1/RIPK3/MLKL Pathway" International Journal of Molecular Sciences 20, no. 10: 2388. https://doi.org/10.3390/ijms20102388
APA StyleLi, H., Wang, Y., Yang, H., Zhang, Y., Xing, L., Wang, J., & Zheng, N. (2019). Furosine, a Maillard Reaction Product, Triggers Necroptosis in Hepatocytes by Regulating the RIPK1/RIPK3/MLKL Pathway. International Journal of Molecular Sciences, 20(10), 2388. https://doi.org/10.3390/ijms20102388







