Knockdown of NANOG Reduces Cell Proliferation and Induces G0/G1 Cell Cycle Arrest in Human Adipose Stem Cells
Abstract
:1. Introduction
2. Results
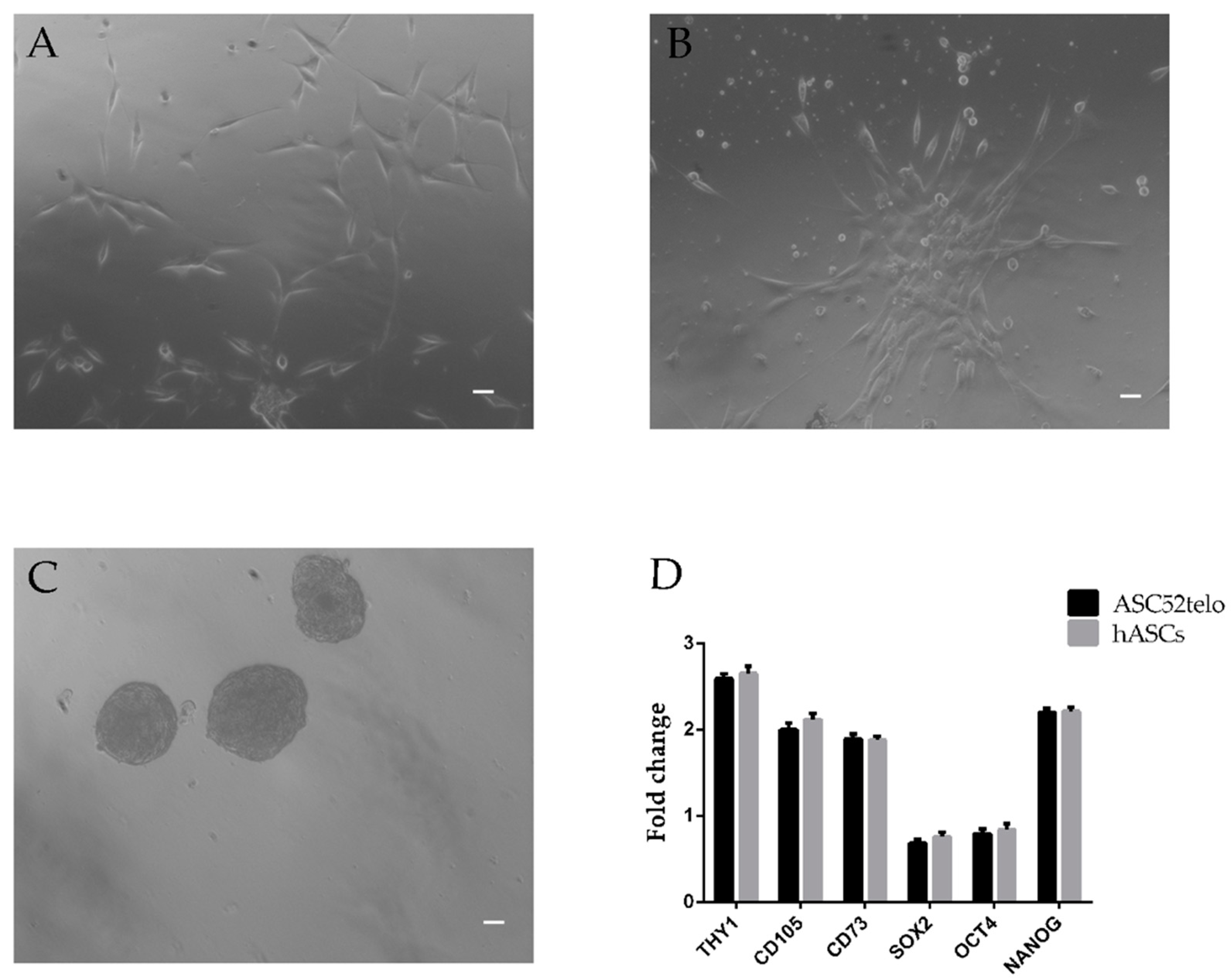
2.1. hASC Isolation and Characterization
2.2. Knockdown of NANOG Silencing by Lentivirus
2.3. Analysis of PREF1
2.4. Downregulation of NANOG Inhibited Cell Proliferation, Increased Population Doubling Time and Reduced Differentiation Potential
2.5. Knockdown of NANOG Inhibited the Expression of CCND1/Cycline D1, Enhanced the Expression of p21 and p27 and Induced Cell Cycle Block in the G0/G1 Phase
2.6. Knockdown of NANOG Regulates DNMT1
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Cell Infection with Lentiviral Particles
4.2. RNA Isolation and Quantitative RT-PCR
4.3. Western Blot Analysis
4.4. Analysis of Cell Cycle Status of MSCs with Lentivirus
4.5. Colony-Forming Assay
4.6. Population Doubling and Cell Proliferation Curve
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MSC | Mesenchymal Stem Cell |
| hASC | human Adipose Stem Cell |
| p21/CDKN1A | Cyclin-dependent kinase inhibitor 1A |
| p27/CDKN1B | Cyclin-dependent kinase inhibitor 1B |
References
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Thomson, J.A. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007, 17, 42–49. [Google Scholar] [CrossRef]
- Kim, J.; Chu, J.; Shen, X.; Wang, J.; Orkin, S.H. An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell 2008, 132, 1049–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darr, H. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development 2006, 133, 1193–1201. [Google Scholar] [CrossRef]
- Hough, S.R.; Clements, I.; Welch, P.J.; Wiederholt, K.A. Differentiation of Mouse Embryonic Stem Cells after RNA Interference-Mediated Silencing of OCT4 and Nanog. Stem Cells 2006, 24, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Hyslop, L.; Stojkovic, M.; Armstrong, L.; Walter, T.; Stojkovic, P.; Przyborski, S.; Herbert, M.; Murdoch, A.; Strachan, T.; Lako, M. Downregulation of NANOG Induces Differentiation of Human Embryonic Stem Cells to Extraembryonic Lineages. Stem Cells 2005, 23, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Oron, E.; Nelson, B.; Razis, S.; Ivanova, N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 2012, 10, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Li, A.I.; Hokugo, A.; Jarrahy, R.; Zuk, P.A. Human adipose tissue as a source of multipotent stem cells. In Stem Cells in Aesthetic Procedures: Art, Science, and Clinical Techniques; Springer-Verlag GmbH: Heidelberg, Germany, 2014. [Google Scholar]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.; Futrell, J.W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar]
- Gronthos, S.; Franklin, D.M.; Leddy, H.A.; Robey, P.G.; Storms, R.W.; Gimble, J.M. Surface protein characterization of human adipose tissue-derived stromal cells. J. Cell. Physiol. 2001, 189, 54–63. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- De Francesco, F.; Tirino, V.; Desiderio, V.; Ferraro, G.; D’Andrea, F.; Giuliano, M.; Libondi, G.; Pirozzi, G.; De Rosa, A.; Papaccio, G. Human CD34+/CD90+ ASCs are capable of growing as sphere clusters, producing high levels of VEGF and forming capillaries. PLoS ONE 2009, 4, e6537. [Google Scholar] [CrossRef] [PubMed]
- Pitrone, M.; Pizzolanti, G.; Tomasello, L.; Coppola, A.; Morini, L.; Pantuso, G.; Ficarella, R.; Guarnotta, V.; Perrini, S.; Giorgino, F.; et al. NANOG plays a hierarchical role in the transcription network regulating the pluripotency and plasticity of adipose tissue-derived stem cells. Int. J. Mol. Sci. 2017, 18, 1107. [Google Scholar] [CrossRef]
- Potdar, P.D.; Sutar, J.P. Establishment and molecular characterization of mesenchymal stem cell lines derived from human visceral & subcutaneous adipose tissues. J. Stem Cells Regen. Med. 2010, 6, 26–35. [Google Scholar] [PubMed]
- Langroudi, L.; Forouzandeh, M.; Soleimani, M.; Atashi, A.; Golestaneh, A.F. Induction of differentiation by down-regulation of Nanog and Rex-1 in cord blood derived unrestricted somatic stem cells. Mol. Biol. Rep. 2013, 40, 4429–4437. [Google Scholar] [CrossRef] [PubMed]
- Perrini, S.; Ficarella, R.; Picardi, E.; Cignarelli, A.; Barbaro, M.; Nigro, P.; Peschechera, A.; Palumbo, O.; Carella, M.; De Fazio, M.; et al. Differences in Gene Expression and Cytokine Release Profiles Highlight the Heterogeneity of Distinct Subsets of Adipose Tissue-Derived Stem Cells in the Subcutaneous and Visceral Adipose Tissue in Humans. PLoS ONE 2013, 8, e57892. [Google Scholar] [CrossRef] [PubMed]
- Lengner, C.J.; Camargo, F.D.; Hochedlinger, K.; Welstead, G.G.; Zaidi, S.; Gokhale, S.; Scholer, H.R.; Tomilin, A.; Jaenisch, R. Oct4 Expression Is Not Required for Mouse Somatic Stem Cell Self-Renewal. Cell Stem Cell 2007, 1, 403–415. [Google Scholar] [CrossRef] [Green Version]
- Rodeheffer, M.S.; Birsoy, K.; Friedman, J.M. Identification of White Adipocyte Progenitor Cells In Vivo. Cell 2008, 135, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Smas, C.M.; Sul, H.S. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 1993, 73, 725–734. [Google Scholar] [CrossRef]
- Tang, W.; Zeve, D.; Suh, J.M.; Bosnakovski, D.; Kyba, M.; Hammer, R.E.; Tallquist, M.D.; Graff, J.M. White fat progenitor cells reside in the adipose vasculature. Science 2008, 322, 583–586. [Google Scholar] [CrossRef]
- Tseng, Y.H.; Butte, A.J.; Kokkotou, E.; Yechoor, V.K.; Taniguchi, C.M.; Kriauciunas, K.M.; Cypess, A.M.; Niinobe, M.; Yoshikawa, K.; Patti, M.E.; et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat. Cell Biol. 2005, 7, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hudak, C.; Sul, H.S. Role of preadipocyte factor 1 in adipocyte differentiation. Clin. Lipidol. 2010, 5, 109–115. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, K.-A.; Kim, J.-H.; Sul, H.S. Pref-1, a Preadipocyte Secreted Factor That Inhibits Adipogenesis. J. Nutr. 2018, 136, 2953–2956. [Google Scholar] [CrossRef] [PubMed]
- Mitterberger, M.C.; Lechner, S.; Mattesich, M.; Kaiser, A.; Probst, D.; Wenger, N.; Pierer, G.; Zwerschke, W. DLK1(PREF1) is a negative regulator of adipogenesis in CD105+/CD90+/CD34+/CD31-/FABP4- adipose-derived stromal cells from subcutaneous abdominal fat pats of adult women. Stem Cell Res. 2012, 9, 35–48. [Google Scholar] [CrossRef]
- Biniszkiewicz, D.; Gribnau, J.; Ramsahoye, B.; Gaudet, F.; Eggan, K.; Humpherys, D.; Mastrangelo, M.-A.; Jun, Z.; Walter, J.; Jaenisch, R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002, 22, 2124–2135. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.C.; Su, P.F.; Huang, Y.F.; Yew, T.L.; Hung, S.C. Oct4 and Nanog Directly Regulate Dnmt1 to Maintain Self-Renewal and Undifferentiated State in Mesenchymal Stem Cells. Mol. Cell 2012, 47, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Wade Harper, J.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- Xiong, Y.; Hannon, G.J.; Zhang, H.; Casso, D.; Kobayashi, R.; Beach, D. P21 is a universal inhibitor of cyclin kinases. Nature 1993, 366, 701–704. [Google Scholar] [CrossRef]
- Aleem, E.; Kiyokawa, H.; Kaldis, P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 2005, 7, 831–836. [Google Scholar] [CrossRef]
- Gartel, A.L.; Tyner, A.L. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp. Cell Res. 1999, 246, 280–289. [Google Scholar] [CrossRef]
- Fouse, S.D.; Shen, Y.; Pellegrini, M.; Cole, S.; Meissner, A.; Van Neste, L.; Jaenisch, R.; Fan, G. Promoter CpG Methylation Contributes to ES Cell Gene Regulation in Parallel with Oct4/Nanog, PcG Complex, and Histone H3 K4/K27 Trimethylation. Cell Stem Cell 2008, 2, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Lei, H.; Oh, S.P.; Okano, M.; Jüttermann, R.; Goss, K.A.; Jaenisch, R.; Li, E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 1996, 122, 3195–3205. [Google Scholar] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Coppola, A.; Tomasello, L.; Pitrone, M.; Cillino, S.; Richiusa, P.; Pizzolanti, G.; Giordano, C. Human limbal fibroblast-like stem cells induce immune-tolerance in autoreactive T lymphocytes from female patients with Hashimoto’s thyroiditis. Stem Cell Res. Ther. 2017, 8, 154. [Google Scholar] [CrossRef] [PubMed]
- Tomasello, L.; Mauceri, R.; Coppola, A.; Pitrone, M.; Pizzo, G.; Campisi, G.; Pizzolanti, G.; Giordano, C. Mesenchymal stem cells derived from inflamed dental pulpal and gingival tissue: A potential application for bone formation. Stem Cell Res. Ther. 2017, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef] [PubMed]





| Primary Antibody/Localization Marker | Code Number | Diluition | Incubation |
|---|---|---|---|
| NANOG, nuclear and cytoplasmatic | sc-293121, Santa Cruz Biotechnology | 1:500 | o/n, 4 °C |
| DNMT1, nuclear | sc-271729, Santa Cruz Biotechnology | 1:500 | o/n, 4 °C |
| B-Actin clone AC-74 | A5316, Sigma Aldrich | 1:10,000 | o/n, 4 °C |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitrone, M.; Pizzolanti, G.; Coppola, A.; Tomasello, L.; Martorana, S.; Pantuso, G.; Giordano, C. Knockdown of NANOG Reduces Cell Proliferation and Induces G0/G1 Cell Cycle Arrest in Human Adipose Stem Cells. Int. J. Mol. Sci. 2019, 20, 2580. https://doi.org/10.3390/ijms20102580
Pitrone M, Pizzolanti G, Coppola A, Tomasello L, Martorana S, Pantuso G, Giordano C. Knockdown of NANOG Reduces Cell Proliferation and Induces G0/G1 Cell Cycle Arrest in Human Adipose Stem Cells. International Journal of Molecular Sciences. 2019; 20(10):2580. https://doi.org/10.3390/ijms20102580
Chicago/Turabian StylePitrone, Maria, Giuseppe Pizzolanti, Antonina Coppola, Laura Tomasello, Stefania Martorana, Gianni Pantuso, and Carla Giordano. 2019. "Knockdown of NANOG Reduces Cell Proliferation and Induces G0/G1 Cell Cycle Arrest in Human Adipose Stem Cells" International Journal of Molecular Sciences 20, no. 10: 2580. https://doi.org/10.3390/ijms20102580
APA StylePitrone, M., Pizzolanti, G., Coppola, A., Tomasello, L., Martorana, S., Pantuso, G., & Giordano, C. (2019). Knockdown of NANOG Reduces Cell Proliferation and Induces G0/G1 Cell Cycle Arrest in Human Adipose Stem Cells. International Journal of Molecular Sciences, 20(10), 2580. https://doi.org/10.3390/ijms20102580





