Abstract
The zebrafish (Danio rerio) is a small vertebrate ideally suited to the modeling of human diseases. Large numbers of genetic alterations have now been modeled and could be used to study organ development by means of a genetic approach. To date, limited attention has been paid to the possible use of the zebrafish toolbox in studying human mitochondrial disorders affecting the nervous system. Here, we review the pertinent scientific literature discussing the use of zebrafish in modeling gene mutations involved in mitochondria-related neurological human diseases. A critical analysis of the literature suggests that the zebrafish not only lends itself to exploration of the pathological consequences of mitochondrial energy output on the nervous system but could also serve as an attractive platform for future drugs in an as yet untreatable category of human disorders.
1. Introduction
The term mitochondrial disease is classically used in reference to defects of the oxidative phosphorylation (OxPhos) system and defects of energy metabolism. Instead, the term mitochondrial medicine serves to group together, in a single category, the ample array of clinical presentations associated with these defects, which range from single organ failure to complex phenotypes, including neurodegenerative diseases and cancers [1]. The human mitochondrial proteome is believed to be made up of over 1500 proteins (Human MitoCarta (www.broadinstitute.org)) [2]. This reflects the intrinsic complexity of the organelle and makes it conceivable that dysfunctions of mitochondria give rise to highly heterogeneous clinical presentations. Patients may be affected at any age and with multisystem involvement, that often affects organs with high energy demands. However, how defects in mitochondria can cause such widespread range of human disorders remains to be unveiled. Mitochondria are dynamic and mobile organelles that play an essential role in several cellular processes, such as the generation and regulation of cellular energy, initiation of apoptosis, iron-sulfur cluster biogenesis, and calcium buffering [3,4]. The OxPhos is the process by which mitochondria produce the adenosine triphosphate (ATP) molecules needed by the cells. This system consists of five multimeric enzyme complexes (EC): Complex I (EC 1.6.5.3) or NADH:ubiquinone reductase, CI; Complex II (EC 1.3.5.1) or succinate dehydrogenase, CII; Complex III (EC 1.10.2.2) or quinol-cytochrome c (cyt c) reductase, CIII; Complex IV (EC 1.9.31) or cyt c oxidase (COX), CIV; Complex V (EC 3.6.14) or FoF1-ATPase, CV; and two electron transport carriers, namely, ubiquinone (coenzyme Q, CoQ) and cyt c [5]. The OxPhos is under a dual genetic control: thirteen of the key structural polypeptides that constitute the multimeric subunits of the respiratory chain complexes and ATP synthase are encoded by the mitochondrial DNA (mtDNA), in addition to two ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs), which are required to perform autochthonous protein synthesis [6]. Conversely, approximately eighty of the remaining proteins making up the OxPhos complexes are encoded by nuclear DNA (nDNA) genes. Although it encodes the basic machinery for protein synthesis, as well as replication, repair, and transcription, human mtDNA remains entirely dependent upon the nucleus for the provision of enzymes and accessory components. Hence, a genetic classification of OxPhos-related mitochondrial diseases distinguishes two broad categories: disorders due to mutations in the mtDNA, and obeying to the rules of mitochondrial genetics, and disorders due to mutations in the nDNA, which are governed by canonic Mendelian genetics [1]. Correct communication between the two genomes is crucial for mtDNA integrity, copy number regulation and mitochondrial protein production. Thus, not only nucleotide variants in the mtDNA itself, but also mutations in nuclear genes involved in mtDNA replication and maintenance might disrupt the integrity of the mitochondrial genome and influence its functional properties [7]. The pathological consequences impact mostly on tissues highly dependent upon ATP production such as heart, muscle, and particularly the central (CNS) and the peripheral nervous systems (PNS) [8].
Producing up to 90% of ATP molecules generated in the brain, the OxPhos is responsible for powering cell signaling and neuronal activity processes. It is not too surprising that mitochondria also play a key role in aging-related neurodegenerative disorders such as Parkinson’s disease (PD), Alzheimer’s disease, Huntington’s disease, and in amyotrophic lateral sclerosis [9,10]. Among the identified genes found to be associated with inherited forms of neurodegenerative disorders, at least half directly or secondarily affect one or several mitochondrial functions [11]. In cells, mitochondria actively fuse and divide (fission), branch, and change their size in a dynamic manner. This process, named mitochondrial network dynamics, seems indispensable to guarantee an appropriate population of healthy and functional mitochondria at every time [12]. Mitochondria network dynamics work in tight collaboration with the mitophagy pathway to ensure the quality control of these organelles [13]. Mitophagy is a selective form of macro-autophagy that degrades damaged or unnecessary mitochondria. Defects in this process have been reported as crucial at the onset and progression of neurodegenerative diseases where autophagic failure is one of the main factors contributing to neuronal cell death [14]. Neurons are particularly vulnerable to autophagic impairment as well as to mitochondrial dysfunction because of their high energy demands and to their post-mitotic non-proliferating nature [15]. Altogether, these considerations lead to the idea that mitochondria, as well as being a focus for a considerable amount of research, provide important clues to be potential target for therapies in a broad range of neurodegenerative diseases.
2. Zebrafish Resources to Mimic Human Diseases
The discovery of a large number of mitochondrial genes causing human diseases prompted the generation of a number of animal models, including simple invertebrate (Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster) and more sophisticated mammalian models (such as Mus musculus), which replicate various aspects of neurological disorders caused by mitochondrial damage, and thus are considered useful to understand the pathophysiological mechanisms and to evaluate putative therapeutic candidates [16,17]. Nevertheless, there remains a need to achieve more efficient high-throughput screenings of vertebrate models in order to fully address the questions of mitochondrial physiology and function to study the effects of mitochondrial damage in individual organs and in the animal behaviors. Recently, the zebrafish has emerged as an excellent organism for studying a broad range of human genetic diseases, including those affecting the different functional structures of the CNS [18,19]. In this view, the zebrafish is attractive for several reasons including its particular biology and genetics. The zebrafish breed with high rates and big numbers of fertilized eggs, which are transparent, tiny, and mobile during the key developmental stages, can be easily obtained daily. The healthy zebrafish embryos develop rapidly and in synchrony, within one day of development, many key features of the CNS are detectable and can be studied up to the larval stage and beyond. Another major advantage of using zebrafish is the simplicity and effectiveness with which genes can be manipulated to allow biological observations in the cells of the developing embryos. Other advantages are the availability of zebrafish genome data [20] and the accessibility of over 80% of its gene structures in several publicly available databases (Ensembl (www.ensembl.org), Entrez (www.ncbi.nlm.nih.gov/gquery), UCSC Genome Browser (https://genome.ucsc.edu/), and ZFIN, the Zebrafish Model Organism Database (zfin.org)). Zebrafish genes show a high degree of synteny across vertebrate species as well as 50–80% homology with most human sequences. Comparison with the human reference genome sequences shows that approximately 70% of human genes have at least one obvious zebrafish ortholog [21]. In addition, molecular pathways are phylogenetically well conserved, and the basic physiology is similar to that of mammals [22]. Orthologs for most human genes can be easily identified by bioinformatics tools and manipulated to mimic human pathologies by gain- and loss-of-function approaches. However, it has to be noticed that zebrafish gene identification can be complicated by the fact that several genes are duplicated as a result of a whole genome duplication event that occurred during teleost fish evolution; this implies that in zebrafish specific functions could be partitioned between the two duplicated genes, or simply lost or disrupted for one of the genes, or even complemented if one of the two copies is disrupted [23]; nevertheless, in some cases duplication events enhance the possibility to obtain information on gain/loss of some gene functions.
A standard technique for studying embryo development is to knockdown a specific gene function by injecting gene-specific phosphorodiamidate morpholino oligonucleotides (MOs) in early embryonic stages [24]. MOs are chemically modified antisense oligonucleotides that can specifically bind messenger RNAs (mRNAs) and transiently block translation (when they are targeted to sequences near the initiation codon) or splicing (when they are targeted to exon-intron boundaries), thus providing gene knockdown in the zebrafish embryo. However, major limitations of this technique are the transient effects of MOs and the multiple off-target effects which fail to impact on the juvenile or adult phenotype [25]. More recently, the successful application of new technologies for genome editing, such as zinc finger nucleases [26,27], transcription activator-like effectors (TALENs) [28], and technologies based on clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (CRISPR-Cas9) systems [29], has allowed more profitable targeted mutagenesis and functional gene ablation in zebrafish. Another approach is the undirected mutation-driven method using irradiation, retroviruses or the most commonly used chemical mutagens such as the DNA alkylating agent N-ethyl-N-nitrosourea (ENU) [30]. In the last years, the reverse genetics approaches, where DNA mutations can be studied without an observable phenotype, have been further improved by recent advances in genomic technologies and next-generation sequencing (NGS) [30,31]. The results of this progress have led to an increase in the number of novel zebrafish mutated lines.
3. Zebrafish Resources to Explore Mitochondrial Physiology
In the last decade, zebrafish modeling has been used to study various aspects of mitochondrial physiology, e.g., the dynamics of the cellular mitochondrial network, the mitochondrial life cycle, and mtDNA metabolism during development. The technologies applied to study these aspects in the zebrafish model have rapidly increased as shown in Table 1 and briefly described below.

Table 1.
Tools used for the investigation of mitochondrial physiology.
Through a balance between fusion and fission, mitochondria maintain a web-shaped network in cells [32,33]. The study of mitochondrial morphology in the zebrafish was initially conducted using molecular probes [34] or immunofluorescent antibodies [35]. In order to study changes in mitochondrial morphology at the organism level in normal or disease conditions in real time, transgenic zebrafish model expressing mitochondrially-targeted fluorescent proteins (green/red) were created [36,37]. Several neurodegenerative diseases have been linked with deficits in mitochondrial axonal transport [38]. To study the “life cycle” of mitochondria and their dynamics in neurons, transgenic lines were generated using Gal4/UAS genetics that allows the expression of transgene labeled mitochondria [39] also in specific neural cells. By using this tool, studies on mitochondrial transport related to neurological disease were conducted in Rohon-Beard sensory neurons [40,41], retinal ganglion cells [42], motor neurons [43], and dopaminergic neurons [44] of zebrafish, developing different transgenic lines with fluorescently labeled mitochondria.
Recently, Mandal and colleagues (2018) have optimized a direct visualization of mitochondria and the analysis of their lifetime, health, and function in axons of the posterior lateral line (pLL) in zebrafish, using a panel of technologies that allows investigating in vivo several mitochondrial aspects in the zebrafish model (Table 1) [45].
Song and colleagues (2009) have investigated on mitochondrial reactive oxygen species (ROS) formation using the oxidative fluorescent dye dihydrorodamine-123 (DHR-123) [46], while the examination of mitochondrially derived ROS was conducted in zebrafish using a live cell redox sensor that identifies mitochondria-generated superoxides (MitoSOXTM) [47]. Biothilos, such as cysteine (Cys) and homocysteine (Hcy) have an important role in ROS homeostasis in mitochondria and are very sensitive to the oxidative stress [48]. The real-time monitoring of Cys and Hcy levels in zebrafish mitochondria allows monitoring their oxidative stress level. For this purpose, Yue et al. (2018) developed a ratiometric two-photon fluorescent probe (Mito-MQ) for measuring Cys/Hcy level in mitochondria and applied this technology in vivo, into 5-day-old zebrafish larvae [48]. All these studies provide a starting point for performing further mitochondrial studies under physiological or disease conditions in this relatively simple organism [49]. Micro-oxygraphy has been used in the whole organism, especially at the early stages of embryo development, to study bioenergetic metabolism and mitochondrial physiology related to neurodegenerative conditions [50]. Embryos have also been shown to lend themselves to the evaluation of the total number of mtDNA genomes at different developmental stages and to the assessment of the spatial expression of genes regulating mtDNA biogenesis and OxPhos complexes [51].
Due to the fact that many mitochondrial defects affect the nervous system, and many structure and function of the zebrafish CNS are very similar to the human ones [52], we recapitulated the scientific literature concerning the use of zebrafish for modelling neurological disorders caused by mitochondrial alterations. A critical review of what has been done to date might be of use for future investigations.
4. Modeling Mitochondrial Defects in Zebrafish
Mutations in mtDNA or nDNA genes that encode mitochondrial proteins cause a heterogeneous group of disorders primarily due to dysfunctions of the OxPhos and electron transport chain. However, other mutations that involve mitochondrial physiology, such as ion channels, quality control system, mitochondrial carriers, or cellular perturbations that affects mitochondria, such as mitochondrial transport cause an energy deficiency that affects high demand tissues including nervous system.
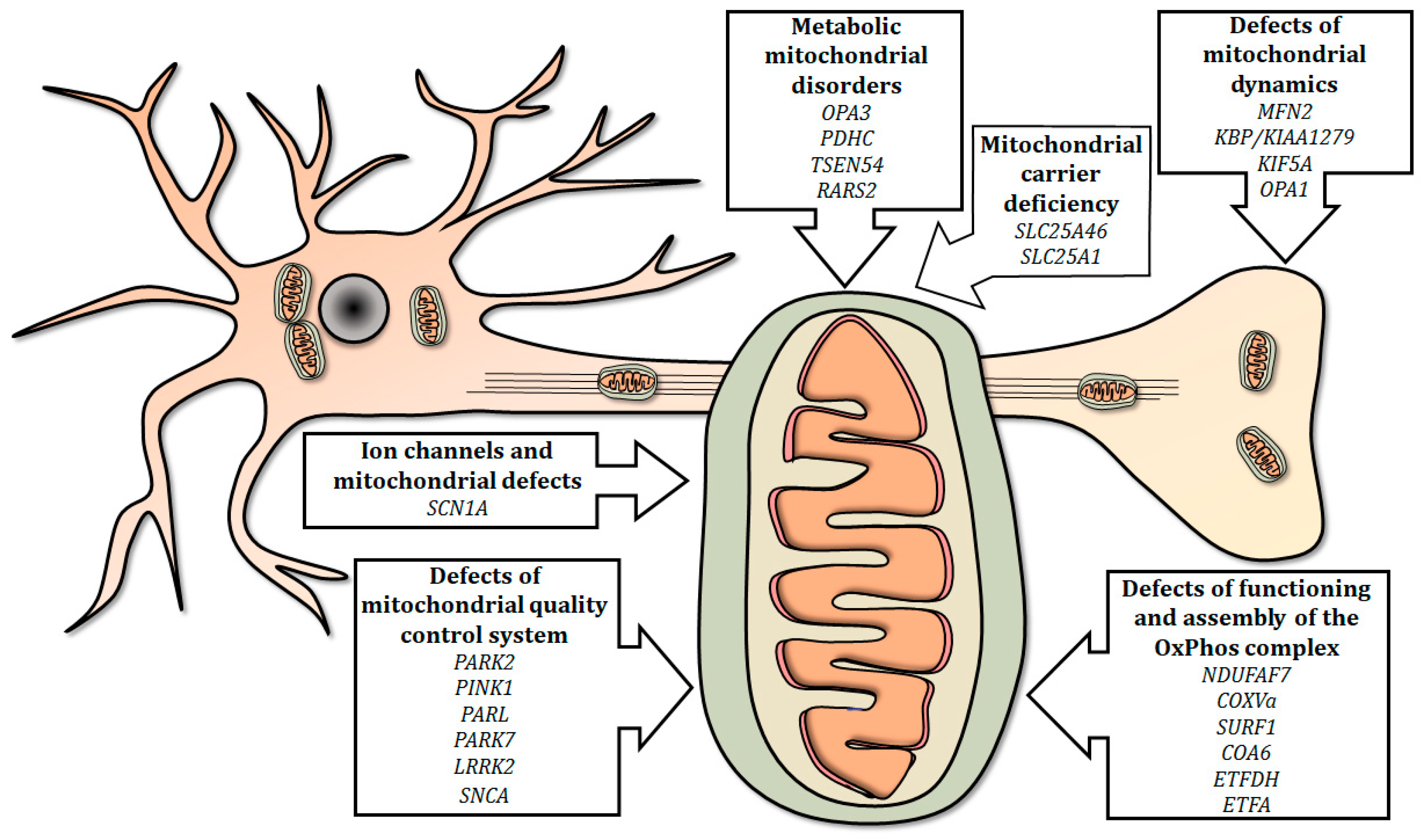
In this review, we summarized the current use of the zebrafish as a relatively simple and efficient tool for studying the effects of gene mutations involved in mitochondria-related human neurological diseases (Figure 1).
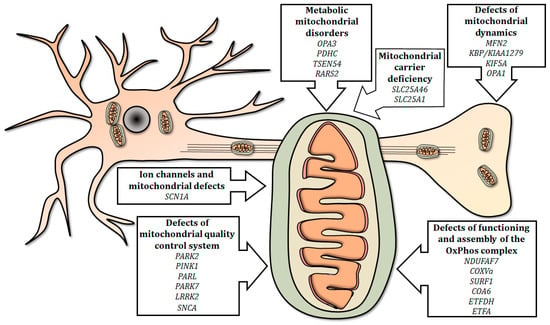
Figure 1.
Summary of mitochondria-related genes involved in neurological human disorders, modeled in mutant zebrafish. Mutations in genes involved in mitochondrial electron transport chain, physiology, quality control, dynamics and metabolism, as well as mutations in selected genes encoding carriers and ion channels, can disrupt the integrity of the mitochondrion and/or influence its functions, thus determining the onset of neurological disorders.
Mutated genes modeled in zebrafish are presented in the following sections, covering six main topics; the list of genes investigated is summarized in Table 2.

Table 2.
Human genes related to mitochondrial disorders affecting the nervous system modeled in zebrafish.
4.1. Defects of Functioning and Assembly of the OxPhos Complex
The term “primary mitochondrial diseases” refers to a wide range of complex syndromes collectively characterized by impaired OxPhos function with clinical expression especially in the brain, muscle, and eyes [78,79]. The deficiency of OxPhos complex assembly factors causes several severe neurological disorders in human such as cardiomyoencephalopathy, leukoencephalopathy, psychomotor delay, seizures, and Leigh syndrome [5]. Currently, very few studies have been conducted on specific gene mutations to investigate these disorders using zebrafish models. Complex I is the largest enzyme of the mitochondrial OxPhos system and its deficiency caused by pathogenic mutations in genes encoding the structural subunits have been identified and associated with mitochondrial disorders [80].
A mutation on the methyltransferase NADH:Ubiquinone Oxidoreductase Complex Assembly Factor 7 (NDUFAF7), which is an assembly factor of complex I, has been found in association with the reduction of intracellular ROS and ATP levels and with the reduction of Complex I activity, and it has also been correlated with pathologic myopia [81]. Zurita Rendon and colleagues investigated the function of NDUFAF7 in models in vivo: Ndufaf7 knockout mice and MO knockdown zebrafish. Delayed hatching times and morphological abnormality resulted by the disruption of the NDUFAF7 paralogue gene, and the steady-state levels of complex I was specifically affected by the MO knockdown in zebrafish [54]. Furthermore, mutations of COX complex have been described in a number of human mitochondrial diseases with peripheral neuropathies. Among the mitochondrial diseases, COX deficiency can present with a number of different infantile clinical phenotypes including classical Leigh syndrome, fatal infantile COX deficiency, and hypertrophic cardiomyopathy and myopathy [82]. Most COX deficiencies in humans are related to defective function of structural or ancillary proteins making up the holocomplex, including the assembly genes SCO2 and SURF1. Zebrafish morphants of the orthologs of either human COXVa or SURF1 showed a profound histochemical defect of COX activity and impaired holoenzyme assembly [59]. As a consequence, morphants showed a dramatic increase in apoptosis in hindbrain and neural tube and exhibited a severe motility defect. By contrast, the heart of mutant zebrafish lacked apoptotic cells but showed increasingly poor performance over time, a phenotype consistent with tissue energy deficiency [55]. More recently, copper supplementation has been shown to rectify the disassembly pattern of the COX holocomplex in a zebrafish line where the COX assembly factor 6 (COA6) was knocked-down [56].
Multiple acyl-CoA dehydrogenase deficiency (MADD) is an autosomal recessive disorder, which is clinically heterogeneous; patients with this disease display multiple defects including neurological impairment. This condition is due to deficiency of any one of three proteins: the alpha (ETFA) and beta (ETFB) subunits of mitochondrial electron transfer flavoprotein, or the electron transfer flavoprotein dehydrogenase (ETFDH). The clinical pictures due to the different enzyme defects appear to be indistinguishable; each defect can lead to a range of mild or severe cases, depending presumably on the location and nature of the intragenic lesion [83]. Inactivation of the etfdh gene (xavier zebrafish mutant) resulted in severe metabolic abnormalities. In particular, there were biochemical abnormalities consistent with mitochondrial dysfunction, and increased neuronal proliferation caused by the activation of the PPARG-ERK pathway [46]. A new mutant strain termed dark Xavier, because of its enlarged, dark and fatty liver [57], was generated. This mutant strain, carrying mutations in etfa, displayed severe neurologic deficits including encephalopathy that is usually seen in patients with MADD, and recapitulated many key clinical and metabolic features seen in MADD patients such as multi-organ defects of the brain, liver and kidney.
4.2. Ion Channels and Mitochondrial Defects
Dravet syndrome (DS), a severe genetic form of epilepsy, has been associated with mutations in the sodium channel protein type 1 subunit alpha (SCN1A) [58]. Starting from the observation of mitochondrial defects in muscle biopsies in DS patients, OxPhos and mitochondrial glycolysis were studied in the scn1Lab zebrafish mutant, a zebrafish model of DS. Even though no defects of OxPhos complexes I–IV were observed in scn1Lab mutants, the authors noted a decreased expression of glycolysis related genes [58]. A decrease of complex I activity was suspected to be induced by the oxidative stress and post-translational oxidative modification caused by the spontaneous seizures, observed in these mutants [58].
4.3. Defects of Mitochondrial Quality Control System
The underlying causes of several neurological disorders converge on impaired mitochondrial physiology and maintenance. PD is a frequent neurological disorder caused by dopaminergic neuronal death in the substantia nigra, resulting in a reduced level of dopamine in the striatum that is the direct cause of motor dysfunction of PD patients [84]. PD-associated neuronal loss is strictly connected with localized mitochondrial dysfunction and oxidative stress before the clinical onset of motor symptoms [85]. Among gene products associated with hereditary PD we find those located within (PARKIN, PINK1, PARL, and DJ-1) or linked to (LRRK2) mitochondria. PARKIN and PINK1 are neuroprotective proteins which act together in a mitochondrial quality control pathway promoting the clearance of damaged mitochondria via autophagy [86].
PARKIN is an E3 ubiquitin ligase and like the human protein, zebrafish parkin is ubiquitously expressed throughout embryonic development and in adult tissues. PARKIN is involved in oxidative stress and the stable overexpression of parkin is able to protect fish against proteotoxic stress preventing cell death [67]. In zebrafish, loss of parkin elicits an approximately 20% loss of dopaminergic neurons in the ventral diencephalon. Morphants do not show any abnormal mitochondrial morphology, but mitochondrial complex I activity is spectrophotometrically reduced [59].
PINK1 is a ubiquitously expressed protein with an N-terminal mitochondrial-targeting motif and a conserved serine⁄threonine kinase domain, and two of its targets have been identified: TNF receptor-associated protein 1, which protects against oxidative stress [87], and DRP1, which promotes mitochondrial fission [88]. PINK1 seems to act similarly to PARKIN in oxidative stress conditions in particular, protecting neurons against stress-induced mitochondrial damage and apoptosis [89]. Zebrafish pink1 is expressed ubiquitously in the brain, and its abrogation results in a selective decline of some important mRNAs, and only distinct groups of dopaminergic neurons are sensitive to loss of pink1 in zebrafish [61]. The pink1 null mutant zebrafish line, referred to as pink−/−, shows mitochondrial dysfunction and loss of dopaminergic neurons [62]. Differential transcriptomic investigations in pink1 morphants [90] revealed global impairment of TGF-β signaling, retinoic acid receptor (RAR) activation, altered biogenesis of mitochondria, and, among the major hits, dysfunction of the hypoxia-induced signaling pathway when compared with a wild-type strain. Soman and co-workers have highlighted that the pharmacological and/or genetic inhibition of the inner mitochondrial membrane calcium uniporter is able to rescue loss of dopaminergic neurons in pink−/− mutants recovering the PD-phenotype [91]. Recently, this transgenic line (pink−/−) has been also exploited to develop new class of stress-dependent autophagy-stimulating drugs to prevent the loss of dopaminergic neurons in PD-zebrafish model [92].
PARL codes for an integral membrane protease of the inner mitochondrial membrane having a role in the normal trafficking and processing of PINK1 and PARKIN within mitochondria [93]. Moreover, PARL cleaves the optic atrophy 1 protein (OPA1) [94] that is a crucial protein involved in the fusion of inner mitochondrial membranes and in the formation of cristae, crucial for proper mitochondrial metabolism in early development [71]. Bioinformatics analyses revealed that in zebrafish there are two paralogues, parla and parlb, whose transcripts are both ubiquitously expressed during embryogenesis [63]. The PD-phenotype caused by the KD of both parl genes can be rescued by the overexpression of human PARL mRNA. Morphants showed that cell death likely occurred because parl is unable to prevent mitochondrial fission or to produce soluble OPA1, which prevents the widening of the cristae junction and inhibits the release of cyt c from the mitochondria [63].
PARK7 encodes DJ-1 that is localized in the mitochondrial matrix; the zebrafish orthologue of DJ-1 is expressed in adult brain, muscle and gut, and is predicted to have high sequence homology of 83% with human DJ-1 protein [95]. Loss of human DJ-1 leads to an altered mitochondrial morphology contributing to the increased cellular sensitivity to oxidative stress [96]. In zebrafish, knockdown of dj-1 does not cause decrease in number of dopaminergic neurons but leads to high susceptibility to programmed cell death activated by p53 [64]. Moreover, it is well known that the astrocytes, which outnumber neurons in the brain, have a key role in neuronal protection controlling redox homeostasis [84] producing neuron-protective factors [85]. The selective astroglial dj-1 overexpression in transgenic zebrafish line Tg(gfap: egfp-2A-flag-zDJ-1) is able to protect fishes from neurological damage probably through the up-regulation of redox and inflammatory proteins regulated by Nrf2 activation [65].
LRRK2 has mainly cytoplasmic localization but associates also with the outer mitochondrial membrane. LRRK2 interacts with Parkin in cultured mammalian neuronal cells [97], and with PRDX3, an important antioxidant scavenger of hydrogen peroxide within mitochondria [98]. In human neuronal cell lines, the overexpression of LRRK2 causes mitochondrial fragmentation associated with increased levels of DLP1, a mitochondrial fission protein [99]. Knockdown of lrrk2 in morphants zebrafish causes neurodegeneration and locomotion defects that can be rescued both by expressing the wild-type LRRK2 and with the administration of L-dopa, a drug commonly used for treating PD patients [66].
Another zebrafish model for PD has been created by overexpression of human α-synuclein (SNCA) in zebrafish peripheral sensory neurons that leads to a moderate cell death, whereas many axons exhibit diffuse or focal swellings [67]. Under basal conditions SNCA does not interact with mitochondrial membranes; nonetheless, ultrastructural studies reveal that a fraction of synuclein normally associates to mitochondria, and it can produce mitochondrial fragmentation and functionality impairment of the complex I activity [100]. The orthologue of human α-synuclein appears not to be present in the zebrafish genome [101]. However, the two synuclein isoforms, β and γ2-synuclein, can compensate the absence of α-synuclein, and their loss induces motor impairments and reduction of dopamine levels [68].
All of these observations lend further support to the notion that mitochondrial dysfunction plays a key role in zebrafish models of PD and that zebrafish represents a proficient model to investigate in vivo the alterations of mitochondrial physiology underlying neurological disorders.
4.4. Defects of Mitochondrial Dynamics
Mitochondrial dynamics, such as fission, fusion and transport, are fundamental processes for neurons because pre- and post-synaptic terminals need high amount of energy and Ca2+ buffering and are particularly susceptible to the deficiency of these systems [102]. When mtDNA and proteins accumulate defects, damaged mitochondria need to be replaced by newly ones assembled in the body of the cell and then transported along the axons. This anterograde and retrograde movement along the microtubules within neurons, vital to maintain energy homeostasis and essential neuronal activities, is mediated by motor/adaptor proteins and cytoskeletal elements [43,102]. Fission is necessary for the transport and to regulate mito-apoptosis of damaged mitochondria, while fusion allows the survival of damaged or senescent mitochondria by exchanging materials among mitochondria themselves [103].
Mutations in the MFN2 (mitofusin 2) and OPA1 genes, both involved in mitochondrial transport and/or fusion, cause two hereditary neurodegenerative disorders, respectively an axonal peripheral neuropathy (Charcot-Marie-Tooth Type 2, CMT2) and the dominant optic atrophy (DOA) [73]. MFN2 codes for a dynamin-like GTPases located in the outer mitochondrial membrane, and its major function is promoting mitochondria movement and fusion. Vettori and colleagues (2011) developed a zebrafish model for the neuromuscular disorder CMT2 [69]. Patients with CMT2A are characterized by muscular atrophy, distal muscle weakness accompanied by distal sensory loss, foot deformities and gait impairment. Zebrafish knockdown of mfn2 showed not dramatic alterations of the mitochondrial network structure but significant motor impairment or unresponsiveness to touch. Additional observations revealed a generalized impairment in the axonal structure of primary motor neurons, accompanied by the presence of shortened or missing axons, altered distribution of acetylcholine receptors with a reduction of the number and size of their clusters, which are important in neuromuscular junctions for the synaptic signals from the motor axon. Moreover, mfn2 morphants displayed also an altered alignment of the myofibers [69]. Additionally, a spontaneous mutant strain harboring a nonsense mutation in mitofusin 2 (mfn2L285X) presented altered mitochondrial dynamics and a change of mitochondrial morphology [70]. The phenotype of this mutant strain revealed also a progressive motor dysfunction and alterations at the neuromuscular junction, associated with defective axonal transport of mitochondria, resembling the progressive motor dysfunction observed in CMT2 patients [70]. In order to study p.L76P and p.R94Q, two missense mutations in MFN2, mutant capped MFN2 mRNAs were injected in the one-cell stage embryos of the new-created zebrafish line Tg(hb9: MTS-Kaede), characterized by the fluorescent labeling of motor neuron mitochondria [43]. Mutant capped MFN2 mRNAs produced a significant reduction in the density of moving mitochondria for both mutations, but a significant reduction of mitochondrial density was observed in p.L76P overexpression in zebrafish motoneurons [43]. On the contrary, a reduction of the transport of mitochondria along the axon was observed in p.R94Q expressing larvae, suggesting a different mechanism of action of the two MFN2 mutations [43].
KIF5A (Kinesin Family Member 5A) is another gene involved in mitochondrial transport, and mutations of this gene have also been associated with CMT2 [38]. KIF5A belongs to the Kinesin superfamily proteins (KIFs), which are microtubule-based molecular motors. Mammals possess three KIF5s: Kif5B that is ubiquitously expressed, and KIF5A and KIF5C that are neuron-specific [104]. In zebrafish there are five kif5 genes (kif5Aa, kif5Ab, kif5Ba, kif5Bb, and kif5C) and kif5Aa, kif5Ab, and kif5C are the most expressed in neural tissues [104]. Campbell and colleagues observed that only the kif5Aa mutated zebrafish line showed larval lethality and sensorimotor deficits similar to CMTs [38].
Kinesin proteins have an important role during neuronal development, but additional factors affect their regulation [37]. Kif1-binding protein (KBP/KIAA1279) binds to the motor domain of the KIF1B and KIF1C, and homozygous mutations of KBP cause Goldberg–Shprintzen syndrome, a severe disorder characterized by neurological symptoms, mental retardation, and disruption of white matter tracts. Lyons and colleagues (2008) investigated the KPB paralogue gene in zebrafish kbpst23 mutants and demonstrated its key role in the regulation of axonal cytoskeleton organization, regulation of axonal outgrowth speed, localization of axonal cargo and maintaining of axonal longer-term integrity. The consequence on mitochondria of the KBP mutation in zebrafish was a mislocalization and a reduction in the number of axonal mitochondria [37].
OPA1, a dynamin-related GTPase, is involved in the fusion of the outer and inner mitochondrial membranes, and gene mutations are associated with the autosomal dominant optic atrophy (ADOA) [71]. ADOA is characterized by progressive loss of visual acuity, desensitization of central visual field, optic nerve pallor, and eventual blindness [71]. In zebrafish, the opa1 morphants morphologically showed a reduction of the eyes size. In morphant 72 hpf (hours-post-fertilization) embryos a reduction of the startle response and of the locomotor activity were also observed associated to bioenergetics defects [71]. Furthermore, morphants showed upregulation of pgc1a, the orthologue of the master regulator of mitochondrial biogenesis, at 24 and 48 hpf, suggesting increased mitochondrial biogenesis [71]. The opa1 morphants allowed elucidating the connection between opa1 depletion, mitochondrial dysfunction, and development [71].
These results implicate that modeling in zebrafish proteins regulating mitochondrial dynamics might serve to study the distribution of mitochondria in early stages of development and to improve knowledge on the role of mitochondrial networks in neurodegeneration.
4.5. Mitochondrial Carrier Deficiency
Mitochondrial carriers (MCS) are transport proteins that allow small molecules to shuttle between mitochondria and cytoplasm playing a central role in mitochondrial function to regulating cellular metabolism [105] (and literature cited therein). Few studies have addressed the role of mitochondrial carriers in neurological disorders and no less the functions of many human carriers still remain better to be characterized. In mammals, MCS comprise the transporters of the 53-member canonical SLC25 family and a lesser number of identified non-canonical transporters.
SLC25A1, the first member of the solute carrier family SLC25, is the mitochondrial citrate carrier that mediates the exchange of mitochondrial citrate/isocitrate with cytosolic malate [106,107]. SLC25A1 is involved in fatty acid and sterol biosynthesis, gluconeogenesis and glycolysis [106], maintenance of chromosome integrity and regulation of autophagy [108,109]. The identification of patients harboring mutations in SLC25A1 and presenting congenital myasthenic syndrome 23 (CMS23) [72] or the more severe combined D-2- and L-2-hydroxyglutaric aciduria (D2L2AD) [110,111] phenotype confirms the relevance of this protein in humans. SLC25A1 dysfunction interferes with brain, eye, and possibly neuromuscular development, in addition to causing a distinctive urinary organic acid profile. Notably, knocking down the two zebrafish SLC25A1 orthologues (i.e., slc25a1a and slc25a1b) by injection of antisense MOs in fertilized eggs mirrors human CMS23 in terms of variable brain, eye, and cardiac involvement, and leads to abnormalities in the neuromuscular junction, regardless of the severity of the phenotype [72]. MO-injected embryos display altered tail morphology, and impaired swimming and touch-evoked escape responses at 48 hpf. Histologically, muscle morphology is normal but neuromuscular junction development is abnormal. Motor axon terminals show short and erratic outgrowth toward the muscle fiber with no evidence of complete synapse formation, suggesting presynaptic nerve terminal abnormalities. Knockdown embryos often show edema of the hindbrain, heart, yolk sac and tail. Abnormal heart development is observed with increased severity of phenotype with reduced blood flow to the tail [72].
SLC25A46, another member of the mitochondrial solute carrier family SLC25, was found to correspond to UGO1, a mitochondrial fusion factor in Saccharomyces cerevisiae. Previously, genome studies had linked SLC25A46 to atopic dermatitis but a high expression of the SLC25A46 transcript had been observed in the spinal cord, cerebellum, and optic chiasm neurons of patients [73]. Severe slc25a46 zebrafish morphants showed a curly-tail morphology while wild-appearing larvae display an altered swimming [73] associated to significantly shorter axon tracts in motor neurons and fewer neuronal processes in spinal cord neuropil of larvae. The dendritic degeneration observed in slc25a46 morphants was consistent with CMT2 pathology. In degenerating neurons, an incomplete mitochondrial fission was observed, and mitochondria resulted aggregated and predominantly in the apical portion of the soma [73]. Interestingly, different mutations in SLC25A46, that cause loss of function of the mutant protein, were also found in patients with the pontocerebellar hypoplasia (PCH) [74]. SCL25A46 seems to act as a pro-fission component regulating mitochondrial dynamics in humans and zebrafish.
4.6. Metabolic Mitochondrial Disorders
Metabolic disorders causing neurological impairment have successfully been modelled in zebrafish. To date, these disorders include Costeff syndrome, Barth syndrome, pyruvate dehydrogenase complex (PDHC) deficiency, and PCH.
Costeff syndrome is a neuro-ophthalmological disorder characterized by increased urinary excretion of 3-methylglutaconic acid and 3-methylglutaric acid, early-onset optic atrophy, and later occurrence of spasticity and extra-pyramidal features [112]. Costeff syndrome is caused by mutations in OPA3 that encodes an inner mitochondrial membrane protein. In zebrafish opa3 mRNA is expressed in the optic nerve and retinal layers, brain, inner ear, heart, liver, intestine, and swim bladder. Morphants do not show vision loss, hyperreflexia or spasticity characteristic of the human disorder but a penetrant behavior resembling the ataxia and extrapyramidal (bradykinesia) features seen in Costeff syndrome [75].
The PDHC is a nuclear-encoded mitochondrial matrix multienzyme complex that catalyzes the irreversible conversion of pyruvate into the acetyl form of coenzyme A (acetyl-CoA), thus playing a central role in linking glycolysis to the tricarboxylic acid cycle and lipogenic pathways [113]. Deficient biochemical activity causes fatal neonatal conditions including congenital lactic acidosis, growth retardation and Leigh syndrome. A spontaneous zebrafish mutant of PDHC deficiency, exhibits phenotypes similar to human patients and the administration of a ketogenic diet resembling the leading therapy proposed for children with PDHC deficiency is able to recover the normal condition [76].
Currently, PCH classification comprises 11 types based on distinct genetic causes and clinical features. The disease is associated with prenatal onset, hypoplasia/atrophy of the cerebellum, hypoplasia of the ventral pons, progressive microcephaly and variable neocortical atrophy [114]. Zebrafish models of PCH have been generated through the knockdown of tsen54 and rars2, two genes encoding proteins associated with tRNA splicing and tRNA aminoacylation. Studies in morphants suggest a common disease pathway may exist between TSEN54- and RARS2-related PCH, which may involve a tRNA processing-related mechanism [77].
All these studies reveal that the zebrafish modeling is sensitive to metabolic insults from embryonic stages, and thus it may be useful in order to understand disease mechanisms and critical to develop therapeutic options in inborn errors of metabolism affecting mitochondrial functions [22].
5. Zebrafish as In Vivo Model to Test New Compounds in Mitochondrial Related Neurological Disorders
The zebrafish represents a valuable model for investigating the molecular mechanisms underlying the neurotoxicity of agents affecting the normal neuronal mitochondrial physiology and for testing potential neuroprotective compounds. As mentioned above, defects in mitochondrial biogenesis, mitochondrial dynamics, and mitochondrial respiratory chain activities lead to a several human diseases; below, we provide several new models for mitochondrial dysfunction which could have great potential for diagnostics and therapies in neuronal mitochondrial-related diseases.
The impairment of the OxPhos system affects directly mitochondrial function causing several disease phenotypes. The inhibition of ATP production can be induced in zebrafish by exposure to 2-4 dinitrophenol (DNP), a weight loss agent with significant toxicity, used in research as an uncoupler of OxPhos [115]. Past mid-embryogenesis embryos treated by DNP show a strong decrease in ATP levels and developmental defects. The neural tissues emerge particularly affected by DNP, which inhibits motor neuron axon arbor outgrowth and proper formation of the retina [116].
COX, the terminal oxidase enzyme of the mitochondrial respiratory chain, needs copper (Cu) as an essential micronutrient required for its assembly and activity. Mutations in genes related to the delivery for Cu to COX leads to a respiratory deficiency and fatal mitochondrial disease. In a zebrafish model of Cu deficiency, low nanomolar quantity of the anticancer drug elesclomol (ES) is able to recover phenotypes associated with Cu deficiency by increasing cellular and mitochondrial Cu content [117].
Pyruvate oxidation defects are also among the causes of mitochondrial diseases mainly due to deficiency of subunits of PDHC. In zebrafish, noa mutants (carrying a mutation in the pdh-e2 subunit) treated with 4-phenylbutyrate (PBA), a histone deacetylase inhibitor usually employed in urea cycle disorders, showed a significant decrease in melanophore expansion, more active locomotion, and lower levels of lactate and pyruvate than untreated mutants. These findings prompted evaluation of PBA in human mutant cells and in a mouse model of PDHC deficiency [118]. Another zebrafish mutant (noir strain, harboring a stop codon mutation in the pdh subunit E1 β gene) showed a retinal phenotype and reduced locomotion and fatigue, and the phenotype improved upon treatment with a ketogenic diet causing an increase in acetyl-CoA production, able to bypass the metabolic block [119].
Recently, the idebenone metabolite 6-(9-carboxynonyl)-2,3-dimethoxy-5-methyl-1,4-benzoquinone (QS10) was used to recover of respiration and to allow zebrafish wild-type embryos survival after the treatment with the neurotoxin Rotenone, an inhibitor of complex I [120]. Currently, idebenone was proposed as a first-line therapy in Leber’s hereditary optic neuropathy (LHON) syndrome because its capacity to improve visual defects in a subset of patients [121,122].
It was widely described [123] that also in zebrafish some structural analogs of dopamine, such as 6-hydroxydopamine (6-OHDA) and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), selectively affect dopaminergic neurons inducing a PD-like phenotype. In particular, the metabolite of MPTP, 1-methyl-4-phenylpyridinium (MPP+), inhibits complex I and impairs mitochondrial respiration [124]. These findings allowed generating numerous zebrafish PD-models to test new potential compounds with neuroprotective activity.
The transgenic zebrafish line Tg(ETvmat2: GFP) showed a significant neurodegeneration of dopaminergic neurons and locomotor dysfunction upon treatment with MPTP [125]. This model has widely been used as an in vivo tool for screening neuroprotective agents for PD, such as Paenolum, that is the main component derived from the bark of the Paeonia suffruticosa, acting as a strong antioxidant and reducing the mitochondrial cell death pathway [126].
Moreover, it has been shown that the exposure of 24 hpf zebrafish embryos with melatonin added together with MPTP or added once MPTP removal, prevents and recovers the parkinsonian phenotype restoring mitochondrial normalcy and the brain function in zebrafish. Melatonin is able to restore in a dose-specific manner the gene expression and the normal function of the PD-related genes park2/pink1/park7 loop, and also the normal motor activity of the embryos [127]. Furthermore, sleep disorders constitute major nonmotor features of PD and several trials indicate that melatonin and its analogs are useful in treating disturbed sleep in PD patients [128,129].
Zhang and co-workers (2017) developed a multifactorial zebrafish drug-screening platform discovering a series of piperazine phenothiazines, with neuroprotective activity that rescued stress-induced mitochondrial dysfunction thus preventing dopaminergic neuron loss in a zebrafish model of pink1 deficiency [92]. In particular, trifluoperazine (TFP) is able to increase autophagic flux acting downstream of, or parallel to, pink1/parkin to restore TFEB translocation, which is a master regulator of the autophagy-lysosomal pathway [92].
Another zebrafish model of PD was generated by treating the fish with 6-OHDA, and the marine-derived compound 11-dehydrosinulariolide (11-de) increases cytosolic or mitochondrial dj-1 expression, and then activates the downstream Akt/PI3K, p-CREB, and Nrf2/HO-1 pathways [130]. Several marine-derived natural compounds have been approved for clinical use [131]. Huang and co-workers summarized those effective for the treatment of PD and that have been approved in clinical trials [132]. All these evidences suggest that zebrafish can be a powerful instrument for a full understanding of mitochondrial pathogenesis related to the nervous system.
6. Materials and Methods
A literature search was performed on original articles published by the 28th November 2018, which were identified from PubMed using the terms <<zebrafish>> AND <<neurological dysfunction [All Fields] >> and <<zebrafish [All Fields]>> AND <<mitochondrion [All Fields]>>as queries. These searches yielded respectively 80 and 503 matches in PubMed. We also performed an active search of the references listed in publications discussing neurological diseases associated with mitochondrial damage and the use of zebrafish. Altogether, we critically re-evaluated 571 articles net of repetitions but 526 were excluded because they did not present direct research on zebrafish, mitochondria and neurological disorders. Overall, 47 articles were included in the review.
7. Conclusions
Mitochondria are dynamic organelles that carry out many cell functions and, in particular, they are known as the “powerhouse” of the cell, generating ATP via OxPhos. The high number/density of mitochondria within neurons to attend their high energy demands provides a rationale for the sensitivity of the CNS to energy deficits due to mitochondrial impairment.
The large number of novel genotypes and the increased number of mutants developed in the last years, as well as the new tools developed for analyzing mitochondrial lifetime, health, and function in zebrafish combined with the use of MO and CRISPR-Cas9 systems, have allowed to investigate and understand the role of several genes involved in human mitochondrial-related neurological disorders.
The list of identified mutations associated with mitochondrial alterations in neurological disorders is still growing thanks to the use of NGS in the diagnostics of neurological human diseases. Zebrafish has proven itself to be an applicable and versatile model to investigate mutations responsible of mitochondrial disorders.
In parallel, one of the key characteristics of the zebrafish is that it can serve as a platform in high-throughput screenings performed to assess drug efficacy or to analyze off-target effects in a whole-organism context. Many drugs for treating human diseases have comparable effects in zebrafish embryos, and it is likely that additional compounds identified by large-molecule screening in zebrafish will be worth testing in mammalian systems. The zebrafish can be exploited as a practical tool for performing more detailed and rapid studies to shed light on the effects and consequences (inert, ameliorating, or detrimental) during development, which have rarely been analyzed in proper clinical trials. In addition, the model can serve while assessing safety and efficacy of drugs in their early stages of development.
Author Contributions
Conceptualization, F.M.S., V.P. and M.M.; methodology, F.M.S. and M.M.; investigation, G.F. and V.N.; resources, writing—original draft preparation, G.F., V.N., writing—review and editing, F.M.S. M.M., G.L, B.F., T.V and A.B.; visualization, M.M..; supervision, F.M.S. and M.M.; project administration, G.F.; funding acquisition, F.M.S., V.P.
Funding
V.P. was supported by 2010 funds from the University of Bari (Fondi ex-60%, 2009–2011). Funding from Ricerca Corrente 2018, projects MITONEXT and MIT-OMICS from Italian Ministry of Health are also acknowledged.
Acknowledgments
We thank Catherine J. Wrenn for her expert editorial assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| 6-OHDA | 6-hydroxydopamine |
| acetyl-CoA | Acetyl form of coenzyme A |
| ADOA | Autosomal dominant optic atrophy |
| Cas9 | CRISPR-associated protein 9 |
| CMT2 | Charcot-Marie-Tooth Type 2 |
| CNS | Central nervous system |
| COA | COX assembly factor |
| CoQ | Coenzyme Q |
| COX | Cytochrome c oxidase |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Cys | Cysteine |
| cyt c | cytochrome c |
| DNP | 2-4 dinitrophenol |
| DOA | Dominant optic atrophy |
| DS | Dravet syndrome |
| EC | Enzyme complexes |
| ENU | N-ethyl-N-nitrosourea |
| Hcy | Homocysteine |
| MADD | Multiple acyl-CoA dehydrogenase deficiency |
| MCS | Mitochondrial carriers |
| MO | Morpholino oligonucleotide |
| MPP+ | 1-methyl-4-phenylpyridinium |
| MPTP | 1-methylmethyl-4-phenylphenyl-1,2,3,6-tetrahydropyridinetetrahydropyridine |
| mRNA | Messenger RNA |
| mtDNA | Mitochondrial DNA |
| nDNA | Nuclear DNA |
| NGS | Next-generation sequencing |
| OxPhos | Oxidative phosphorylation |
| PBA | 4-phenylbutyrate |
| PCH | Pontocerebellar hypoplasia |
| PD | Parkinson’s disease |
| PDHC | Pyruvate dehydrogenase complex |
| pLL | Posterior lateral line |
| PNS | Peripheral nervous system |
| ROS | Reactive oxygen species |
| rRNA | Ribosomal RNA |
| TALEN | Transcription activator-like effector |
| tRNA | Transfer RNA |
References
- DiMauro, S.; Schon, E.A.; Carelli, V.; Hirano, M. The clinical maze of mitochondrial neurology. Nat. Rev. Neurol. 2013, 9, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Lill, R. The role of mitochondria in cellular iron-sulfur protein biogenesis: Mechanisms, connected processes, and diseases. Cold Spring Harb. Perspect. Biol. 2013, 5, a011312. [Google Scholar] [CrossRef] [PubMed]
- Higgins, G.C.; Beart, P.M.; Shin, Y.S.; Chen, M.J.; Cheung, N.S.; Nagley, P. Oxidative stress: Emerging mitochondrial and cellular themes and variations in neuronal injury. J. Alzheimers Dis. 2010, 20, 453–473. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, D.; Zeviani, M. Human diseases associated with defects in assembly of OXPHOS complexes. Essays. Biochem. 2018, 62, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Spinazzola, A.; Zeviani, M. Mitochondrial diseases: A cross-talk between mitochondrial and nuclear genomes. Adv. Exp. Med. Biol. 2009, 652, 69–84. [Google Scholar] [PubMed]
- Farrar, G.J.; Chadderton, N.; Kenna, P.F.; Millington-Ward, S. Mitochondrial disorders: Aetiologies, models systems, and candidate therapies. Trends Genet. 2013, 29, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Bergman, O.; Ben-Shachar, D. Mitochondrial Oxidative Phosphorylation System (OXPHOS) Deficits in Schizophrenia: Possible Interactions with Cellular Processes. Can. J. Psychiatry 2016, 61, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Cardaioli, E.; da Pozzo, P.; Formichi, P.; Gallus, G.N.; Radi, E. Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 2012, 322, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Schon, E.A.; Przedborski, S. Mitochondria: The next (neurode)generation. Neuron 2011, 70, 1033–1053. [Google Scholar] [CrossRef]
- Busch, K.B.; Kowald, A.; Spelbrink, J.N. Quality matters: How does mitochondrial network dynamics and quality control impact on mtDNA integrity? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014, 369, 20130442. [Google Scholar] [CrossRef] [PubMed]
- Ploumi, C.; Daskalaki, I.; Tavernarakis, N. Mitochondrial biogenesis and clearance: A balancing act. FEBS J. 2017, 284, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.C.; Holzbaur, E.L. Autophagosome dynamics in neurodegeneration at a glance. J. Cell Sci. 2015, 128, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Vicente, M. Neuronal Mitophagy in Neurodegenerative Diseases. Front. Mol. Neurosci. 2017, 10, 64. [Google Scholar] [CrossRef]
- Ruzzenente, B.; Rotig, A.; Metodiev, M.D. Mouse models for mitochondrial diseases. Hum. Mol. Genet. 2016, 25, 115–122. [Google Scholar] [CrossRef]
- Spradling, A.; Ganetsky, B.; Hieter, P.; Johnston, M.; Olson, M.; Orr-Weaver, T.; Rossant, J.; Sanchez, A.; Waterston, R. New roles for model genetic organisms in understanding and treating human disease: Report from the 2006 Genetics Society of America meeting. Genetics 2006, 172, 2025–2032. [Google Scholar] [PubMed]
- Kabashi, E.; Brustein, E.; Champagne, N.; Drapeau, P. Zebrafish models for the functional genomics of neurogenetic disorders. Biochim. Biophys. Acta. 2011, 1812, 335–345. [Google Scholar] [CrossRef]
- Steele, S.L.; Prykhozhij, S.V.; Berman, J.N. Zebrafish as a model system for mitochondrial biology and diseases. Transl. Res. 2014, 163, 79–98. [Google Scholar] [CrossRef] [PubMed]
- Broughton, R.E.; Milam, J.E.; Roe, B.A. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001, 11, 1958–1967. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Wager, K.; Mahmood, F.; Russell, C. Modelling inborn errors of metabolism in zebrafish. J. Inherit. Metab. Dis. 2014, 37, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Postlethwait, J.; Ruotti, V.; Carvan, M.J.; Tonellato, P.J. Automated analysis of conserved syntenies for the zebrafish genome. Methods Cell Biol. 2004, 77, 255–271. [Google Scholar] [PubMed]
- Nasevicius, A.; Ekker, S.C. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Gerety, S.S.; Wilkinson, D.G. Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev. Biol. 2011, 350, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Doyon, Y.; McCammon, J.M.; Miller, J.C.; Faraji, F.; Ngo, C.; Katibah, G.E.; Amora, R.; Hocking, T.D.; Zhang, L.; Rebar, E.J.; et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 702–708. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Noyes, M.B.; Zhu, L.J.; Lawson, N.D.; Wolfe, S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008, 26, 695–701. [Google Scholar] [CrossRef]
- Zu, Y.; Tong, X.; Wang, Z.; Liu, D.; Pan, R.; Li, Z.; Hu, Y.; Luo, Z.; Huang, P.; Wu, Q.; et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 2013, 10, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed]
- Varshney, G.K.; Burgess, S.M. Mutagenesis and phenotyping resources in zebrafish for studying development and human disease. Brief Funct. Genom. 2014, 13, 82–94. [Google Scholar] [CrossRef]
- Rafferty, S.A.; Quinn, T.A. A beginner’s guide to understanding and implementing the genetic modification of zebrafish. Prog. Biophys. Mol. Biol. 2018, 138, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.C. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu. Rev. Genet. 2012, 46, 265–287. [Google Scholar] [CrossRef] [PubMed]
- Chlystun, M.; Campanella, M.; Law, A.L.; Duchen, M.R.; Fatimathas, L.; Levine, T.P.; Gerke, V.; Moss, S.E. Regulation of mitochondrial morphogenesis by annexin A6. PLoS ONE 2013, 8, e53774. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.Y.; Horng, J.L.; Kunkel, J.G.; Hwang, P.P. Proton pump-rich cell secretes acid in skin of zebrafish larvae. Am. J. Physiol. Cell Physiol. 2006, 290, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Wada, Y.; Kawamura, S. ES1 is a mitochondrial enlarging factor contributing to form mega-mitochondria in zebrafish cones. Sci. Rep. 2016, 6, 22360. [Google Scholar] [CrossRef] [PubMed]
- Drerup, C.M.; Herbert, A.L.; Monk, K.R.; Nechiporuk, A.V. Regulation of mitochondria-dynactin interaction and mitochondrial retrograde transport in axons. Elife 2017, 6, e22234. [Google Scholar] [CrossRef] [PubMed]
- Lyons, D.A.; Naylor, S.G.; Mercurio, S.; Dominguez, C.; Talbot, W.S. KBP is essential for axonal structure, outgrowth and maintenance in zebrafish, providing insight into the cellular basis of Goldberg-Shprintzen syndrome. Development 2008, 135, 599–608. [Google Scholar] [CrossRef]
- Campbell, P.D.; Shen, K.; Sapio, M.R.; Glenn, T.D.; Talbot, W.S.; Marlow, F.L. Unique function of Kinesin Kif5A in localization of mitochondria in axons. J. Neurosci. 2014, 34, 14717–14732. [Google Scholar] [CrossRef]
- Halpern, M.E.; Rhee, J.; Goll, M.G.; Akitake, C.M.; Parsons, M.; Leach, S.D. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 2008, 5, 97–110. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.C.; Vargas, M.E.; Sagasti, A. WldS and PGC-1alpha regulate mitochondrial transport and oxidation state after axonal injury. J. Neurosci. 2013, 33, 14778–14790. [Google Scholar] [CrossRef]
- Plucinska, G.; Paquet, D.; Hruscha, A.; Godinho, L.; Haass, C.; Schmid, B.; Misgeld, T. In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. J. Neurosci. 2012, 32, 16203–16212. [Google Scholar] [CrossRef]
- Auer, T.O.; Xiao, T.; Bercier, V.; Gebhardt, C.; Duroure, K.; Concordet, J.P.; Wyart, C.; Suster, M.; Kawakami, K.; Wittbrodt, J.; et al. Deletion of a kinesin I motor unmasks a mechanism of homeostatic branching control by neurotrophin-3. Elife 2015, 4, e05061. [Google Scholar] [CrossRef] [PubMed]
- Bergamin, G.; Cieri, D.; Vazza, G.; Argenton, F.; Mostacciuolo, M.L. Zebrafish Tg(hb9:MTS-Kaede): A new in vivo tool for studying the axonal movement of mitochondria. Biochim. Biophys. Acta. 2016, 1860, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Dukes, A.A.; Bai, Q.; van Laar, V.S.; Zhou, Y.Z.; Ilin, V.; David, C.N.; Agim, Z.S.; Bonkowsky, J.L.; Cannon, J.R.; Watkins, S.C.; et al. Live imaging of mitochondrial dynamics in CNS dopaminergic neurons in vivo demonstrates early reversal of mitochondrial transport following MPP+ exposure. Neurobiol. of Disease 2016, 95, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Pinter, K.; Drerup, C.M. Analyzing Neuronal Mitochondria in vivo Using Fluorescent Reporters in Zebrafish. Front. Cell Dev. Biol. 2018, 6, 144. [Google Scholar] [CrossRef]
- Song, Y.; Selak, M.A.; Watson, C.T.; Coutts, C.; Scherer, P.C.; Panzer, J.A.; Gibbs, S.; Scott, M.O.; Willer, G.; Gregg, R.G.; et al. Mechanisms underlying metabolic and neural defects in zebrafish and human multiple acyl-CoA dehydrogenase deficiency (MADD). PLoS ONE 2009, 4, e8329. [Google Scholar] [CrossRef]
- Dowling, J.J.; Arbogast, S.; Hur, J.; Nelson, D.D.; McEvoy, A.; Waugh, T.; Marty, I.; Lunardi, J.; Brooks, S.V.; Kuwada, J.Y.; et al. Oxidative stress and successful antioxidant treatment in models of RYR1-related myopathy. Brain 2012, 135, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Yue, P.; Yang, X.; Ning, P.; Xi, X.; Yu, H.; Feng, Y.; Shao, R.; Meng, X. A mitochondria-targeted ratiometric two-photon fluorescent probe for detecting intracellular cysteine and homocysteine. Talanta 2018, 178, 24–30. [Google Scholar] [CrossRef]
- Bourdineaud, J.P.; Rossignol, R.; Brethes, D. Zebrafish: A model animal for analyzing the impact of environmental pollutants on muscle and brain mitochondrial bioenergetics. Int. J. Biochem. Cell Biol. 2013, 45, 16–22. [Google Scholar] [CrossRef]
- Stackley, K.D.; Beeson, C.C.; Rahn, J.J.; Chan, S.S. Bioenergetic profiling of zebrafish embryonic development. PLoS ONE 2011, 6, e25652. [Google Scholar] [CrossRef]
- Artuso, L.; Romano, A.; Verri, T.; Domenichini, A.; Argenton, F.; Santorelli, F.M.; Petruzzella, V. Mitochondrial DNA metabolism in early development of zebrafish (Danio rerio). Biochim. Biophys. Acta. 2012, 1817, 1002–1011. [Google Scholar] [CrossRef]
- Lowery, L.A.; de Rienzo, G.; Gutzman, J.H.; Sive, H. Characterization and classification of zebrafish brain morphology mutants. Anat. Rec. 2009, 292, 94–106. [Google Scholar] [CrossRef]
- Kim, M.J.; Kang, K.H.; Kim, C.H.; Choi, S.Y. Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. Biotechniques 2008, 45, 331–334. [Google Scholar]
- Rendón, O.Z.; Neiva, L.S.; Sasarman, F.; Shoubridge, E.A. The arginine methyltransferase NDUFAF7 is essential for complex I assembly and early vertebrate embryogenesis. Hum. Mol. Genet. 2014, 23, 5159–5170. [Google Scholar] [CrossRef]
- Baden, K.N.; Murray, J.; Capaldi, R.A.; Guillemin, K. Early developmental pathology due to cytochrome c oxidase deficiency is revealed by a new zebrafish model. J. Biol. Chem. 2007, 282, 34839–34849. [Google Scholar] [CrossRef]
- Ghosh, A.; Trivedi, P.P.; Timbalia, S.A.; Griffin, A.T.; Rahn, J.J.; Chan, S.S.; Gohil, V.M. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 2014, 23, 3596–3606. [Google Scholar] [CrossRef]
- Kim, S.H.; Scott, S.A.; Bennett, M.J.; Carson, R.P.; Fessel, J.; Brown, H.A.; Ess, K.C. Multi-organ abnormalities and mTORC1 activation in zebrafish model of multiple acyl-CoA dehydrogenase deficiency. PLoS Genet. 2013, 9, e1003563. [Google Scholar] [CrossRef]
- Kumar, M.G.; Rowley, S.; Fulton, R.; Dinday, M.T.; Baraban, S.C.; Patel, M. Altered Glycolysis and Mitochondrial Respiration in a Zebrafish Model of Dravet Syndrome. eNeuro 2016, 3. [Google Scholar] [CrossRef]
- Flinn, L.; Mortiboys, H.; Volkmann, K.; Koster, R.W.; Ingham, P.W.; Bandmann, O. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain 2009, 132, 1613–1623. [Google Scholar] [CrossRef]
- Petrucelli, L.; O’Farrell, C.; Lockhart, P.J.; Baptista, M.; Kehoe, K.; Vink, L.; Choi, P.; Wolozin, B.; Farrer, M.; Hardy, J.; et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: Proteasome dysfunction selectively affects catecholaminergic neurons. Neuron 2002, 36, 1007–1019. [Google Scholar] [CrossRef]
- Sallinen, V.; Kolehmainen, J.; Priyadarshini, M.; Toleikyte, G.; Chen, Y.C.; Panula, P. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol. Dis. 2010, 40, 93–101. [Google Scholar] [CrossRef]
- Flinn, L.J.; Keatinge, M.; Bretaud, S.; Mortiboys, H.; Matsui, H.; de Felice, E.; Woodroof, H.I.; Brown, L.; McTighe, A.; Soellner, R.; et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann. Neurol. 2013, 74, 837–847. [Google Scholar] [CrossRef]
- Noble, S.; Ismail, A.; Godoy, R.; Xi, Y.; Ekker, M. Zebrafish Parla- and Parlb-deficiency affects dopaminergic neuron patterning and embryonic survival. J. Neurochem. 2012, 122, 196–207. [Google Scholar] [CrossRef]
- Bretaud, S.; Allen, C.; Ingham, P.W.; Bandmann, O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J. Neurochem. 2007, 100, 1626–1635. [Google Scholar] [CrossRef]
- Froyset, A.K.; Edson, A.J.; Gharbi, N.; Khan, E.A.; Dondorp, D.; Bai, Q.; Tiraboschi, E.; Suster, M.L.; Connolly, J.B.; Burton, E.A.; et al. Astroglial DJ-1 over-expression up-regulates proteins involved in redox regulation and is neuroprotective in vivo. Redox. Biol. 2018, 16, 237–247. [Google Scholar] [CrossRef]
- Sheng, D.; Qu, D.; Kwok, K.H.; Ng, S.S.; Lim, A.Y.; Aw, S.S.; Lee, C.W.; Sung, W.K.; Tan, E.K.; Lufkin, T.; et al. Deletion of the WD40 domain of LRRK2 in Zebrafish causes Parkinsonism-like loss of neurons and locomotive defect. PLoS Genet. 2010, 6, e1000914. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.C.; Lulla, A.; Stahl, M.C.; Wheat, N.D.; Bronstein, J.M.; Sagasti, A. Axon degeneration and PGC-1alpha-mediated protection in a zebrafish model of alpha-synuclein toxicity. Dis. Model Mech. 2014, 7, 571–582. [Google Scholar] [CrossRef]
- Milanese, C.; Sager, J.J.; Bai, Q.; Farrell, T.C.; Cannon, J.R.; Greenamyre, J.T.; Burton, E.A. Hypokinesia and reduced dopamine levels in zebrafish lacking beta- and gamma1-synucleins. J. Biol. Chem. 2012, 287, 2971–2983. [Google Scholar] [CrossRef]
- Vettori, A.; Bergamin, G.; Moro, E.; Vazza, G.; Polo, G.; Tiso, N.; Argenton, F.; Mostacciuolo, M.L. Developmental defects and neuromuscular alterations due to mitofusin 2 gene (MFN2) silencing in zebrafish: A new model for Charcot-Marie-Tooth type 2A neuropathy. Neuromuscul. Disord. 2011, 21, 58–67. [Google Scholar] [CrossRef]
- Chapman, A.L.; Bennett, E.J.; Ramesh, T.M.; de Vos, K.J.; Grierson, A.J. Axonal Transport Defects in a Mitofusin 2 Loss of Function Model of Charcot-Marie-Tooth Disease in Zebrafish. PLoS ONE 2013, 8, e67276. [Google Scholar] [CrossRef]
- Rahn, J.J.; Stackley, K.D.; Chan, S.S. Opa1 is required for proper mitochondrial metabolism in early development. PLoS ONE 2013, 8, e59218. [Google Scholar] [CrossRef]
- Chaouch, A.; Porcelli, V.; Cox, D.; Edvardson, S.; Scarcia, P.; de Grassi, A.; Pierri, C.L.; Cossins, J.; Laval, S.H.; Griffin, H.; et al. Mutations in the Mitochondrial Citrate Carrier SLC25A1 are Associated with Impaired Neuromuscular Transmission. J. Neuromuscul. Dis. 2014, 1, 75–90. [Google Scholar]
- Abrams, A.J.; Hufnagel, R.B.; Rebelo, A.; Zanna, C.; Patel, N.; Gonzalez, M.A.; Campeanu, I.J.; Griffin, L.B.; Groenewald, S.; Strickland, A.V.; et al. Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat. Genet. 2015, 47, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Steffen, J.; Yourshaw, M.; Mamsa, H.; Andersen, E.; Rudnik-Schoneborn, S.; Pope, K.; Howell, K.B.; McLean, C.A.; Kornberg, A.J.; et al. Loss of function of SLC25A46 causes lethal congenital pontocerebellar hypoplasia. Brain 2016, 139, 2877–2890. [Google Scholar] [CrossRef]
- Pei, W.; Kratz, L.E.; Bernardini, I.; Sood, R.; Yokogawa, T.; Dorward, H.; Ciccone, C.; Kelley, R.I.; Anikster, Y.; Burgess, H.A.; et al. A model of Costeff Syndrome reveals metabolic and protective functions of mitochondrial OPA3. Development 2010, 137, 2587–2596. [Google Scholar] [CrossRef][Green Version]
- Taylor, M.R.; Hurley, J.B.; van Epps, H.A.; Brockerhoff, S.E. A zebrafish model for pyruvate dehydrogenase deficiency: Rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc. Natl. Acad. Sci. USA 2004, 101, 4584–4589. [Google Scholar] [CrossRef]
- Kasher, P.R.; Namavar, Y.; van Tijn, P.; Fluiter, K.; Sizarov, A.; Kamermans, M.; Grierson, A.J.; Zivkovic, D.; Baas, F. Impairment of the tRNA-splicing endonuclease subunit 54 (tsen54) gene causes neurological abnormalities and larval death in zebrafish models of pontocerebellar hypoplasia. Hum. Mol. Genet. 2011, 20, 1574–1584. [Google Scholar] [CrossRef]
- Wallace, D.C. Mitochondrial diseases in man and mouse. Science 1999, 283, 1482–1488. [Google Scholar] [CrossRef]
- Koopman, W.J.; Distelmaier, F.; Smeitink, J.A.; Willems, P.H. OXPHOS mutations and neurodegeneration. EMBO J. 2013, 32, 9–29. [Google Scholar] [CrossRef]
- Rodenburg, R.J. Mitochondrial complex I-linked disease. Biochim. Biophys. Acta. 2016, 1857, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, Y.; Chen, S.; Wu, Y.; Lin, S.; Duan, Y.; Zheng, K.; Zhang, L.; Gu, X.; Hong, W.; et al. A Novel Potentially Causative Variant of NDUFAF7 Revealed by Mutation Screening in a Chinese Family With Pathologic Myopia. Invest. Ophthalmol. Vis. Sci. 2017, 58, 4182–4192. [Google Scholar] [CrossRef]
- DiMauro, S.; Tanji, K.; Schon, E.A. The many clinical faces of cytochrome c oxidase deficiency. Adv. Exp. Med. Biol. 2012, 748, 341–357. [Google Scholar]
- Olsen, R.K.; Andresen, B.S.; Christensen, E.; Bross, P.; Skovby, F.; Gregersen, N. Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency. Hum. Mutat. 2003, 22, 12–23. [Google Scholar] [CrossRef]
- Dauer, W.; Przedborski, S. Parkinson’s disease: Mechanisms and models. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef]
- Jones, N. Mitochondrial dysfunction occurs early in PD. Nat. Rev. Neurol. 2010, 6, 60. [Google Scholar] [CrossRef]
- Seirafi, M.; Kozlov, G.; Gehring, K. Parkin structure and function. FEBS J. 2015, 282, 2076–2088. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Olzmann, J.A.; Chin, L.S.; Li, L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007, 5, e172. [Google Scholar] [CrossRef]
- Kashatus, J.A.; Nascimento, A.; Myers, L.J.; Sher, A.; Byrne, F.L.; Hoehn, K.L.; Counter, C.M.; Kashatus, D.F. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 2015, 57, 537–551. [Google Scholar] [CrossRef]
- Xi, Y.; Noble, S.; Ekker, M. Modeling neurodegeneration in zebrafish. Curr. Neurol. Neurosci. Rep. 2011, 11, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshini, M.; Tuimala, J.; Chen, Y.C.; Panula, P. A zebrafish model of PINK1 deficiency reveals key pathway dysfunction including HIF signaling. Neurobiol. Dis. 2013, 54, 127–138. [Google Scholar] [CrossRef]
- Soman, S.; Keatinge, M.; Moein, M.; da Costa, M.; Mortiboys, H.; Skupin, A.; Sugunan, S.; Bazala, M.; Kuznicki, J.; Bandmann, O. Inhibition of the mitochondrial calcium uniporter rescues dopaminergic neurons in pink1(−/−) zebrafish. Eur. J. Neurosci. 2017, 45, 528–535. [Google Scholar] [CrossRef]
- Zhang, Y.; Nguyen, D.T.; Olzomer, E.M.; Poon, G.P.; Cole, N.J.; Puvanendran, A.; Phillips, B.R.; Hesselson, D. Rescue of Pink1 Deficiency by Stress-Dependent Activation of Autophagy. Cell Chem. Biol. 2017, 24, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Lee, J.R.; Grimes, D.A.; Racacho, L.; Ye, D.; Yang, H.; Ross, O.A.; Farrer, M.; McQuibban, G.A.; Bulman, D.E. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum. Mol. Genet. 2011, 20, 1966–1974. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Rogozin, I.B.; Davidovic, L.; Letellier, M.C.; Pellegrini, L. The rhomboids: A nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003, 4, 19. [Google Scholar] [CrossRef]
- Bai, Q.; Mullett, S.J.; Garver, J.A.; Hinkle, D.A.; Burton, E.A. Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 2006, 1113, 33–44. [Google Scholar] [CrossRef]
- Irrcher, I.; Aleyasin, H.; Seifert, E.L.; Hewitt, S.J.; Chhabra, S.; Phillips, M.; Lutz, A.K.; Rousseaux, M.W.; Bevilacqua, L.; Jahani-Asl, A.; et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010, 19, 3734–3746. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.W.; Pei, Z.; Jiang, H.; Moore, D.J.; Liang, Y.; West, A.B.; Dawson, V.L.; Dawson, T.M.; Ross, C.A. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 18676–18681. [Google Scholar] [CrossRef] [PubMed]
- Angeles, D.C.; Gan, B.H.; Onstead, L.; Zhao, Y.; Lim, K.L.; Dachsel, J.; Melrose, H.; Farrer, M.; Wszolek, Z.K.; Dickson, D.W.; et al. Mutations in LRRK2 increase phosphorylation of peroxiredoxin 3 exacerbating oxidative stress-induced neuronal death. Hum. Mutat. 2011, 32, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, M.H.; Fujioka, H.; Liu, J.; Wilson-Delfosse, A.; Chen, S.G.; Perry, G.; Casadesus, G.; Zhu, X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet. 2012, 21, 1931–1944. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K. alpha-Synuclein and mitochondria: Partners in crime? Neurotherapeutics 2013, 10, 391–399. [Google Scholar] [CrossRef]
- Sun, Z.; Gitler, A.D. Discovery and characterization of three novel synuclein genes in zebrafish. Dev. Dyn. 2008, 237, 2490–2495. [Google Scholar] [CrossRef] [PubMed]
- Lovas, J.R.; Wang, X. The meaning of mitochondrial movement to a neuron’s life. Biochim. Biophys. Acta. 2013, 1833, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Rankin, J.; Brown, R.; Dobyns, W.B.; Harington, J.; Patel, J.; Quinn, M.; Brown, G. Pontocerebellar hypoplasia type 6: A British case with PEHO-like features. Am. J. Med. Genet. A. 2010, 152, 2079–2084. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.D.; Marlow, F.L. Temporal and tissue specific gene expression patterns of the zebrafish kinesin-1 heavy chain family, kif5s, during development. Gene. Expr. Patterns. 2013, 13, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.B. Functional Properties of the Mitochondrial Carrier System. Trends Cell Biol. 2017, 27, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.S.; Mayor, J.A.; Wood, D.O. The mitochondrial tricarboxylate transport protein. cDNA cloning, primary structure, and comparison with other mitochondrial transport proteins. J. Biol. Chem. 1993, 268, 13682–13690. [Google Scholar] [PubMed]
- Moraes, T.F.; Reithmeier, R.A. Membrane transport metabolons. Biochim. Biophys. Acta. 2012, 1818, 2687–2706. [Google Scholar] [CrossRef] [PubMed]
- Morciano, P.; Carrisi, C.; Capobianco, L.; Mannini, L.; Burgio, G.; Cestra, G.; de Benedetto, G.E.; Corona, D.F.; Musio, A.; Cenci, G. A conserved role for the mitochondrial citrate transporter Sea/SLC25A1 in the maintenance of chromosome integrity. Hum. Mol. Genet. 2009, 18, 4180–4188. [Google Scholar] [CrossRef] [PubMed]
- Catalina-Rodriguez, O.; Kolukula, V.K.; Tomita, Y.; Preet, A.; Palmieri, F.; Wellstein, A.; Byers, S.; Giaccia, A.J.; Glasgow, E.; Albanese, C.; et al. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget 2012, 3, 1220–1235. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Porcelli, V.; Jalas, C.; Soiferman, D.; Kellner, Y.; Shaag, A.; Korman, S.H.; Pierri, C.L.; Scarcia, P.; Fraenkel, N.D.; et al. Agenesis of corpus callosum and optic nerve hypoplasia due to mutations in SLC25A1 encoding the mitochondrial citrate transporter. J. Med. Genet. 2013, 50, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Nota, B.; Struys, E.A.; Pop, A.; Jansen, E.E.; Fernandez Ojeda, M.R.; Kanhai, W.A.; Kranendijk, M.; van Dooren, S.J.; Bevova, M.R.; Sistermans, E.A.; et al. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined D-2- and L-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 2013, 92, 627–631. [Google Scholar] [CrossRef]
- Powell, K.A.; Davies, J.R.; Taylor, E.; Wride, M.A.; Votruba, M. Mitochondrial localization and ocular expression of mutant Opa3 in a mouse model of 3-methylglutaconicaciduria type III. Invest. Ophthalmol. Vis. Sci. 2011, 52, 4369–4380. [Google Scholar] [CrossRef]
- Patel, M.S.; Roche, T.E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990, 4, 3224–3233. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, T.; Baas, F.; Barth, P.G.; Poll-The, B.T. What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet. J. Rare Dis. 2018, 13, 92. [Google Scholar] [CrossRef] [PubMed]
- Grundlingh, J.; Dargan, P.I.; El-Zanfaly, M.; Wood, D.M. 2,4-dinitrophenol (DNP): A weight loss agent with significant acute toxicity and risk of death. J. Med. Toxicol. 2011, 7, 205–212. [Google Scholar] [CrossRef]
- Bestman, J.E.; Stackley, K.D.; Rahn, J.J.; Williamson, T.J.; Chan, S.S. The cellular and molecular progression of mitochondrial dysfunction induced by 2,4-dinitrophenol in developing zebrafish embryos. Differentiation 2015, 89, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Soma, S.; Latimer, A.J.; Chun, H.; Vicary, A.C.; Timbalia, S.A.; Boulet, A.; Rahn, J.J.; Chan, S.S.L.; Leary, S.C.; Kim, B.E.; et al. Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc. Natl. Acad. Sci. USA 2018, 115, 8161–8166. [Google Scholar] [CrossRef]
- Ferriero, R.; Manco, G.; Lamantea, E.; Nusco, E.; Ferrante, M.I.; Sordino, P.; Stacpoole, P.W.; Lee, B.; Zeviani, M.; Brunetti-Pierri, N. Phenylbutyrate therapy for pyruvate dehydrogenase complex deficiency and lactic acidosis. Sci. Transl. Med. 2013, 5, 175ra31. [Google Scholar] [CrossRef]
- Maurer, C.M.; Schonthaler, H.B.; Mueller, K.P.; Neuhauss, S.C. Distinct retinal deficits in a zebrafish pyruvate dehydrogenase-deficient mutant. J. Neurosci. 2010, 30, 11962–11972. [Google Scholar] [CrossRef] [PubMed]
- Giorgio, V.; Schiavone, M.; Galber, C.; Carini, M.; da Ros, T.; Petronilli, V.; Argenton, F.; Carelli, V.; Acosta Lopez, M.J.; Salviati, L.; et al. The idebenone metabolite QS10 restores electron transfer in complex I and coenzyme Q defects. Biochim. Biophys. Acta. Bioenerg. 2018, 1859, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Chang, M. Leber’s hereditary optic neuropathy misdiagnosed as optic neuritis and Lyme disease in a patient with multiple sclerosis. BMJ Case Rep. 2018, 11, e227109. [Google Scholar] [CrossRef] [PubMed]
- Klopstock, T.; Yu-Wai-Man, P.; Dimitriadis, K.; Rouleau, J.; Heck, S.; Bailie, M.; Atawan, A.; Chattopadhyay, S.; Schubert, M.; Garip, A.; et al. A randomized placebo-controlled trial of idebenone in Leber’s hereditary optic neuropathy. Brain 2011, 134, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.S.; Geng, W.S.; Jia, J.J. Neurotoxin-Induced Animal Models of Parkinson Disease: Pathogenic Mechanism and Assessment. ASN Neuro 2018, 10, 1759091418777438. [Google Scholar] [CrossRef] [PubMed]
- Bove, J.; Perier, C. Neurotoxin-based models of Parkinson’s disease. Neuroscience 2012, 211, 51–76. [Google Scholar] [CrossRef]
- Wen, L.; Wei, W.; Gu, W.; Huang, P.; Ren, X.; Zhang, Z.; Zhu, Z.; Lin, S.; Zhang, B. Visualization of monoaminergic neurons and neurotoxicity of MPTP in live transgenic zebrafish. Dev. Biol. 2008, 314, 84–92. [Google Scholar] [CrossRef]
- Lu, X.L.; Lin, Y.H.; Wu, Q.; Su, F.J.; Ye, C.H.; Shi, L.; He, B.X.; Huang, F.W.; Pei, Z.; Yao, X.L. Paeonolum protects against MPP+-induced neurotoxicity in zebrafish and PC12 cells. BMC Complement. Altern. Med. 2015, 15, 137. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Casado, M.E.; Lima, E.; Garcia, J.A.; Doerrier, C.; Aranda, P.; Sayed, R.K.; Guerra-Librero, A.; Escames, G.; Lopez, L.C.; Acuna-Castroviejo, D. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J. Pineal. Res. 2016, 61, 96–107. [Google Scholar] [CrossRef]
- Srinivasan, V.; Cardinali, D.P.; Srinivasan, U.S.; Kaur, C.; Brown, G.M.; Spence, D.W.; Hardeland, R.; Pandi-Perumal, S.R. Therapeutic potential of melatonin and its analogs in Parkinson’s disease: Focus on sleep and neuroprotection. Ther. Adv. Neurol. Disord. 2011, 4, 297–317. [Google Scholar] [CrossRef] [PubMed]
- Loddo, G.; Calandra-Buonaura, G.; Sambati, L.; Giannini, G.; Cecere, A.; Cortelli, P.; Provini, F. The Treatment of Sleep Disorders in Parkinson’s Disease: From Research to Clinical Practice. Front. Neurol. 2017, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.W.; Hung, H.C.; Huang, S.Y.; Chen, C.H.; Chen, Y.R.; Chen, C.Y.; Yang, S.N.; Wang, H.D.; Sung, P.J.; Sheu, J.H.; et al. Neuroprotective Effect of the Marine-Derived Compound 11-Dehydrosinulariolide through DJ-1-Related Pathway in In Vitro and In Vivo Models of Parkinson’s Disease. Mar. Drugs 2016, 14, 187. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the past and charting the future of marine natural products drug discovery and chemical biology. Chem. Biol. 2012, 19, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhang, Z.; Cui, W. Marine-Derived Natural Compounds for the Treatment of Parkinson’s Disease. Mar. Drugs 2019, 17, 221. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).