Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion
Abstract
1. Introduction
2. Results
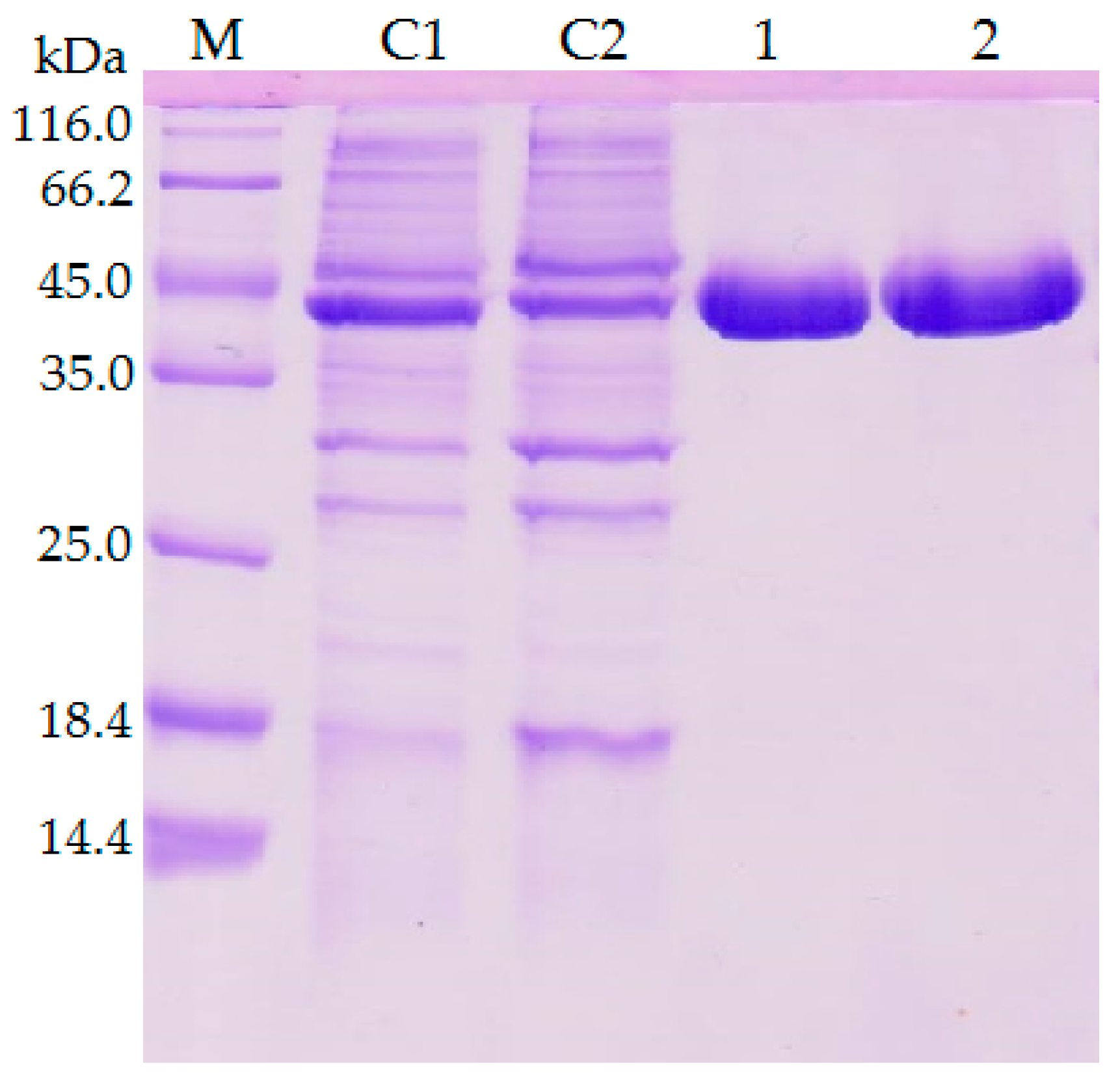
2.1. Construction of Mutant Lipase and Purification
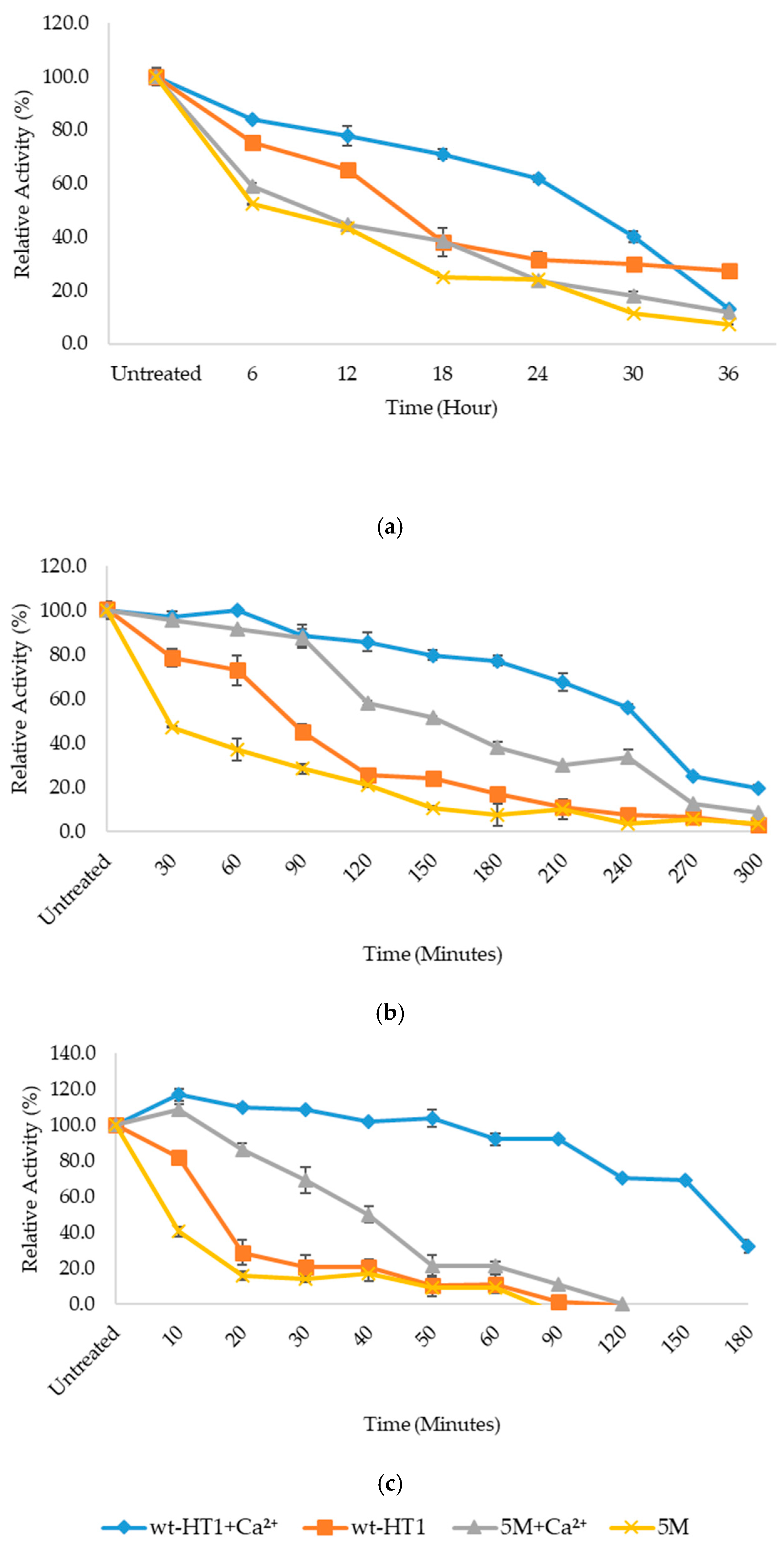
2.2. Optimum Temperature and Thermostability of Lipases
2.3. Optimum pH and pH Stability of Lipases
2.4. Effect of Metal Ions on Lipase Stability
2.5. Thermostability of Mutant in the Presence of Calcium Ion
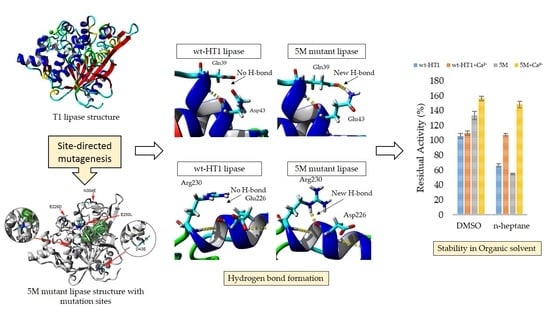
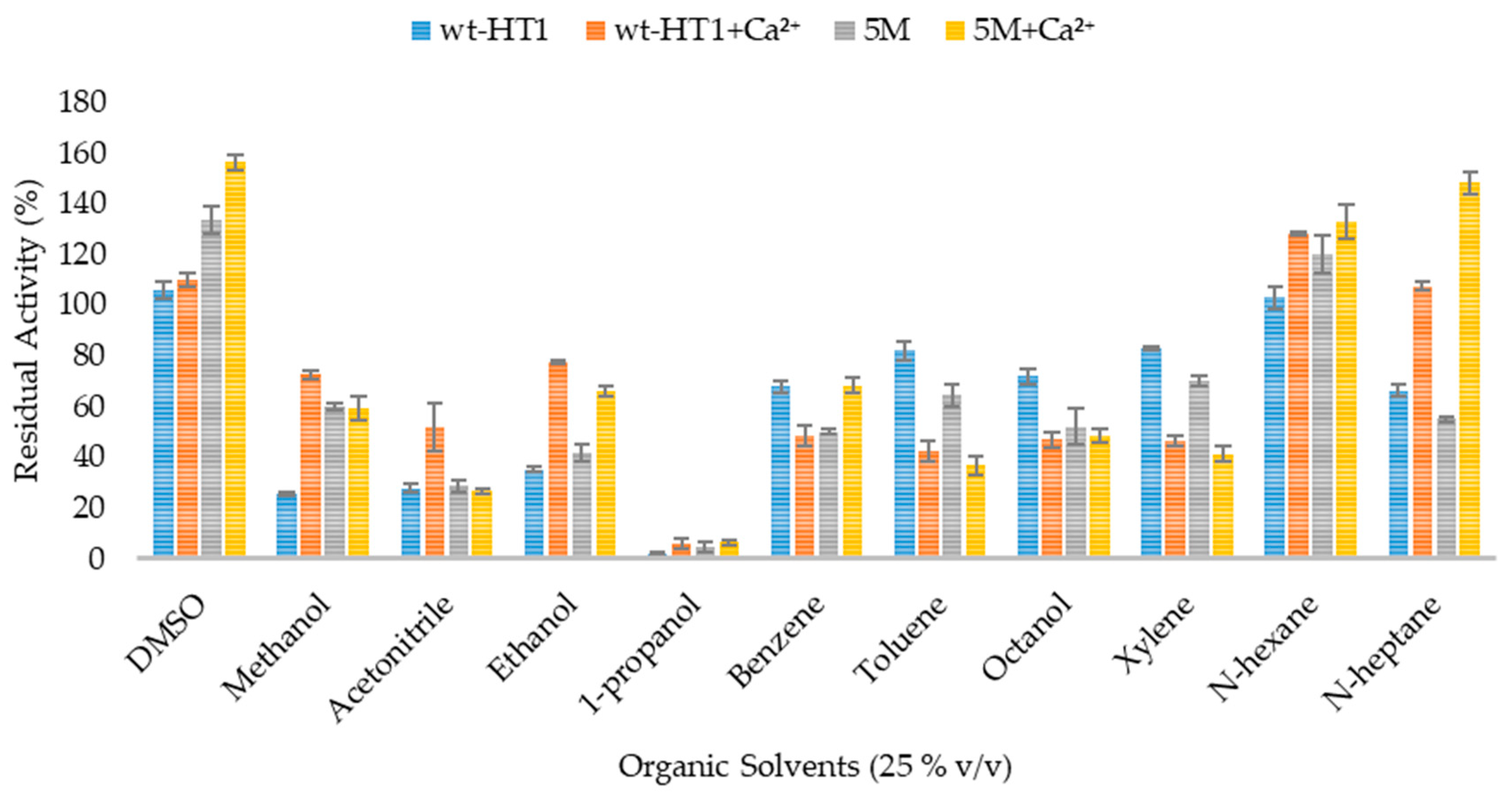
2.6. Effect of Organic Solvent and Surfactants in Lipase Activity in the Presence of Calcium Ions
2.7. Secondary Structure and Melting Point Analysis
2.8. Homology Modeling and Structural Analysis
3. Discussion
4. Materials and Methods
4.1. Construction of Mutant Lipase
4.2. Protein Preparation and Purification
4.3. Lipase Assay and Protein Estimation
4.4. Optimum Temperature and Thermostability
4.5. Optimum pH and pH Stability
4.6. Effect of Metal Ions on Lipase Activity
4.7. Thermostability of Mutant in the Presence of Calcium Ion
4.8. Effect of Organic Solvents on Lipase Activity in the Presence of Calcium Ions
4.9. Circular Dichroism Spectral Analysis
4.10. Homology Modeling and Structural Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miele, A.E.; Federici, L.; Sciara, G.; Draghi, F.; Brunori, M.; Vallone, B. Analysis of the effect of microgravity on protein crystal quality: The case of a myoglobin triple mutant. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 982. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Pan, J.; Niu, X.; Zhou, Y.; Bi, R. Influence of microgravity on protein crystal structures. Chin. Sci. Bull. 2000, 45, 1002–1006. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Linssen, A.B.; Berendsen, H.J.; Janssen, D.B. Molecular dynamics simulations as a tool for improving protein stability. Protein Eng. 2002, 15, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Law, R.J.; Capener, C.; Baaden, M.; Bond, P.J.; Campbell, J.; Patargias, G.; Arinaminpathy, Y.; Sansom, M.S.P. Membrane protein structure quality in molecular dynamics simulation. J. Mol. Graph. Model. 2005, 24, 157–165. [Google Scholar] [CrossRef]
- Kamaraj, B.; Purohit, R. In silico screening and molecular dynamics simulation of disease-associated nsSNP in TYRP1 gene and its structural consequences in OCA3. BioMed Res. Int. 2013, 2013, 697051. [Google Scholar] [CrossRef]
- Rathi, P.C.; Fulton, A.; Jaeger, K.E.; Gohlke, H. Application of Rigidity Theory to the Thermostabilization of Lipase A from Bacillus subtilis. PLoS Comput. Biol. 2016, 12, e1004754. [Google Scholar] [CrossRef]
- Singh, B.; Bulusu, G.; Mitra, A. Understanding the thermostability and activity of Bacillus subtilis lipase mutants: Insight from molecular dynamics simulations. J. Phys. Chem. B. 2015, 119, 392–409. [Google Scholar] [CrossRef]
- Park, H.J.; Joo, J.C.; Park, K.; Kim, Y.H.; Yoo, Y.J. Prediction of the solvent affecting site and the computational design of stable Candida antarctica lipase B in a hydrophilic organic solvent. J. Biotechnol. 2013, 163, 346–352. [Google Scholar] [CrossRef]
- Ni, Z.; Jin, R.; Chen, H.; Lin, X. Just an additional hydrogen bond can dramatically reduce the catalytic activity of Bacillus subtilis lipase A I12T mutant: An integration of computational modeling and experimental analysis. Comput. Biol. Med. 2013, 43, 1882–1888. [Google Scholar] [CrossRef]
- Lan, D.; Wang, Q.; Xu, J.; Zhou, P.; Yang, B.; Wang, Y. Residue Asn277 affects the stability and substrate specificity of the SMG1 lipase from Malassezia globose. Int. J. Mol. Sci. 2015, 16, 7273–7288. [Google Scholar] [CrossRef]
- Bertoldo, J.B.; Razzera, G.; Vernal, J.; Brod, F.C.A.; Arisi, A.C.M.; Terenzi, H. Structural stability of Staphylococcus xylosus lipase is modulated by Zn2+ ions. Biochim. Biophys. Acta 2011, 1814, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Vogt, G.; Woell, S.; Argos, P. Protein thermal Stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 1997, 269, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Kar, T.; Scheiner, S. Comparison of cooperativity in CH-O and OH-O hydrogen bonds. J. Phys. Chem. A 2006, 108, 9161–9168. [Google Scholar] [CrossRef]
- Ragone, R. Hydrogen-bonding classes in proteins and their contribution to the unfolding reaction. Protein Sci. 2001, 10, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Goomber, S.; Kumar, R.; Singh, R.; Mishra, N.; Kaur, J. Point Mutation Gln121-Arg increased temperature optima of Bacillus lipase (1.4 subfamily) by fifteen degrees. Int. J. Biol. Macromol. 2016, 88, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Joo, J.C.; Park, K.; Kim, Y.H.; Yoo, Y.J. Stabilization of Candida antarctica Lipase B in Hydrophilic Organic Solvent by Rational Design of Hydrogen Bond. Biotechnol. Bioprocess Eng. 2012, 17, 722–728. [Google Scholar] [CrossRef]
- Kawata, T.; Ogino, H. Amino acid residues involved in organic solvent-stability of the LST-03 lipase. Biochem. Biophys. Res. Commun. 2010, 400, 384–388. [Google Scholar] [CrossRef]
- Yedavalli, P.; Rao, N.M. Engineering the loops in a lipase for stability in DMSO. Protein Eng. Des. Sel. 2013, 26, 317–324. [Google Scholar] [CrossRef]
- Sharma, S.; Kanwar, S.S. Organic Solvent Tolerant Lipases and Applications. Sci. World J. 2014, 2014, 625258. [Google Scholar] [CrossRef]
- Choi, W.C.; Kim, M.H.; Ro, H.S.; Ryu, S.R.; Oh, T.K.; Lee, J.K. Zinc in lipase L1 from Geobacillus stearothermophilus L1 and structural implications on thermal stability. FEBS Lett. 2005, 579, 3461–3466. [Google Scholar] [CrossRef]
- Simons, J.W.F.A.; van Kampen, M.D.; Ubarretxena-Belandia, I.; Cox, R.C.; Alves dos Santos, C.M.; Egmond, M.R.; Verheij, H.M. Identification of a Calcium Binding Site in Staphylococcus hyicus Lipase: Generation of Calcium-Independent Variants. Biochemistry 1999, 38, 2–10. [Google Scholar] [CrossRef]
- Lotrakulab, P.; Dharmsthitiab, S. Purification and characterization of lipase from Aeromonas sobria LP004. J. Biotechnol. 1997, 54, 113–120. [Google Scholar] [CrossRef]
- Tayyab, M.I.; Rashid, N.; Akhtar, M. Isolation and identification of lipase producing thermophilic Geobacillus sp. SBS-4S: cloning and characterization of the lipase. J. Biosci Bioeng. 2011, 111, 272–278. [Google Scholar] [CrossRef]
- Shibata, H.; Kato, H.; Oda, J. Calcium Ion-Dependent Reactivation of a Pseudomonas Lipase by Its Specific Modulating Protein, LipB1. J. Biochem. 1998, 123, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, H.K.; Lee, J.K.; Park, S.Y.; Oh, T.K. Thermostable lipase of Bacillus stearothermophilus: High-level Production, Purification, and Calcium-dependent Thermostability. Biosci. Biotechnol. Biochem. 2000, 64, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Sabri, S.; Rahman, R.N.Z.R.A.; Leow, T.C.; Basri, M.; Salleh, A.B. Secretory expression and characterization of a highly Ca2+-activated thermostable L2 lipase. Protein Expr. Purif. 2009, 68, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Leow, T.C.; Rahman, R.N.Z.R.A.; Basri, M. A thermoalkaliphilic lipase of Geobacillus sp. T1. Extremophiles 2007, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Hussain, C.H.A.C. Molecular Modification for Effective Purification, Scaling up Production and Separation of T1 Lipase. Ph.D. Thesis, Universiti Putra Malaysia, Serdang, Selangor, 2017. [Google Scholar]
- Matsumura, H.; Yamamoto, T.; Leow, T.C.; Mori, T.; Salleh, A.B.; Basri, M.; Inoue, T.; Kai, Y.; Rahman, R.N.Z.A. Novel cation-p interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins 2008, 70, 592–598. [Google Scholar] [CrossRef]
- Aris, S.N.A.M.; Leow, T.C.; Ali, M.S.M.; Mahiran, B.; Salleh, A.B.; Rahman, R.N.Z.A. Crystallographic Analysis of Ground and Space Thermostable T1 Lipase Crystal Obtained via Counter Diffusion Method Approach. BioMed Res. Int. 2014, 1–8. [Google Scholar]
- Ishak, S.N.H.I.; Aris, S.N.A.M.; Halim, K.B.A.; Ali, M.S.M.; Leow, T.C.; Kamarudin, N.H.A.; Masomian, M.; Rahman, R.N.Z.R.A. Molecular Dynamic Simulation of Space and Earth-Grown Crystal Structures of Thermostable T1 Lipase Geobacillus zalihae Revealed a Better Structure. Molecules 2017, 22, 1574. [Google Scholar] [CrossRef] [PubMed]
- Maiangwa, J.; Ali, M.S.M.; Salleh, A.B.; Rahman, R.N.Z.R.A.; Normi, Y.M.; Shariff, F.M.; Leow, T.C. Lid opening and conformational stability of T1 Lipase is mediated by increasing chain length polar solvents. PeerJ 2017, 5, e3341. [Google Scholar] [CrossRef]
- Choo, D. Analysis of the Structure-stability Relationship of Cold-adapted Lipase PsLip1 from Homology Modeling. Genom. Inform. 2011, 9, 79–84. [Google Scholar] [CrossRef]
- Pace, C.N.; Fu, H.; Fryar, K.L.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K.; et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef]
- Wu, J.P.; Li, M.; Zhou, Y.; Yang, L.R.; Xu, G. Introducing a salt bridge into the lipase of Stenotrophomonas maltophilia results in a very large increase in thermal stability. Biotechnol. Lett. 2015, 37, 403–407. [Google Scholar] [CrossRef]
- Dror, A.; Shemesh, E.; Dayan, N.; Fishman, A. Protein engineering by random mutagenesis and structure-guided consensus of Geobacillus stearothermophilus lipase T6 for enhanced stability in methanol. Appl. Environ. Microbiol. 2014, 80, 1515. [Google Scholar] [CrossRef]
- Sharma, M.; Kumar, R.; Singh, R.; Kaur, J. Thirty-degree shift in optimum temperature of a thermophilic lipase by a single-point mutation: effect of serine to threonine mutation on structural flexibility. Mol. Cell Biochem. 2017, 430, 21–30. [Google Scholar] [CrossRef]
- Rakesh, K.; Nisha, C.; Ranvir, S.; Pushpender, K.S.; Jagdeep, K. Engineering of A Lipase towards thermostability: Studies on Additive Effect of the two Thermo-Stabilising Mutations at Protein Surface. Adv. Genet Eng. 2015, 4, 126. [Google Scholar]
- Santarossa, G.; Lafranconi, P.G.; Alquati, C.; DeGioia, L.; Alberghina, L.; Fantucci, P.; Lotti, M. Mutations in the ‘‘lid’’ region affect chain length specificity and thermostability of a Pseudomonas fragi lipase. FEBS Lett. 2005, 579, 2383–2386. [Google Scholar] [CrossRef]
- Veno, J.; Kamarudin, N.H.A.; Ali, M.S.M.; Masomian, M.; Rahman, R.N.Z.R.A. Directed evolution of recombinant C-terminal truncated Staphylococcus epidermidis lipase AT2 for enhancement of thermostability. Int. J. Mol. Sci. 2017, 18, 2202. [Google Scholar] [CrossRef]
- Khaleghinejad, S.H.; Motalleb, G.; Karkhane, A.A.; Aminzadeh, S.; Yakhchali, B. Study the effect of F17S mutation on the chimeric Bacillus thermocatenulatus lipase. J. Genet. Eng. Biotechnol. 2016, 14, 83–89. [Google Scholar] [CrossRef]
- Singh, B.; Bulusu, G.; Mitra, A. Effects of point mutations on the thermostability of B. subtilis lipase: investigating nonadditivity. J. Comput. Aided Mol. Des. 2016, 30, 899–916. [Google Scholar] [CrossRef] [PubMed]
- Ghori, M.I.; Iqbal, M.J.; Hameed, A. Characterization of a Novel Lipase From Bacillus sp. Isolated From Tannery Wastes. Braz. J. Microbiol. 2011, 42, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Gao, R.; Diao, N.; Xie, G.; Gao, G.; Cao, S. Cloning, expression and characterization of an organic solvent tolerant lipase from Pseudomonas fluorescens JCM5963. J. Mol. Catal. B Enzym. 2009, 56, 78–84. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhou, X.; Chen, Z. Cloning, expression, and biochemical characterization of a thermostable lipase from Geobacillus stearothermophilus JC. World J. Microbiol. Biotechnol. 2010, 26, 747–751. [Google Scholar] [CrossRef]
- Kambourova, M.; Kirilova, N.; Mandeva, R.; Derekova, A. Purification and properties of thermostable lipase from a thermophilic Bacillus stearothermophilus MC 7. J. Mol. Catal. B Enzym. 2003, 22, 307–313. [Google Scholar] [CrossRef]
- Lopes, M.F.S.; Leitão, A.L.; Regalla, M.; Marques, J.J.F.; Carrondo, M.J.T.; Crespo, M.T.B. Characterization of a highly thermostable extracellular lipase from Lactobacillus plantarum. Int. J. Food Microbiol. 2002, 76, 107–115. [Google Scholar] [CrossRef]
- Kukreja, V.; Bera, M.B. Lipase from Pseudomonas aeruginosa MTCC 2488: Partial purification, characterization and calcium dependent thermostability. Indian J. Biotechnol. 2004, 4, 222–226. [Google Scholar]
- Sakinc, T.; Kleine, B.; Gatermann, S.G. Biochemical characterization of the surface-associated lipase of Staphylococcus saprophyticus. FEMS Microbiol. Lett. 2007, 274, 335–341. [Google Scholar] [CrossRef]
- Rashid, N.; Shimada, Y.; Ezaki, S.; Atomi, H.; Imanaka, T. Low-Temperature Lipase from Psychrotrophic Pseudomonas sp. Strain KB700A. Appl. Environm. Microbiol. 2001, 67, 4064–4069. [Google Scholar] [CrossRef]
- Li, S.X.; Ma, Q. Essential Role of Gly33 in a Novel Organic Solvent-Tolerant Lipase from Serratia marcescens ECU1010 as Determined by Site-Directed Mutagenesis. Appl. Biochem. Biotechnol. 2014, 172, 2945–2954. [Google Scholar] [CrossRef]
- Hertadi, R.; Widhyastuti, H. Effect of Ca2+ Ion to The Activity and Stability of Lipase Isolated from Chromohalobacter japonicas BK-AB18. International Symposium on Applied Chemistry 2015. Procedia Chem. 2015, 16, 306–313. [Google Scholar] [CrossRef]
- Shirazi, N.H.; Ranjbar, B.; Khajeh, K.; Moghadam, T.T. Structure–function analysis of a new bacterial lipase: Effect of local structure reorganization on lipase activity. Int. J. Biol. Macromol. 2013, 54, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, H. Calcium ion-induced formation of β-sheet/-turn structure leading to alteration of osteogenic activity of bone morphogenetic protein-2. Sci. Rep. 2015, 5, 12694. [Google Scholar] [CrossRef] [PubMed]
- Zarzhitsky, S.; Edri, H.; Azoulay, Z.; Cohen, I.; Ventura, Y.; Gitelman, A.; Rapaport, H. The Effect of pH and Calcium Ions on the Stability of Amphiphilic and Anionic β-Sheet Peptide Hydrogels. Biopolymers 2013, 100, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.Z.; Ma, B.L. Influence of metal ions on folding pathway and conformational stability of bovine serum albumin. J. Mol. Struct. 2008, 877, 44–49. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Rhee, J.S. A simple and rapid colorimetric method for determination of free fatty acids for lipase assay. J. Am. Oil Chem. Soc. 1986, 63, 89. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Analysis of the kinetics of folding of proteins and peptides using circular dichroism. Nat. Protoc. 2007, 1, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.W.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by C-alpha geometry: phi, psi and C-beta deviation. Proteins Struct. Funct. Genet. 2002, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Lüthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef] [PubMed]




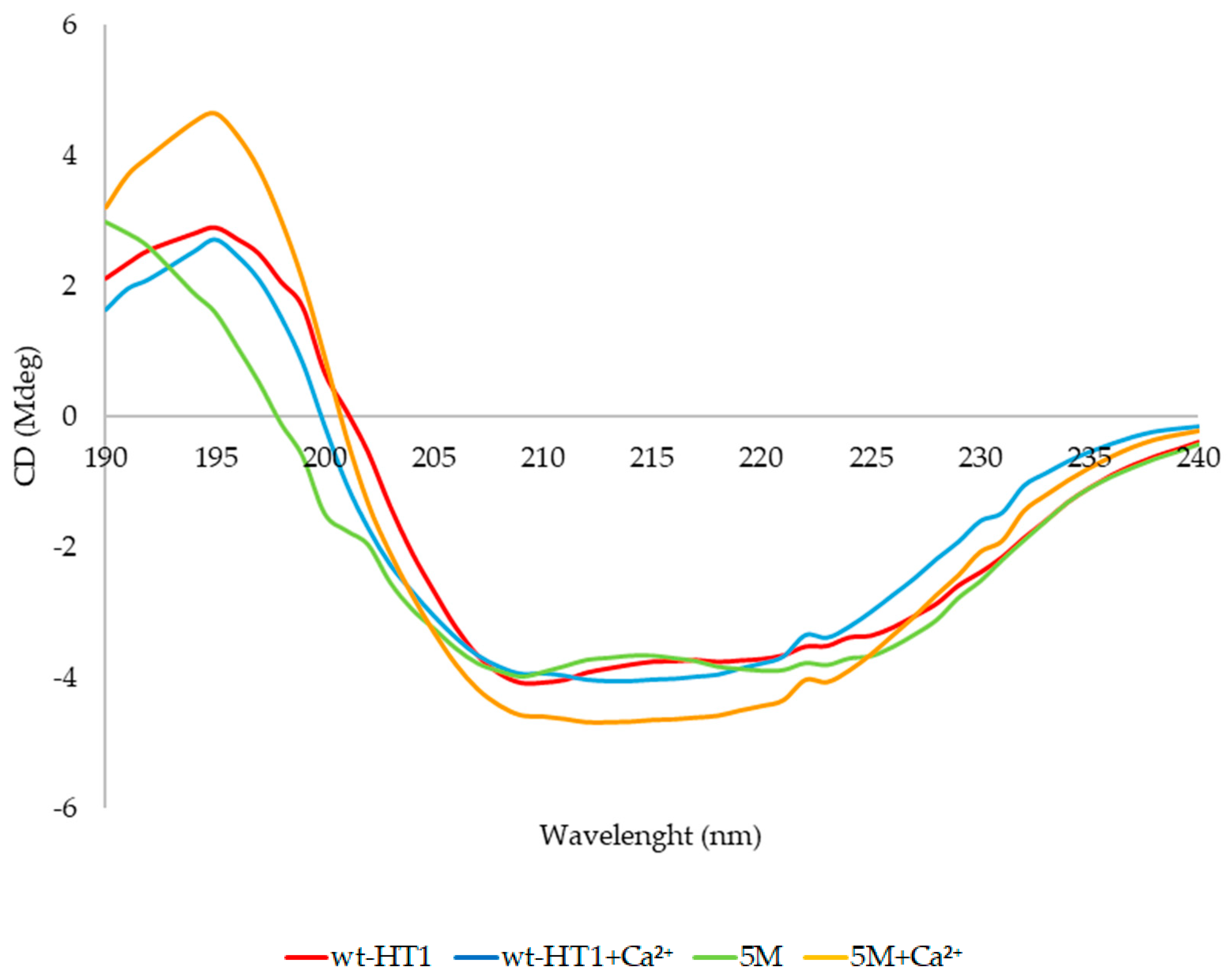
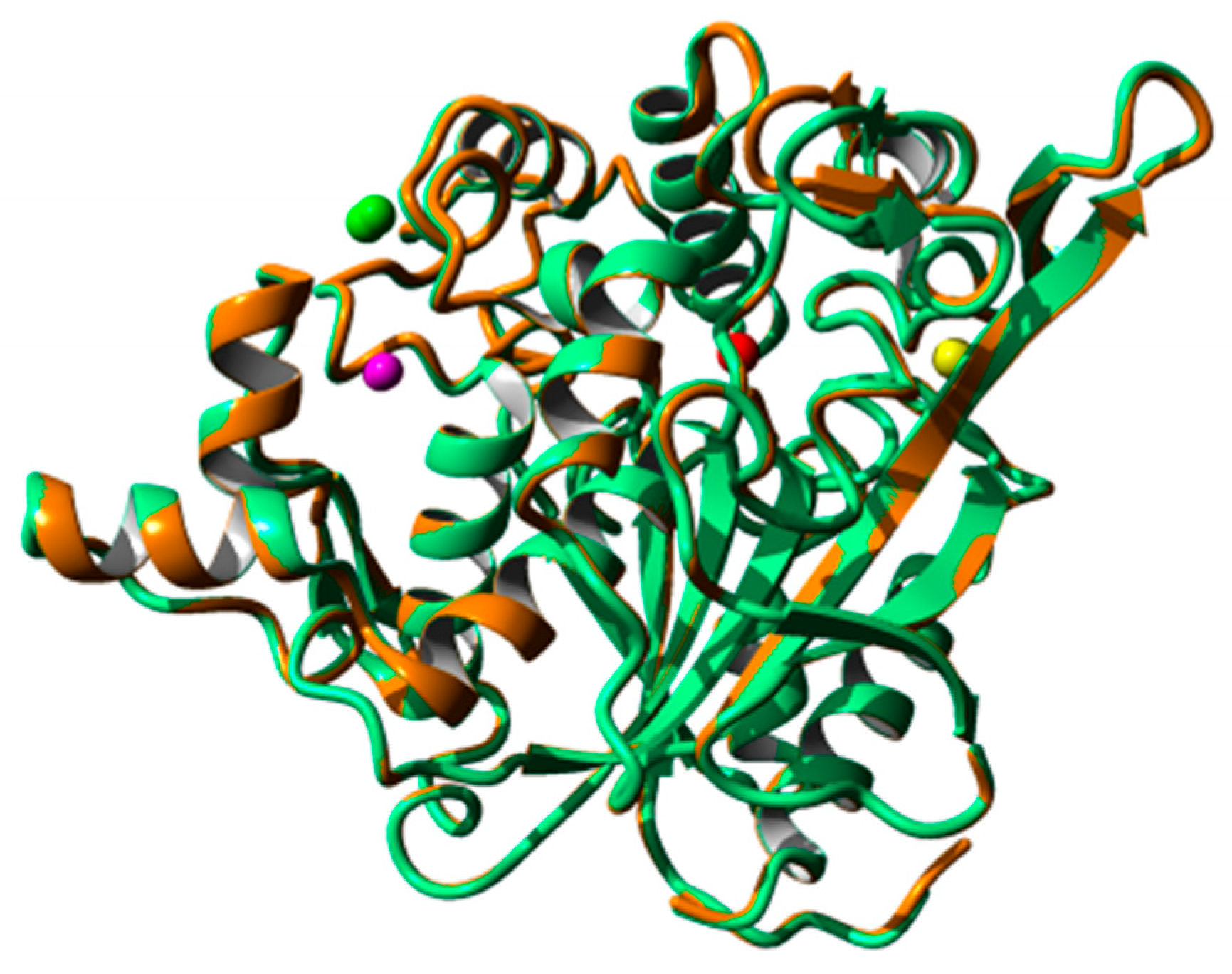
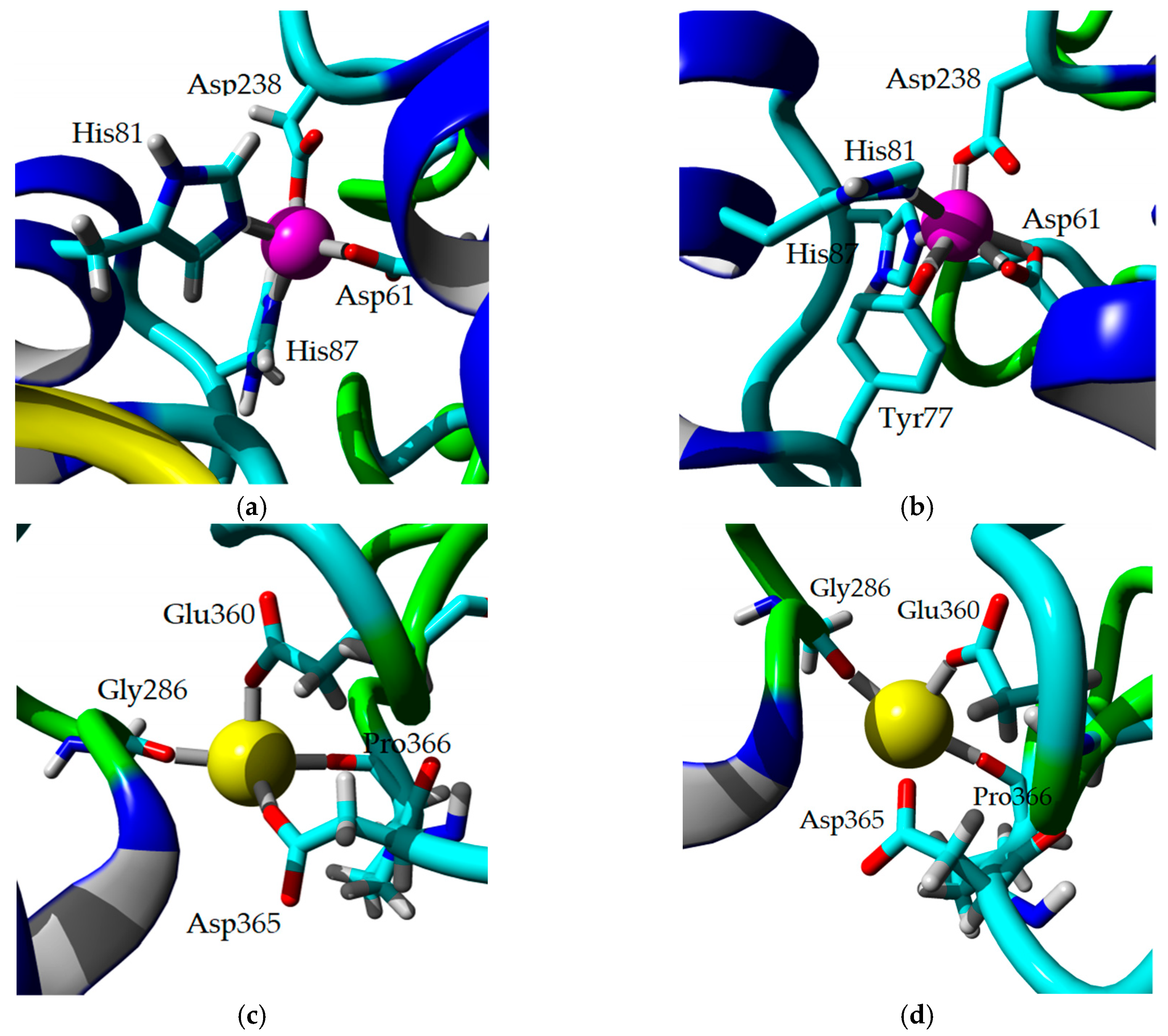


| Lipase | Concentration (mM) | Na2+ | Ca2+ | Mg2+ | Fe3 | Ni2+ | Cu2+ | Zn2+ |
|---|---|---|---|---|---|---|---|---|
| 5M | 1 | 91.5 ± 6.7 | 150.0 ± 2.6 | 59.6 ± 2.2 | 18.8 ± 5.9 | 86.4 ± 5 | 15.3 ± 2.3 | 75.0 ± 0.7 |
| 5 | 76.7 ± 0.4 | 193.0 ± 5.0 | 53.7 ± 1.8 | 7.8 ± 2.1 | 78.0 ± 8.9 | 0.0 ± 0.0 | 21.8 ± 1.2 | |
| wt-HT1 | 1 | 102.6 ± 4.1 | 122.0 ± 1.7 | 81.6 ± 5.2 | 55.9 ± 1.3 | 106.0 ± 2.4 | 51.2 ± 4.2 | 78.7 ± 1.2 |
| 5 | 103.3 ± 0.6 | 123.0 ± 4.3 | 101.0 ± 3.8 | 9.9 ± 2.9 | 93.0 ± 6.5 | 15.6 ± 4.0 | 25.2 ± 5.4 |
| Lipases with and without Ca2⁺ | α-Helix | β-Sheet | Turn | Random |
|---|---|---|---|---|
| wt-HT1 + Ca2⁺ | 15.30% | 49.90% | 7.60% | 27.20% |
| 5M + Ca2⁺ | 20.30% | 46.30% | 8.10% | 25.30% |
| wt-HT1 | 28.70% | 17.60% | 22.40% | 31.30% |
| 5M | 23.90% | 18.30% | 18.00% | 39.70% |
| Lipases with and without Ca2⁺ | Melting Temperature (Tm) |
|---|---|
| wt-HT1 + Ca2⁺ | 83.0 °C ± 0.2 |
| 5M + Ca2⁺ | 76.0 °C ± 0.7 |
| wt-HT1 | 70.9 °C ± 0.1 |
| 5M | 67.7 °C ± 0.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishak, S.N.H.; Masomian, M.; Kamarudin, N.H.A.; Ali, M.S.M.; Leow, T.C.; Rahman, R.N.Z.R.A. Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion. Int. J. Mol. Sci. 2019, 20, 2561. https://doi.org/10.3390/ijms20102561
Ishak SNH, Masomian M, Kamarudin NHA, Ali MSM, Leow TC, Rahman RNZRA. Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion. International Journal of Molecular Sciences. 2019; 20(10):2561. https://doi.org/10.3390/ijms20102561
Chicago/Turabian StyleIshak, Siti Nor Hasmah, Malihe Masomian, Nor Hafizah Ahmad Kamarudin, Mohd Shukuri Mohamad Ali, Thean Chor Leow, and Raja Noor Zaliha Raja Abd. Rahman. 2019. "Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion" International Journal of Molecular Sciences 20, no. 10: 2561. https://doi.org/10.3390/ijms20102561
APA StyleIshak, S. N. H., Masomian, M., Kamarudin, N. H. A., Ali, M. S. M., Leow, T. C., & Rahman, R. N. Z. R. A. (2019). Changes of Thermostability, Organic Solvent, and pH Stability in Geobacillus zalihae HT1 and Its Mutant by Calcium Ion. International Journal of Molecular Sciences, 20(10), 2561. https://doi.org/10.3390/ijms20102561