The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia
Abstract
1. Introduction
2. Epigenetics and Normal Placental Development
2.1. Description of the Placenta and Placental Cells
2.2. Human Placental Development
2.3. Epigenetics Mechanisms in Placental Development
2.3.1. DNA Methylation
Differentiation of Stem Cells
Regulation of Homeotic Genes
Placental Development and Cancer Pathways
2.3.2. Non-coding RNAs and Epigenetic Regulation of Placenta Development
Definition
MiRNA and Normal Human Placental Development
lncRNA and Normal Human Placental Development
2.3.3. Histone Modifications in the Developing Placenta
2.3.4. Imprinting and Placental Development
Placentation and the Materno-Fetal Conflict
Definition of Imprinted Genes and Links with Viviparity
Example of the H19-IGF2 Cluster; Cross Species Conservation of Imprinted Genes
3. Epigenetic Alterations in Preeclampsia
3.1. DNA Methylation Alterations in Preeclampsia
3.1.1. Methylation Alterations in the Preeclamptic Placenta
Common Alterations of Gene Expression in PE are Associated to Methylation Alterations
Limits of the Genome-Wide, Multicellular Approach for Preeclampsia Methylation Profiling
Single-Cell Analysis, the Next Frontier to Methylation Epigenomic Approaches
An Example of Specific Gene Alterations of Methylation: Regulation of Invasion
3.1.2. Maternal Blood Epigenetic Marks in Preeclampsia
3.1.3. Maternal Endothelial Cells
3.1.4. Cord Blood Cells
3.2. Non Coding RNAs
3.2.1. LncRNAs in Preeclampsia
MALAT-1
MEG3
RNA-ATB
PVT1, TUG1 and DIAPH2-AS1: Regulating Gene Expression through Recruitment of Chromatin Remodeling Complexes
3.2.2. micro RNA and Preeclampsia
microRNAs in Preeclampsia
miR-210
miR-155
Circulating miR-155
miR-155 in Endothelial Cells
miR-155 in Vascular Smooth Muscle Cells
Potential Biomarkers: microRNAs Circulating in Maternal Plasma
3.2.3. Additional Considerations on the Analysis of lncRNA Functions
Possible Caveats of the Current Trophoblast In Vitro Models
What about the Syncytiotrophoblast?
3.3. Histone Modifications
3.4. Imprinting
4. Perspectives and Conclusions
Supplementary Materials
Funding
Conflicts of Interest
References
- Steegers, E.A.; von Dadelszen, P.; Duvekot, J.J.; Pijnenborg, R. Pre-eclampsia. Lancet 2010, 376, 631–644. [Google Scholar] [CrossRef]
- Roland, C.S.; Hu, J.; Ren, C.E.; Chen, H.; Li, J.; Varvoutis, M.S.; Leaphart, L.W.; Byck, D.B.; Zhu, X.; Jiang, S.W. Morphological changes of placental syncytium and their implications for the pathogenesis of preeclampsia. Cell. Mol. Life Sci. 2016, 73, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Redman, C.W.; Sargent, I.L. Latest advances in understanding preeclampsia. Science 2005, 308, 1592–1594. [Google Scholar] [CrossRef] [PubMed]
- Fisher, S.J. Why is placentation abnormal in preeclampsia? Am. J. Obstet. Gynecol. 2015, 213, S115–S122. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, B. Placental origins of preeclampsia: Challenging the current hypothesis. Hypertension 2008, 51, 970–975. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, B. The Critical Role of Abnormal Trophoblast Development in the Etiology of Preeclampsia. Curr. Pharm. Biotechnol. 2018, 19, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Doridot, L.; Passet, B.; Mehats, C.; Rigourd, V.; Barbaux, S.; Ducat, A.; Mondon, F.; Vilotte, M.; Castille, J.; Breuiller-Fouche, M.; et al. Preeclampsia-like symptoms induced in mice by fetoplacental expression of STOX1 are reversed by aspirin treatment. Hypertension 2013, 61, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Sibley, C.P.; Pardi, G.; Cetin, I.; Todros, T.; Piccoli, E.; Kaufmann, P.; Huppertz, B.; Bulfamante, G.; Cribiu, F.M.; Ayuk, P.; et al. Pathogenesis of intrauterine growth restriction (IUGR)-conclusions derived from a European Union Biomed 2 Concerted Action project ‘Importance of Oxygen Supply in Intrauterine Growth Restricted Pregnancies’-a workshop report. Placenta 2002, 23 (Suppl. A), S75–S79. [Google Scholar] [CrossRef]
- Possomato-Vieira, J.S.; Khalil, R.A. Mechanisms of Endothelial Dysfunction in Hypertensive Pregnancy and Preeclampsia. Adv. Pharmacol. 2016, 77, 361–431. [Google Scholar]
- Nelissen, E.C.; van Montfoort, A.P.; Dumoulin, J.C.; Evers, J.L. Epigenetics and the placenta. Hum. Reprod. Update 2011, 17, 397–417. [Google Scholar] [CrossRef]
- Vaiman, D. Genes, epigenetics and miRNA regulation in the placenta. Placenta 2017, 52, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.P.; Price, E.M. The human placental methylome. Cold Spring Harb. Perspect. Med. 2015, 5, a023044. [Google Scholar] [CrossRef] [PubMed]
- Januar, V.; Desoye, G.; Novakovic, B.; Cvitic, S.; Saffery, R. Epigenetic regulation of human placental function and pregnancy outcome: Considerations for causal inference. Am. J. Obstet. Gynecol. 2015, 213, S182–S196. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [PubMed]
- Burton, G.J.; Fowden, A.L. The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef]
- James, J.L.; Carter, A.M.; Chamley, L.W. Human placentation from nidation to 5 weeks of gestation. Part I: What do we know about formative placental development following implantation? Placenta 2012, 33, 327–334. [Google Scholar] [CrossRef]
- Knofler, M. Critical growth factors and signalling pathways controlling human trophoblast invasion. Int. J. Dev. Biol. 2010, 54, 269–280. [Google Scholar] [CrossRef]
- Knofler, M.; Pollheimer, J. Human placental trophoblast invasion and differentiation: A particular focus on Wnt signaling. Front. Genet. 2013, 4, 190. [Google Scholar] [CrossRef]
- Rouault, C.; Clement, K.; Guesnon, M.; Henegar, C.; Charles, M.-A.; Heude, B.; Evain-Brion, D.; Degrelle, S.A.; Fournier, T. Transcriptomic signatures of villous cytotrophoblast and syncytiotrophoblast in term human placenta. Placenta 2016, 44, 83–90. [Google Scholar] [CrossRef]
- Khan, M.A.; Manna, S.; Malhotra, N.; Sengupta, J.; Ghosh, D. Expressional regulation of genes linked to immunity & programmed development in human early placental villi. Indian J. Med. Res. 2014, 139, 125–140. [Google Scholar]
- Henikoff, S.; Greally, J.M. Epigenetics, cellular memory and gene regulation. Curr. Biol. 2016, 26, R644–R648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Pradhan, S. Mammalian epigenetic mechanisms. IUBMB Life 2014, 66, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Rahat, B.; Sharma, R.; Bagga, R.; Hamid, A.; Kaur, J. Imbalance between matrix metalloproteinases and their tissue inhibitors in preeclampsia and gestational trophoblastic diseases. Reproduction 2016, 152, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Dokras, A.; Gardner, L.M.; Kirschmann, D.A.; Seftor, E.A.; Hendrix, M.J. The tumour suppressor gene maspin is differentially regulated in cytotrophoblasts during human placental development. Placenta 2002, 23, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Dokras, A.; Coffin, J.; Field, L.; Frakes, A.; Lee, H.; Madan, A.; Nelson, T.; Ryu, G.Y.; Yoon, J.G.; Madan, A. Epigenetic regulation of maspin expression in the human placenta. Mol. Hum. Reprod. 2006, 12, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Camolotto, S.A.; Racca, A.C.; Ridano, M.E.; Genti-Raimondi, S.; Panzetta-Dutari, G.M. PSG gene expression is up-regulated by lysine acetylation involving histone and nonhistone proteins. PLoS ONE 2013, 8, e55992. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.C.; Chang, C.W.; Chang, G.D.; Yao, T.P.; Chen, H. Histone deacetylase 3 binds to and regulates the GCMa transcription factor. Nucleic Acids Res. 2006, 34, 1459–1469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abell, A.N.; Jordan, N.V.; Huang, W.; Prat, A.; Midland, A.A.; Johnson, N.L.; Granger, D.A.; Mieczkowski, P.A.; Perou, C.M.; Gomez, S.M.; et al. MAP3K4/CBP-regulated H2B acetylation controls epithelial-mesenchymal transition in trophoblast stem cells. Cell Stem Cell 2011, 8, 525–537. [Google Scholar] [CrossRef]
- Ellery, P.M.; Cindrova-Davies, T.; Jauniaux, E.; Ferguson-Smith, A.C.; Burton, G.J. Evidence for transcriptional activity in the syncytiotrophoblast of the human placenta. Placenta 2009, 30, 329–334. [Google Scholar] [CrossRef]
- Fogarty, N.M.; Burton, G.J.; Ferguson-Smith, A.C. Different epigenetic states define syncytiotrophoblast and cytotrophoblast nuclei in the trophoblast of the human placenta. Placenta 2015, 36, 796–802. [Google Scholar] [CrossRef]
- Xu, Y.; Ge, Z.; Zhang, E.; Zuo, Q.; Huang, S.; Yang, N.; Wu, D.; Zhang, Y.; Chen, Y.; Xu, H.; et al. The lncRNA TUG1 modulates proliferation in trophoblast cells via epigenetic suppression of RND3. Cell Death Dis. 2017, 8, e3104. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Rui, C.; Meng, L.; Zhang, R.; Shen, R.; Ding, H.; Li, J.; Li, J.; Long, W. Long non-coding RNA RPAIN regulates the invasion and apoptosis of trophoblast cell lines via complement protein C1q. Oncotarget 2017, 8, 7637–7646. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Meng, T.; Liu, X.; Sun, M.; Tong, C.; Liu, J.; Wang, H.; Du, J. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int. J. Clin. Exp. Pathol. 2015, 8, 12718–12727. [Google Scholar] [PubMed]
- Zhang, Y.; Zou, Y.; Wang, W.; Zuo, Q.; Jiang, Z.; Sun, M.; De, W.; Sun, L. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J. Cell. Biochem. 2015, 116, 542–550. [Google Scholar] [CrossRef] [PubMed]
- Muys, B.R.; Lorenzi, J.C.; Zanette, D.L.; Lima e Bueno Rde, B.; de Araujo, L.F.; Dinarte-Santos, A.R.; Alves, C.P.; Ramao, A.; de Molfetta, G.A.; Vidal, D.O.; et al. Placenta-Enriched LincRNAs MIR503HG and LINC00629 Decrease Migration and Invasion Potential of JEG-3 Cell Line. PLoS ONE 2016, 11, e0151560. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Jiang, Z.; Yu, X.; Sun, M.; Zhang, Y.; Zuo, Q.; Zhou, J.; Yang, N.; Han, P.; Ge, Z.; et al. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PLoS ONE 2013, 8, e79598. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Huang, S.; Zou, Y.; Xu, Y.; Jiang, Z.; Zou, S.; Xu, H.; Sun, L. The Lnc RNA SPRY4-IT1 Modulates Trophoblast Cell Invasion and Migration by Affecting the Epithelial-Mesenchymal Transition. Sci. Rep. 2016, 6, 37183. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.L.; Chang, K.; Lu, L.S.; Zhao, D.; Han, J.; Zheng, Y.R.; Yan, Y.H.; Yi, P.; Guo, J.X.; Zhou, Y.G.; et al. Lentivirus-mediated RNA interference targeting the H19 gene inhibits cell proliferation and apoptosis in human choriocarcinoma cell line JAR. BMC Cell Biol. 2013, 14, 26. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Chakraborty, S.; Bhattacharya, A.; Biswas, A.; Ain, R. MicroRNA regulation of Transthyretin in trophoblast differentiation and Intra-Uterine Growth Restriction. Sci. Rep. 2017, 7, 16548. [Google Scholar] [CrossRef]
- Umemura, K.; Ishioka, S.; Endo, T.; Ezaka, Y.; Takahashi, M.; Saito, T. Roles of microRNA-34a in the pathogenesis of placenta accreta. J. Obstet. Gynaecol. Res. 2013, 39, 67–74. [Google Scholar] [CrossRef]
- Doridot, L.; Houry, D.; Gaillard, H.; Chelbi, S.T.; Barbaux, S.; Vaiman, D. miR-34a expression, epigenetic regulation, and function in human placental diseases. Epigenetics 2014, 9, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Qiu, Z.; Diao, Z.; Shen, L.; Xue, P.; Sun, H.; Hu, Y. MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta 2012, 33, 824–829. [Google Scholar] [CrossRef]
- Kumar, P.; Luo, Y.; Tudela, C.; Alexander, J.M.; Mendelson, C.R. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol. Cell. Biol. 2013, 33, 1782–1796. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.L.; Liu, M.; Yang, Y.; Yang, H.; Liao, Q.; Bai, Y.; Li, Y.X.; Li, D.; Peng, C.; Wang, Y.L. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1). RNA Biol. 2012, 9, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Mouillet, J.F.; Chu, T.; Parks, W.T.; Sadovsky, E.; Knofler, M.; Sadovsky, Y. C19MC microRNAs regulate the migration of human trophoblasts. Endocrinology 2014, 155, 4975–4985. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Fournier, T.; Harris, L.K.; James, J.; Roberts, C.T.; Yong, H.E.J.; Kalionis, B.; Evain-Brion, D.; Ebeling, P.R.; Wallace, E.M.; et al. Increased methylation and decreased expression of homeobox genes TLX1, HOXA10 and DLX5 in human placenta are associated with trophoblast differentiation. Sci. Rep. 2017, 7, 4523. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.C.; Novakovic, B.; Weinrich, B.; Dewi, C.; Andronikos, R.; Sibson, M.; Macrae, F.; Morley, R.; Pertile, M.D.; Craig, J.M.; et al. Methylation of the adenomatous polyposis coli (APC) gene in human placenta and hypermethylation in choriocarcinoma cells. Cancer Lett. 2008, 268, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Liu, H.; Cao, J.; Liu, Q.; Tang, G.; Liu, W.; Liu, H.; Deng, D.; Qiao, F.; Wu, Y. Promoter Hypomethylation of Maspin Inhibits Migration and Invasion of Extravillous Trophoblast Cells during Placentation. PLoS ONE 2015, 10, e0135359. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.W.; Chim, S.S.; Wong, I.H.; Wong, C.S.; Lee, W.S.; To, K.F.; Tong, J.H.; Yuen, R.K.; Shum, A.S.; Chan, J.K.; et al. Hypermethylation of RASSF1A in human and rhesus placentas. Am. J. Pathol. 2007, 170, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.I.; Blair, J.D.; Lott, P.; Yu, H.O.; Hong, D.; Crary, F.; Ashwood, P.; Walker, C.; Korf, I.; Robinson, W.P.; et al. The human placenta methylome. Proc. Natl. Acad. Sci. USA 2013, 110, 6037–6042. [Google Scholar] [CrossRef] [PubMed]
- Nordor, A.V.; Nehar-Belaid, D.; Richon, S.; Klatzmann, D.; Bellet, D.; Dangles-Marie, V.; Fournier, T.; Aryee, M.J. The early pregnancy placenta foreshadows DNA methylation alterations of solid tumors. Epigenetics 2017, 12, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Yuen, R.K.; Chen, B.; Blair, J.D.; Robinson, W.P.; Nelson, D.M. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 2013, 8, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Shankar, K.; Kang, P.; Zhong, Y.; Borengasser, S.J.; Wingfield, C.; Saben, J.; Gomez-Acevedo, H.; Thakali, K.M. Transcriptomic and epigenomic landscapes during cell fusion in BeWo trophoblast cells. Placenta 2015, 36, 1342–1351. [Google Scholar] [CrossRef] [PubMed]
- James, J.L.; Hurley, D.G.; Gamage, T.K.; Zhang, T.; Vather, R.; Pantham, P.; Murthi, P.; Chamley, L.W. Isolation and characterisation of a novel trophoblast side-population from first trimester placentae. Reproduction 2015, 150, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Gamage, T.K.; Schierding, W.; Hurley, D.; Tsai, P.; Ludgate, J.L.; Bhoothpur, C.; Chamley, L.W.; Weeks, R.J.; Macaulay, E.C.; James, J.L. The role of DNA methylation in human trophoblast differentiation. Epigenetics 2018. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.K.; Dean, W.; Dawson, C.; Lucifero, D.; Madeja, Z.; Reik, W.; Hemberger, M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008, 10, 1280–1290. [Google Scholar] [CrossRef] [PubMed]
- Senner, C.E.; Krueger, F.; Oxley, D.; Andrews, S.; Hemberger, M. DNA methylation profiles define stem cell identity and reveal a tight embryonic-extraembryonic lineage boundary. Stem Cells 2012, 30, 2732–2745. [Google Scholar] [CrossRef]
- Murray, R.; Bryant, J.; Titcombe, P.; Barton, S.J.; Inskip, H.; Harvey, N.C.; Cooper, C.; Lillycrop, K.; Hanson, M.; Godfrey, K.M. DNA methylation at birth within the promoter of ANRIL predicts markers of cardiovascular risk at 9 years. Clin. Epigenet. 2016, 8, 90. [Google Scholar] [CrossRef]
- Santos, J.; Pereira, C.F.; Di-Gregorio, A.; Spruce, T.; Alder, O.; Rodriguez, T.; Azuara, V.; Merkenschlager, M.; Fisher, A.G. Differences in the epigenetic and reprogramming properties of pluripotent and extra-embryonic stem cells implicate chromatin remodelling as an important early event in the developing mouse embryo. Epigenet. Chromatin 2010, 3, 1. [Google Scholar] [CrossRef]
- Bianco-Miotto, T.; Mayne, B.T.; Buckberry, S.; Breen, J.; Rodriguez Lopez, C.M.; Roberts, C.T. Recent progress towards understanding the role of DNA methylation in human placental development. Reproduction 2016, 152, R23–R30. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Wang, H.; Qu, D.; Chen, Y.; Gao, J.; Sun, H. Epigenome-wide DNA methylation assay reveals placental epigenetic markers for noninvasive fetal single-nucleotide polymorphism genotyping in maternal plasma. Transfusion 2014, 54, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, J.; Li, Q.; Zhou, X.; Wang, T.; Xu, M.; Xia, S.; Xing, Q.; Wang, L.; He, L.; et al. DNA methylome profiling of maternal peripheral blood and placentas reveal potential fetal DNA markers for non-invasive prenatal testing. Mol. Hum. Reprod. 2014, 20, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Yuen, R.K.; Gordon, L.; Penaherrera, M.S.; Sharkey, A.; Moffett, A.; Craig, J.M.; Robinson, W.P.; Saffery, R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genom. 2011, 12, 529. [Google Scholar] [CrossRef] [PubMed]
- Rakyan, V.K.; Down, T.A.; Thorne, N.P.; Flicek, P.; Kulesha, E.; Graf, S.; Tomazou, E.M.; Backdahl, L.; Johnson, N.; Herberth, M.; et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res. 2008, 18, 1518–1529. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Helmer, R.A.; Smith, L.A.; Martinez-Zaguilan, R.; Dufour, J.M.; Chilton, B.S. Alternative splicing of helicase-like transcription factor (Hltf): Intron retention-dependent activation of immune tolerance at the feto-maternal interface. PLoS ONE 2018, 13, e0200211. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Lax, E.; Zhou, R.; Cheishvili, D.; Ruder, A.M.; Ludiro, A.; Lapert, F.; Macedo da Cruz, A.; Sandrini, P.; Calzoni, T.; et al. Fetal glucocorticoid receptor (Nr3c1) deficiency alters the landscape of DNA methylation of murine placenta in a sex-dependent manner and is associated to anxiety-like behavior in adulthood. Transl. Psychiatry 2019, 9, 23. [Google Scholar] [CrossRef]
- Price, E.M.; Cotton, A.M.; Penaherrera, M.S.; McFadden, D.E.; Kobor, M.S.; Robinson, W. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics 2012, 7, 652–663. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Sun, Y.; Mao, C.; Yang, S.; Huang, M.; Deng, M.; Ding, N.; Yang, X.; Zhang, M.; Jin, S.; et al. Folate Protects Hepatocytes of Hyperhomocysteinemia Mice From Apoptosis via Cystic Fibrosis Transmembrane Conductance Regulator (CFTR)-Activated Endoplasmic Reticulum Stress. J. Cell. Biochem. 2017, 118, 2921–2932. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Mora, J.R.; Sanchez-Delgado, M.; Petazzi, P.; Moran, S.; Esteller, M.; Iglesias-Platas, I.; Monk, D. Profiling of oxBS-450K 5-hydroxymethylcytosine in human placenta and brain reveals enrichment at imprinted loci. Epigenetics 2018, 13, 182–191. [Google Scholar] [CrossRef]
- Lim, Y.C.; Li, J.; Ni, Y.; Liang, Q.; Zhang, J.; Yeo, G.S.H.; Lyu, J.; Jin, S.; Ding, C. A complex association between DNA methylation and gene expression in human placenta at first and third trimesters. PLoS ONE 2017, 12, e0181155. [Google Scholar] [CrossRef] [PubMed]
- Roost, M.S.; Slieker, R.C.; Bialecka, M.; van Iperen, L.; Gomes Fernandes, M.M.; He, N.; Suchiman, H.E.D.; Szuhai, K.; Carlotti, F.; de Koning, E.J.P.; et al. DNA methylation and transcriptional trajectories during human development and reprogramming of isogenic pluripotent stem cells. Nat. Commun. 2017, 8, 908. [Google Scholar] [CrossRef] [PubMed]
- Decato, B.E.; Lopez-Tello, J.; Sferruzzi-Perri, A.N.; Smith, A.D.; Dean, M.D. DNA Methylation Divergence and Tissue Specialization in the Developing Mouse Placenta. Mol. Biol. Evol. 2017, 34, 1702–1712. [Google Scholar] [CrossRef] [PubMed]
- Green, B.B.; Houseman, E.A.; Johnson, K.C.; Guerin, D.J.; Armstrong, D.A.; Christensen, B.C.; Marsit, C.J. Hydroxymethylation is uniquely distributed within term placenta, and is associated with gene expression. FASEB J. 2016, 30, 2874–2884. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Macaulay, E.C.; Rodger, E.J.; Stockwell, P.A.; Parry, M.F.; Roberts, H.E.; Slatter, T.L.; Hung, N.A.; Devenish, C.J.; Morison, I.M. Placental Hypomethylation Is More Pronounced in Genomic Loci Devoid of Retroelements. G3 (Bethesda) 2016, 6, 1911–1921. [Google Scholar] [CrossRef] [PubMed]
- Branco, M.R.; King, M.; Perez-Garcia, V.; Bogutz, A.B.; Caley, M.; Fineberg, E.; Lefebvre, L.; Cook, S.J.; Dean, W.; Hemberger, M.; et al. Maternal DNA Methylation Regulates Early Trophoblast Development. Dev. Cell 2016, 36, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Penaherrera, M.S.; Saadeh, H.; Andrews, S.; McFadden, D.E.; Kelsey, G.; Robinson, W.P. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res. 2016, 26, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Blair, J.D.; Yuen, R.K.; Robinson, W.P.; von Dadelszen, P. Genome-wide DNA methylation identifies trophoblast invasion-related genes: Claudin-4 and Fucosyltransferase IV control mobility via altering matrix metalloproteinase activity. Mol. Hum. Reprod. 2015, 21, 452–465. [Google Scholar] [CrossRef]
- Mahadevan, S.; Wen, S.; Wan, Y.W.; Peng, H.H.; Otta, S.; Liu, Z.; Iacovino, M.; Mahen, E.M.; Kyba, M.; Sadikovic, B.; et al. NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Hum. Mol. Genet. 2014, 23, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Novakovic, B.; Gordon, L.; Wong, N.C.; Moffett, A.; Manuelpillai, U.; Craig, J.M.; Sharkey, A.; Saffery, R. Wide-ranging DNA methylation differences of primary trophoblast cell populations and derived cell lines: Implications and opportunities for understanding trophoblast function. Mol. Hum. Reprod. 2011, 17, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Amorim, R.P.; Araujo, M.G.L.; Valero, J.; Lopes-Cendes, I.; Pascoal, V.D.B.; Malva, J.O.; da Silva Fernandes, M.J. Silencing of P2X7R by RNA interference in the hippocampus can attenuate morphological and behavioral impact of pilocarpine-induced epilepsy. Purinergic Signal. 2017, 13, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Oudejans, C.B.; Pannese, M.; Simeone, A.; Meijer, C.J.; Boncinelli, E. The three most downstream genes of the Hox-3 cluster are expressed in human extraembryonic tissues including trophoblast of androgenetic origin. Development 1990, 108, 471–477. [Google Scholar] [PubMed]
- Chui, A.; Pathirage, N.A.; Johnson, B.; Cocquebert, M.; Fournier, T.; Evain-Brion, D.; Roald, B.; Manuelpillai, U.; Brennecke, S.P.; Kalionis, B.; et al. Homeobox gene distal-less 3 is expressed in proliferating and differentiating cells of the human placenta. Placenta 2010, 31, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Grati, F.R.; Sirchia, S.M.; Gentilin, B.; Rossella, F.; Ramoscelli, L.; Antonazzo, P.; Cavallari, U.; Bulfamante, G.; Cetin, I.; Simoni, G.; et al. Biparental expression of ESX1L gene in placentas from normal and intrauterine growth-restricted pregnancies. Eur. J. Hum. Genet. 2004, 12, 272–278. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quinn, L.M.; Johnson, B.V.; Nicholl, J.; Sutherland, G.R.; Kalionis, B. Isolation and identification of homeobox genes from the human placenta including a novel member of the Distal-less family, DLX4. Gene 1997, 187, 55–61. [Google Scholar] [CrossRef]
- Rajaraman, G.; Murthi, P.; Quinn, L.; Brennecke, S.P.; Kalionis, B. Homeodomain protein HLX is expressed primarily in cytotrophoblast cell types in the early pregnancy human placenta. Reprod. Fertil. Dev. 2008, 20, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.I.; LaSalle, J.M. How has the study of the human placenta aided our understanding of partially methylated genes? Epigenomics 2013, 5, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [PubMed]
- Sadovsky, Y.; Mouillet, J.F.; Ouyang, Y.; Bayer, A.; Coyne, C.B. The Function of TrophomiRs and Other MicroRNAs in the Human Placenta. Cold Spring Harb. Perspect. Med. 2015, 5, a023036. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Donker, R.B.; Mouillet, J.F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012, 18, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, Y.Q.; Su, B. Molecular evolution of a primate-specific microRNA family. Mol. Biol. Evol. 2008, 25, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Noguer-Dance, M.; Abu-Amero, S.; Al-Khtib, M.; Lefevre, A.; Coullin, P.; Moore, G.E.; Cavaille, J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010, 19, 3566–3582. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Kao, H.W.; Chen, H.C.; Chen, S.J.; Lin, W.C. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics 2009, 4, 587–592. [Google Scholar] [CrossRef]
- Bar, M.; Wyman, S.K.; Fritz, B.R.; Qi, J.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; Bendoraite, A.; Mitchell, P.S.; Nelson, A.M.; et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 2008, 26, 2496–2505. [Google Scholar] [CrossRef] [PubMed]
- Laurent, L.C.; Chen, J.; Ulitsky, I.; Mueller, F.J.; Lu, C.; Shamir, R.; Fan, J.B.; Loring, J.F. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells 2008, 26, 1506–1516. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Jin, P.; Wang, E.; Marincola, F.M.; Stroncek, D.F. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J. Transl. Med. 2009, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Stadler, B.; Ivanovska, I.; Mehta, K.; Song, S.; Nelson, A.; Tan, Y.; Mathieu, J.; Darby, C.; Blau, C.A.; Ware, C.; et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010, 19, 935–950. [Google Scholar] [CrossRef]
- Morales-Prieto, D.M.; Ospina-Prieto, S.; Chaiwangyen, W.; Schoenleben, M.; Markert, U.R. Pregnancy-associated miRNA-clusters. J. Reprod. Immunol. 2013, 97, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ridzon, D.; Wong, L.; Chen, C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007, 8, 166. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Forbes, K.; Farrokhnia, F.; Aplin, J.D.; Westwood, M. Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 2012, 33, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Doridot, L.; Miralles, F.; Barbaux, S.; Vaiman, D. Trophoblasts, invasion, and microRNA. Front. Genet. 2013, 4, 248. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Meng, T. MicroRNA-431 affects trophoblast migration and invasion by targeting ZEB1 in preeclampsia. Gene 2019, 683, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- McAninch, D.; Roberts, C.T.; Bianco-Miotto, T. Mechanistic Insight into Long Noncoding RNAs and the Placenta. Int. J. Mol. Sci. 2017, 18, 1371. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Platas, I.; Martin-Trujillo, A.; Petazzi, P.; Guillaumet-Adkins, A.; Esteller, M.; Monk, D. Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum. Mol. Genet. 2014, 23, 6275–6285. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Jinno, Y.; Ikeda, Y.; Yun, K.; Maw, M.; Masuzaki, H.; Fukuda, H.; Inuzuka, K.; Fujishita, A.; Ohtani, Y.; Okimoto, T.; et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat. Genet. 1995, 10, 318–324. [Google Scholar] [CrossRef]
- Yu, L.; Chen, M.; Zhao, D.; Yi, P.; Lu, L.; Han, J.; Zheng, X.; Zhou, Y.; Li, L. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta 2009, 30, 443–447. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Mellor, J.; Dudek, P.; Clynes, D. A glimpse into the epigenetic landscape of gene regulation. Curr. Opin. Genet. Dev. 2008, 18, 116–122. [Google Scholar] [CrossRef]
- Grewal, S.I.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef]
- Torres-Padilla, M.E.; Parfitt, D.E.; Kouzarides, T.; Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 2007, 445, 214–218. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001, 13, 167–171. [Google Scholar] [CrossRef]
- Charron, C.E.; Chou, P.C.; Coutts, D.J.; Kumar, V.; To, M.; Akashi, K.; Pinhu, L.; Griffiths, M.; Adcock, I.M.; Barnes, P.J.; et al. Hypoxia-inducible factor 1alpha induces corticosteroid-insensitive inflammation via reduction of histone deacetylase-2 transcription. J. Biol. Chem. 2009, 284, 36047–36054. [Google Scholar] [CrossRef] [PubMed]
- Maltepe, E.; Krampitz, G.W.; Okazaki, K.M.; Red-Horse, K.; Mak, W.; Simon, M.C.; Fisher, S.J. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development 2005, 132, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Pollard, P.J.; Loenarz, C.; Mole, D.R.; McDonough, M.A.; Gleadle, J.M.; Schofield, C.J.; Ratcliffe, P.J. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem. J. 2008, 416, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Wellmann, S.; Bettkober, M.; Zelmer, A.; Seeger, K.; Faigle, M.; Eltzschig, H.K.; Buhrer, C. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 2008, 372, 892–897. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Yao, L.; Zhang, Q.; Wang, F.; Mei, H.; Guo, X.; Huang, W. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget 2017, 8, 19795–19802. [Google Scholar] [CrossRef] [PubMed]
- Maltepe, E.; Bakardjiev, A.I.; Fisher, S.J. The placenta: Transcriptional, epigenetic, and physiological integration during development. J. Clin. Investig. 2010, 120, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Franasiak, J.M.; Scott, R.T. Contribution of immunology to implantation failure of euploid embryos. Fertil. Steril. 2017, 107, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Griffith, O.W.; Chavan, A.R.; Protopapas, S.; Maziarz, J.; Romero, R.; Wagner, G.P. Embryo implantation evolved from an ancestral inflammatory attachment reaction. Proc. Natl. Acad. Sci. USA 2017, 114, E6566–E6575. [Google Scholar] [CrossRef] [PubMed]
- Hansen, V.L.; Faber, L.S.; Salehpoor, A.A.; Miller, R.D. A pronounced uterine pro-inflammatory response at parturition is an ancient feature in mammals. Proc. Biol. Sci. 2017, 284, 20171694. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Funk, M.; Vernochet, C.; Leal, F.; Tarazona, O.A.; Meurice, G.; Heidmann, O.; Dupressoir, A.; Miralles, A.; Ramirez-Pinilla, M.P.; et al. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. USA 2017, 114, E10991–E11000. [Google Scholar] [CrossRef] [PubMed]
- McKinnell, Z.; Wessel, G. Ligers and tigons and.....what?....oh my! Mol. Reprod. Dev. 2012, 79, Fm i. [Google Scholar] [CrossRef] [PubMed]
- Surani, M.A.; Barton, S.C.; Norris, M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984, 308, 548–550. [Google Scholar] [CrossRef] [PubMed]
- Wake, N.; Arima, T.; Matsuda, T. Involvement of IGF2 and H19 imprinting in choriocarcinoma development. Int. J. Gynaecol. Obstet. 1998, 60 (Suppl. 1), S1–S8. [Google Scholar] [CrossRef]
- Warren, W.C.; Hillier, L.W.; Marshall Graves, J.A.; Birney, E.; Ponting, C.P.; Grutzner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 2008, 453, 175–183. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Suzuki, S.; Shaw, G.; Kaneko-Ishino, T.; Ishino, F.; Renfree, M.B. The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol. Evol. 2011, 3, 1276–1283. [Google Scholar] [CrossRef] [PubMed]
- Renfree, M.B.; Suzuki, S.; Kaneko-Ishino, T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120151. [Google Scholar] [CrossRef] [PubMed]
- Fresard, L.; Leroux, S.; Servin, B.; Gourichon, D.; Dehais, P.; Cristobal, M.S.; Marsaud, N.; Vignoles, F.; Bed’hom, B.; Coville, J.L.; et al. Transcriptome-wide investigation of genomic imprinting in chicken. Nucleic Acids Res. 2014, 42, 3768–3782. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Lamont, S.J.; Abasht, B. RNA-Seq Analyses Identify Frequent Allele Specific Expression and No Evidence of Genomic Imprinting in Specific Embryonic Tissues of Chicken. Sci. Rep. 2017, 7, 11944. [Google Scholar] [CrossRef] [PubMed]
- Piedrahita, J.A. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res. A Clin. Mol. Teratol. 2011, 91, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Constancia, M.; Hemberger, M.; Hughes, J.; Dean, W.; Ferguson-Smith, A.; Fundele, R.; Stewart, F.; Kelsey, G.; Fowden, A.; Sibley, C.; et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 2002, 417, 945–948. [Google Scholar] [CrossRef] [PubMed]
- Ripoche, M.A.; Kress, C.; Poirier, F.; Dandolo, L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997, 11, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Goodyer, C.G.; Deal, C.; Polychronakos, C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem. Biophys. Res. Commun. 1993, 197, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Cheong, C.Y.; Chng, K.; Ng, S.; Chew, S.B.; Chan, L.; Ferguson-Smith, A.C. Germline and somatic imprinting in the nonhuman primate highlights species differences in oocyte methylation. Genome Res. 2015, 25, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Monk, D.; Arnaud, P.; Apostolidou, S.; Hills, F.A.; Kelsey, G.; Stanier, P.; Feil, R.; Moore, G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. USA 2006, 103, 6623–6628. [Google Scholar] [CrossRef]
- Barbaux, S.; Gascoin-Lachambre, G.; Buffat, C.; Monnier, P.; Mondon, F.; Tonanny, M.B.; Pinard, A.; Auer, J.; Bessieres, B.; Barlier, A.; et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics 2012, 7, 1079–1090. [Google Scholar] [CrossRef]
- Allach El Khattabi, L.; Backer, S.; Pinard, A.; Dieudonne, M.N.; Tsatsaris, V.; Vaiman, D.; Dandolo, L.; Bloch-Gallego, E.; Jammes, H.; Barbaux, S. A genome-wide search for new imprinted genes in the human placenta identifies DSCAM as the first imprinted gene on chromosome 21. Eur. J. Hum. Genet. 2019, 27, 49–60. [Google Scholar] [CrossRef]
- Marjonen, H.; Auvinen, P.; Kahila, H.; Tsuiko, O.; Koks, S.; Tiirats, A.; Viltrop, T.; Tuuri, T.; Soderstrom-Anttila, V.; Suikkari, A.M.; et al. rs10732516 polymorphism at the IGF2/H19 locus associates with genotype-specific effects on placental DNA methylation and birth weight of newborns conceived by assisted reproductive technology. Clin. Epigenet. 2018, 10, 80. [Google Scholar] [CrossRef]
- Peters, J. The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef]
- Monk, D. Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 2015, 213, S152–S162. [Google Scholar] [CrossRef]
- Christians, J.K.; Leavey, K.; Cox, B.J. Associations between imprinted gene expression in the placenta, human fetal growth and preeclampsia. Biol. Lett. 2017, 13, 20170643. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Sadovsky, Y. The function of miR-519d in cell migration, invasion, and proliferation suggests a role in early placentation. Placenta 2016, 48, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Petre, G.; Lores, P.; Sartelet, H.; Truffot, A.; Poreau, B.; Brandeis, S.; Martinez, G.; Satre, V.; Harbuz, R.; Ray, P.F.; et al. Genomic duplication in the 19q13.42 imprinted region identified as a new genetic cause of intrauterine growth restriction. Clin. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Vaiman, D.; Calicchio, R.; Miralles, F. Landscape of transcriptional deregulations in the preeclamptic placenta. PLoS ONE 2013, 8, e65498. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.Z.; Zhang, X.; Hu, P.; Liu, X.M.; Hua, X.D.; Wang, X.; Ding, H.J. Screening for differential methylation status in human placenta in preeclampsia using a CpG island plus promoter microarray. Int. J. Mol. Med. 2012, 30, 133–141. [Google Scholar]
- Anton, L.; Olarerin-George, A.O.; Schwartz, N.; Srinivas, S.; Bastek, J.; Hogenesch, J.B.; Elovitz, M.A. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am. J. Pathol. 2013, 183, 1437–1445. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, X.; Rong, C.; Rui, C.; Ji, H.; Qian, Y.J.; Jia, R.; Sun, L. Distinct DNA methylomes of human placentas between pre-eclampsia and gestational diabetes mellitus. Cell. Physiol. Biochem. 2014, 34, 1877–1889. [Google Scholar] [CrossRef]
- Yeung, K.R.; Chiu, C.L.; Pidsley, R.; Makris, A.; Hennessy, A.; Lind, J.M. DNA methylation profiles in preeclampsia and healthy control placentas. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H1295–H1303. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, X.; Wang, J.; Li, Y.; Yang, Y.; Yang, T.; Ma, H.; Wang, L.; Zhang, G.; Cho, W.C.; et al. Placental mesenchymal stem cells of fetal origin deposit epigenetic alterations during long-term culture under serum-free condition. Expert Opin. Biol. Ther. 2015, 15, 163–180. [Google Scholar] [CrossRef]
- Leavey, K.; Wilson, S.L.; Bainbridge, S.A.; Robinson, W.P.; Cox, B.J. Epigenetic regulation of placental gene expression in transcriptional subtypes of preeclampsia. Clin. Epigenet. 2018, 10, 28. [Google Scholar] [CrossRef]
- Calicchio, R.; Doridot, L.; Miralles, F.; Mehats, C.; Vaiman, D. DNA methylation, an epigenetic mode of gene expression regulation in reproductive science. Curr. Pharm. Des. 2014, 20, 1726–1750. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, A.; Hayashi, T.; Kikuchi, N.; Hayashi, A.; Fuseya, C.; Shiozawa, T.; Konishi, I. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1alpha to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int. J. Cancer 2012, 131, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Aouache, R.; Biquard, L.; Vaiman, D.; Miralles, F. Oxidative Stress in Preeclampsia and Placental Diseases. Int. J. Mol. Sci. 2018, 19, 1496. [Google Scholar] [CrossRef] [PubMed]
- Biron-Shental, T.; Sukenik Halevy, R.; Goldberg-Bittman, L.; Kidron, D.; Fejgin, M.D.; Amiel, A. Telomeres are shorter in placental trophoblasts of pregnancies complicated with intrauterine growth restriction (IUGR). Early Hum. Dev. 2010, 86, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Sukenik-Halevy, R.; Amiel, A.; Kidron, D.; Liberman, M.; Ganor-Paz, Y.; Biron-Shental, T. Telomere homeostasis in trophoblasts and in cord blood cells from pregnancies complicated with preeclampsia. Am. J. Obstet. Gynecol. 2016, 214, 283.e1–283.e7. [Google Scholar] [CrossRef]
- Farladansky-Gershnabel, S.; Gal, H.; Kidron, D.; Krizhanovsky, V.; Amiel, A.; Sukenik-Halevy, R.; Biron-Shental, T. Telomere Homeostasis and Senescence Markers Are Differently Expressed in Placentas From Pregnancies With Early- Versus Late-Onset Preeclampsia. Reprod. Sci. 2018, 1933719118811644. [Google Scholar] [CrossRef]
- Cindrova-Davies, T.; Fogarty, N.M.E.; Jones, C.J.P.; Kingdom, J.; Burton, G.J. Evidence of oxidative stress-induced senescence in mature, post-mature and pathological human placentas. Placenta 2018, 68, 15–22. [Google Scholar] [CrossRef]
- Londero, A.P.; Orsaria, M.; Marzinotto, S.; Grassi, T.; Fruscalzo, A.; Calcagno, A.; Bertozzi, S.; Nardini, N.; Stella, E.; Lelle, R.J.; et al. Placental aging and oxidation damage in a tissue micro-array model: An immunohistochemistry study. Histochem. Cell Biol. 2016, 146, 191–204. [Google Scholar] [CrossRef]
- Burton, G.J.; Yung, H.W.; Murray, A.J. Mitochondrial—Endoplasmic reticulum interactions in the trophoblast: Stress and senescence. Placenta 2017, 52, 146–155. [Google Scholar] [CrossRef]
- Chu, T.; Bunce, K.; Shaw, P.; Shridhar, V.; Althouse, A.; Hubel, C.; Peters, D. Comprehensive analysis of preeclampsia-associated DNA methylation in the placenta. PLoS ONE 2014, 9, e107318. [Google Scholar] [CrossRef]
- Blair, J.D.; Yuen, R.K.; Lim, B.K.; McFadden, D.E.; von Dadelszen, P.; Robinson, W.P. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol. Hum. Reprod. 2013, 19, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Yung, H.W.; Atkinson, D.; Campion-Smith, T.; Olovsson, M.; Charnock-Jones, D.S.; Burton, G.J. Differential activation of placental unfolded protein response pathways implies heterogeneity in causation of early- and late-onset pre-eclampsia. J. Pathol. 2014, 234, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Lv, R.; Kong, L.; Cheng, H.; Lan, F.; Li, X. Genome-Wide Mapping of 5mC and 5hmC Identified Differentially Modified Genomic Regions in Late-Onset Severe Preeclampsia: A Pilot Study. PLoS ONE 2015, 10, e0134119. [Google Scholar] [CrossRef] [PubMed]
- Anton, L.; Brown, A.G.; Bartolomei, M.S.; Elovitz, M.A. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS ONE 2014, 9, e100148. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Zhang, K.; Wang, L.; Ou, G.; Zhu, H.; Gao, W.Q. Transcription factor STOX1 regulates proliferation of inner ear epithelial cells via the AKT pathway. Cell Prolif. 2015, 48, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Guibert, S.; Weber, M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr. Top. Dev. Biol. 2013, 104, 47–83. [Google Scholar] [PubMed]
- Bellido, M.L.; Radpour, R.; Lapaire, O.; De Bie, I.; Hosli, I.; Bitzer, J.; Hmadcha, A.; Zhong, X.Y.; Holzgreve, W. MALDI-TOF mass array analysis of RASSF1A and SERPINB5 methylation patterns in human placenta and plasma. Biol. Reprod. 2010, 82, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.M.; Ralph, J.L.; Wright, M.L.; Linggi, B.; Ohm, J.E. DNA methylation as a biomarker for preeclampsia. Biol. Res. Nurs. 2014, 16, 409–420. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, A.; Fang, M.; Fang, R.; Ge, J.; Jiang, Y.; Zhang, H.; Han, C.; Ye, X.; Yu, D.; et al. Methylation levels at IGF2 and GNAS DMRs in infants born to preeclamptic pregnancies. BMC Genom. 2013, 14, 472. [Google Scholar] [CrossRef]
- Xiang, Y.; Zhang, X.; Li, Q.; Xu, J.; Zhou, X.; Wang, T.; Xing, Q.; Liu, Y.; Wang, L.; He, L.; et al. Promoter hypomethylation of TIMP3 is associated with pre-eclampsia in a Chinese population. Mol. Hum. Reprod. 2013, 19, 153–159. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Fei, M.; Xue, G.; Zhou, Q.; Jia, Y.; Li, L.; Xin, H.; Sun, S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: New insights into molecular mechanisms for the disease. J. Cell. Mol. Med. 2012, 16, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.L.; Leavey, K.; Cox, B.; Robinson, W.P. Mining DNA methylation alterations towards a classification of placental pathologies. Hum. Mol. Genet. 2018, 27, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Chelbi, S.T.; Wilson, M.L.; Veillard, A.C.; Ingles, S.A.; Zhang, J.; Mondon, F.; Gascoin-Lachambre, G.; Doridot, L.; Mignot, T.M.; Rebourcet, R.; et al. Genetic and epigenetic mechanisms collaborate to control SERPINA3 expression and its association with placental diseases. Hum. Mol. Genet. 2012, 21, 1968–1978. [Google Scholar] [CrossRef] [PubMed]
- Hogg, K.; Blair, J.D.; McFadden, D.E.; von Dadelszen, P.; Robinson, W.P. Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS ONE 2013, 8, e62969. [Google Scholar] [CrossRef] [PubMed]
- Xirong, X.; Tao, X.; Wang, Y.; Zhu, L.; Ye, Y.; Liu, H.; Zhou, Q.; Li, X.; Xiong, Y. Hypomethylation of tissue factor pathway inhibitor 2 in human placenta of preeclampsia. Thrombosis Res. 2017, 152, 7–13. [Google Scholar]
- Sundrani, D.P.; Reddy, U.S.; Joshi, A.A.; Mehendale, S.S.; Chavan-Gautam, P.M.; Hardikar, A.A. Differential placental methylation and expression of VEGF, FLT-1 and KDR genes in human term and preterm preeclampsia. Clin. Epigenet. 2013, 5. [Google Scholar] [CrossRef]
- Ching, T.; Ha, J.; Song, M.A.; Tiirikainen, M.; Molnar, J.; Berry, M.J.; Towner, D.; Garmire, L.X. Genome-scale hypomethylation in the cord blood DNAs associated with early onset preeclampsia. Clin. Epigenet. 2015, 7, 21. [Google Scholar] [CrossRef]
- Ye, W.; Shen, L.; Xiong, Y.; Zhou, Y.; Gu, H.; Yang, Z. Preeclampsia is Associated with Decreased Methylation of the GNA12 Promoter. Ann. Hum. Genet. 2016, 80, 7–10. [Google Scholar] [CrossRef]
- Yuen, R.K.; Penaherrera, M.S.; von Dadelszen, P.; McFadden, D.E.; Robinson, W.P. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur. J. Hum. Genet. 2010, 18, 1006–1012. [Google Scholar] [CrossRef]
- Hogg, K.; Blair, J.D.; von Dadelszen, P.; Robinson, W.P. Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset pre-eclampsia. Mol. Cell. Endocrinol. 2013, 367, 64–73. [Google Scholar] [CrossRef]
- Xiang, Y.; Cheng, Y.; Li, X.; Li, Q.; Xu, J.; Zhang, J.; Liu, Y.; Xing, Q.; Wang, L.; He, L.; et al. Up-regulated expression and aberrant DNA methylation of LEP and SH3PXD2A in pre-eclampsia. PLoS ONE 2013, 8, e59753. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Weng, X.; Dong, M.; Liu, Y.; Li, W.; Huang, H. Alteration in methylation level at 11β-hydroxysteroid dehydrogenase type 2 gene promoter in infants born to preeclamptic women. BMC Genet. 2014, 15, 96. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, Y. Promoter Methylation Status of WNT2 in Placenta from Patients with Preeclampsia. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 5294–5301. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhou, Q.-J.; Xiong, Y.; Li, B.; Li, X.-T. Preeclampsia is associated with hypermethylation of IGF-1 promoter mediated by DNMT1. Am. J. Transl. Res. 2018, 10, 16–39. [Google Scholar] [PubMed]
- Rahat, B.; Thakur, S.; Bagga, R.; Kaur, J. Epigenetic regulation of STAT5A and its role as fetal DNA epigenetic marker during placental development and dysfunction. Placenta 2016, 44, 46–53. [Google Scholar] [CrossRef]
- Choux, C.; Carmignac, V.; Bruno, C.; Sagot, P.; Vaiman, D.; Fauque, P. The placenta: Phenotypic and epigenetic modifications induced by Assisted Reproductive Technologies throughout pregnancy. Clin. Epigenet. 2015, 7, 87. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, H.; Li, H.; Peng, T.; Gu, W.; Li, X. Hypermethylation of the HLA-G promoter is associated with preeclampsia. Mol. Hum. Reprod. 2015, 21, 736–744. [Google Scholar] [CrossRef]
- Konwar, C.; Del Gobbo, G.; Yuan, V.; Robinson, W.P. Considerations when processing and interpreting genomics data of the placenta. Placenta 2019. [Google Scholar] [CrossRef]
- Tsang, J.C.H.; Vong, J.S.L.; Ji, L.; Poon, L.C.Y.; Jiang, P.; Lui, K.O.; Ni, Y.B.; To, K.F.; Cheng, Y.K.Y.; Chiu, R.W.K.; et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. USA 2017, 114, E7786–E7795. [Google Scholar] [CrossRef]
- Anacker, J.; Segerer, S.E.; Hagemann, C.; Feix, S.; Kapp, M.; Bausch, R.; Kammerer, U. Human decidua and invasive trophoblasts are rich sources of nearly all human matrix metalloproteinases. Mol. Hum. Reprod. 2011, 17, 637–652. [Google Scholar] [CrossRef]
- Vettraino, I.M.; Roby, J.; Tolley, T.; Parks, W.C. Collagenase-I, stromelysin-I, and matrilysin are expressed within the placenta during multiple stages of human pregnancy. Placenta 1996, 17, 557–563. [Google Scholar] [CrossRef]
- Kocarslan, S.; Incebiyik, A.; Guldur, M.E.; Ekinci, T.; Ozardali, H.I. What is the role of matrix metalloproteinase-2 in placenta percreta? J. Obstet. Gynaecol. Res. 2015, 41, 1018–1022. [Google Scholar] [CrossRef] [PubMed]
- Espino, Y.S.S.; Flores-Pliego, A.; Espejel-Nunez, A.; Medina-Bastidas, D.; Vadillo-Ortega, F.; Zaga-Clavellina, V.; Estrada-Gutierrez, G. New Insights into the Role of Matrix Metalloproteinases in Preeclampsia. Int. J. Mol. Sci. 2017, 18, 1448. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, C.; Shen, Y.; Wang, K.; Tang, L.; Zhou, M.; Yang, M.; Pan, T.; Liu, X.; Xu, W. Ten-eleven translocation 2 demethylates the MMP9 promoter, and its down-regulation in preeclampsia impairs trophoblast migration and invasion. J. Biol. Chem. 2018, 293, 10059–10070. [Google Scholar] [CrossRef] [PubMed]
- White, W.M.; Brost, B.; Sun, Z.; Rose, C.; Craici, I.; Wagner, S.J.; Turner, S.T.; Garovic, V.D. Genome-wide methylation profiling demonstrates hypermethylation in maternal leukocyte DNA in preeclamptic compared to normotensive pregnancies. Hypertens. Pregnancy 2013, 32, 257–269. [Google Scholar] [CrossRef] [PubMed]
- White, W.M.; Sun, Z.; Borowski, K.S.; Brost, B.C.; Davies, N.P.; Rose, C.H.; Garovic, V.D. Preeclampsia/Eclampsia candidate genes show altered methylation in maternal leukocytes of preeclamptic women at the time of delivery. Hypertens. Pregnancy 2016, 35, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.J.; Maynard, S.E.; Qian, C.; Lim, K.H.; England, L.J.; Yu, K.F.; Schisterman, E.F.; Thadhani, R.; Sachs, B.P.; Epstein, F.H.; et al. Circulating angiogenic factors and the risk of preeclampsia. N. Engl. J. Med. 2004, 350, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Taglauer, E.S.; Wilkins-Haug, L.; Bianchi, D.W. Review: Cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta 2014, 35 (Suppl.), S64–S68. [Google Scholar] [CrossRef]
- Chim, S.S.; Tong, Y.K.; Chiu, R.W.; Lau, T.K.; Leung, T.N.; Chan, L.Y.; Oudejans, C.B.; Ding, C.; Lo, Y.M. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proc. Natl. Acad. Sci. USA 2005, 102, 14753–14758. [Google Scholar] [CrossRef]
- Qi, Y.H.; Teng, F.; Zhou, Q.; Liu, Y.X.; Wu, J.F.; Yu, S.S.; Zhang, X.; Ma, M.Y.; Zhou, N.; Chen, L.J. Unmethylated-maspin DNA in maternal plasma is associated with severe preeclampsia. Acta Obstet. Gynecol. Scand. 2015, 94, 983–988. [Google Scholar] [CrossRef]
- Tsui, D.W.; Chan, K.C.; Chim, S.S.; Chan, L.W.; Leung, T.Y.; Lau, T.K.; Lo, Y.M.; Chiu, R.W. Quantitative aberrations of hypermethylated RASSF1A gene sequences in maternal plasma in pre-eclampsia. Prenat. Diagn. 2007, 27, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Salvianti, F.; Inversetti, A.; Smid, M.; Valsecchi, L.; Candiani, M.; Pazzagli, M.; Cremonesi, L.; Ferrari, M.; Pinzani, P.; Galbiati, S. Prospective evaluation of RASSF1A cell-free DNA as a biomarker of pre-eclampsia. Placenta 2015, 36, 996–1001. [Google Scholar] [CrossRef] [PubMed]
- Mousa, A.A.; Archer, K.J.; Cappello, R.; Estrada-Gutierrez, G.; Isaacs, C.R.; Strauss, J.F., 3rd; Walsh, S.W. DNA methylation is altered in maternal blood vessels of women with preeclampsia. Reprod. Sci. 2012, 19, 1332–1342. [Google Scholar] [CrossRef] [PubMed]
- Mousa, A.A.; Cappello, R.E.; Estrada-Gutierrez, G.; Shukla, J.; Romero, R.; Strauss, J.F., 3rd; Walsh, S.W. Preeclampsia is associated with alterations in DNA methylation of genes involved in collagen metabolism. Am. J. Pathol. 2012, 181, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Mousa, A.A.; Strauss, J.F., 3rd; Walsh, S.W. Reduced methylation of the thromboxane synthase gene is correlated with its increased vascular expression in preeclampsia. Hypertension 2012, 59, 1249–1255. [Google Scholar]
- Nomura, Y.; Lambertini, L.; Rialdi, A.; Lee, M.; Mystal, E.Y.; Grabie, M.; Manaster, I.; Huynh, N.; Finik, J.; Davey, M.; et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod. Sci. 2014, 21, 131–137. [Google Scholar] [CrossRef]
- Chen, J.; Steegers-Theunissen, R.P.M.; van Meurs, J.B.; Felix, J.F.; Eggink, A.J.; Herzog, E.M.; Wijnands, K.P.J.; Stubbs, A.; Slieker, R.C.; van der Spek, P.J.; et al. Early- and late-onset preeclampsia and the tissue-specific epigenome of the placenta and newborn. Placenta 2017, 58, 122–132. [Google Scholar]
- Novielli, C.; Mando, C.; Tabano, S.; Anelli, G.M.; Fontana, L.; Antonazzo, P.; Miozzo, M.; Cetin, I. Mitochondrial DNA content and methylation in fetal cord blood of pregnancies with placental insufficiency. Placenta 2017, 55, 63–70. [Google Scholar] [CrossRef]
- Qiu, C.; Hevner, K.; Enquobahrie, D.A.; Williams, M.A. A case-control study of maternal blood mitochondrial DNA copy number and preeclampsia risk. Int. J. Mol. Epidemiol. Genet. 2012, 3, 237–244. [Google Scholar]
- Vishnyakova, P.A.; Volodina, M.A.; Tarasova, N.V.; Marey, M.V.; Tsvirkun, D.V.; Vavina, O.V.; Khodzhaeva, Z.S.; Kan, N.E.; Menon, R.; Vysokikh, M.Y.; et al. Mitochondrial role in adaptive response to stress conditions in preeclampsia. Sci. Rep. 2016, 6, 32410. [Google Scholar] [CrossRef]
- Doridot, L.; Chatre, L.; Ducat, A.; Vilotte, J.L.; Lombes, A.; Mehats, C.; Barbaux, S.; Calicchio, R.; Ricchetti, M.; Vaiman, D. Nitroso-redox balance and mitochondrial homeostasis are regulated by STOX1, a pre-eclampsia-associated gene. Antioxid. Redox Signal. 2014, 21, 819–834. [Google Scholar] [CrossRef] [PubMed]
- Brodowski, L.; Zindler, T.; von Hardenberg, S.; Schroder-Heurich, B.; von Kaisenberg, C.S.; Frieling, H.; Hubel, C.A.; Dork, T.; von Versen-Hoynck, F. Preeclampsia-Associated Alteration of DNA Methylation in Fetal Endothelial Progenitor Cells. Front. Cell Dev. Biol. 2019, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, Y.; Du, L.; Li, X.; Li, X.; Chen, D. Evaluation of circulating placenta-related long noncoding RNAs as potential biomarkers for preeclampsia. Exp. Ther. Med. 2018, 15, 4309–4317. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Kuang, L.Y.; He, F.; Du, L.L.; Li, Q.L.; Sun, W.; Zhou, Y.M.; Li, X.M.; Li, X.Y.; Chen, D.J. The Role and Molecular Mechanism of Long Nocoding RNA-MEG3 in the Pathogenesis of Preeclampsia. Reprod. Sci. 2018, 25, 1619–1628. [Google Scholar] [CrossRef] [PubMed]
- Jairajpuri, D.S.; Malalla, Z.H.; Mahmood, N.; Almawi, W.Y. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 2017, 627, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Lykoudi, A.; Kolialexi, A.; Lambrou, G.I.; Braoudaki, M.; Siristatidis, C.; Papaioanou, G.K.; Tzetis, M.; Mavrou, A.; Papantoniou, N. Dysregulated placental microRNAs in Early and Late onset Preeclampsia. Placenta 2018, 61, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Purwosunu, Y.; Sekizawa, A.; Okazaki, S.; Farina, A.; Wibowo, N.; Nakamura, M.; Rizzo, N.; Saito, H.; Okai, T. Prediction of preeclampsia by analysis of cell-free messenger RNA in maternal plasma. Am. J. Obstet. Gynecol. 2009, 200, 386.e1–386.e7. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhao, W.; Lv, H.; Li, W.P.; Chen, Z.J.; Zhang, C. Transcriptomic Profiling in Human Decidua of Severe Preeclampsia Detected by RNA Sequencing. J. Cell. Biochem. 2018, 119, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Chen, L.L. Life without A tail: New formats of long noncoding RNAs. Int. J. Biochem. Cell Biol. 2014, 54, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- He, X.; He, Y.; Xi, B.; Zheng, J.; Zeng, X.; Cai, Q.; OuYang, Y.; Wang, C.; Zhou, X.; Huang, H.; et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS ONE 2013, 8, e81437. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Rui, C.; Song, X.; Dai, X.; Xue, X.; Lu, Y.; Shen, R.; Li, J.; Li, J.; Ding, H. Distinct expression profiles of lncRNAs between early-onset preeclampsia and preterm controls. Clin. Chim. Acta 2016, 463, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol. Cancer 2017, 16, 107. [Google Scholar] [CrossRef] [PubMed]
- Amigorena, S. © 1998 Nature Publishing Group. Nature Medicine. 1998. Available online: http://www.nature.com/naturemedicine (accessed on 3 May 2019).
- Mullen, C.A. Review: Analogies between trophoblastic and malignant cells. Am. J. Reprod. Immunol. 1998, 39, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, C.; Bruni, L.; Dangles-Marie, V.; Pecking, A.P.; Bellet, D. Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 2007, 13, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Genbacev, O.; Zhou, Y.; Ludlow, J.W.; Fisher, S.J. Regulation of human placental development by oxygen tension. Science 1997, 277, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, R.; Tano, K.; Mizuno, R.; Nakamura, Y.; Ijiri, K.; Rakwal, R.; Shibato, J.; Masuo, Y.; Mayeda, A.; Hirose, T.; et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 2012, 18, 738–751. [Google Scholar] [CrossRef]
- Tseng, J.-J.; Hsieh, Y.-T.; Hsu, S.-L.; Chou, M.-M. Metastasis associated lung adenocarcinoma transcript 1 is up-regulated in placenta previa increta/percreta and strongly associated with trophoblast-like cell invasion in vitro. Mol. Hum. Reprod. 2009, 15, 725–731. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Liu, F.; Liu, D.; Miao, H.; Ren, J.; Xu, J.; Ding, L.; Hu, Y.; Wang, Z.; et al. Long Non-Coding RNA MALAT1 Promotes Proliferation, Angiogenesis, and Immunosuppressive Properties of Mesenchymal Stem Cells by Inducing VEGF and IDO. J. Cell. Biochem. 2017, 118, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocinol. 2012, 48, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Davatzikos, C.; Rathore, S.; Bakas, S.; Pati, S.; Bergman, M.; Kalarot, R.; Sridharan, P.; Gastounioti, A.; Jahani, N.; Cohen, E.; et al. Cancer imaging phenomics toolkit: Quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 2018, 5, 011018. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.C.; Jiang, Y.; Yang, M.M.; He, S.N.; Xi, X.; Xu, Y.T.; Hu, W.S.; Luo, Q. Hypermethylation of delta-like homolog 1/maternally expressed gene 3 loci in human umbilical veins: Insights into offspring vascular dysfunction born after preeclampsia. J. Hypertens. 2019, 37, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A Long Noncoding RNA Activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, H.; Kong, W.; Zhang, Y.; Cao, L.; Gao, L.; Zhou, R. Down-regulated long non-coding RNA-ATB in preeclampsia and its effect on suppressing migration, proliferation, and tube formation of trophoblast cells. Placenta 2017, 49, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, D.; Yang, Y.U.; Cui, X.; Sun, J.; Liang, C.; Qin, H.; Yang, X.; Liu, S.; Yan, Q. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and α1,3-fucosylation. Cell Death Differ. 2017, 24, 2161–2172. [Google Scholar] [CrossRef] [PubMed]
- Renthal, N.E.; Chen, C.-C.; Williams, K.C.; Gerard, R.D.; Prange-Kiel, J.; Mendelson, C.R. miR-200 family and targets, ZEB1 and ZEB2, modulate uterine quiescence and contractility during pregnancy and labor. Proc. Natl. Acad. Sci. USA 2010, 107, 20828–20833. [Google Scholar] [CrossRef]
- Paysan, L.; Piquet, L.; Saltel, F.; Moreau, V. Rnd3 in Cancer: A Review of the Evidence for Tumor Promoter or Suppressor. Mol. Cancer Res. 2016, 14, 1033–1044. [Google Scholar] [CrossRef]
- Xu, Y.; Lian, Y.; Zhang, Y.; Huang, S.; Zuo, Q.; Yang, N.; Chen, Y.; Wu, D.; Sun, L. The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. J. Cell. Mol. Med. 2018, 22, 1272–1282. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, J.; He, Y.; Zhang, H.; Jiang, M.; Cao, D.; Wang, A. HOXD8/DIAPH2-AS1 epigenetically regulates PAX3 and impairs HTR-8/SVneo cell function under hypoxia. Biosci. Rep. 2019, 39, BSR20182022. [Google Scholar] [CrossRef] [PubMed]
- Loupe, J.M.; Miller, P.J.; Bonner, B.P.; Maggi, E.C.; Vijayaraghavan, J.; Crabtree, J.S.; Taylor, C.M.; Zabaleta, J.; Hollenbach, A.D. Comparative transcriptomic analysis reveals the oncogenic fusion protein PAX3-FOXO1 globally alters mRNA and miRNA to enhance myoblast invasion. Oncogenesis 2016, 5, e246. [Google Scholar] [CrossRef] [PubMed]
- Pineles, B.L.; Romero, R.; Montenegro, D.; Tarca, A.L.; Han, Y.M.; Kim, Y.M.; Draghici, S.; Espinoza, J.; Kusanovic, J.P.; Mittal, P.; et al. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am. J. Obstet. Gynecol. 2007, 196, e261. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.m.; Han, T.; Sargent, I.L.; Yin, G.w.; Yao, Y.q. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 2009, 200, e661. [Google Scholar] [CrossRef]
- Biró, O.; Nagy, B.; Rigó, J. Identifying miRNA regulatory mechanisms in preeclampsia by systems biology approaches. Hypertens. Pregnancy 2017, 36, 90–99. [Google Scholar] [CrossRef]
- Zhou, C.; Zou, Q.Y.; Li, H.; Wang, R.F.; Liu, A.X.; Magness, R.R.; Zheng, J. Preeclampsia Downregulates MicroRNAs in Fetal Endothelial Cells: Roles of miR-29a/c-3p in Endothelial Function. J. Clin. Endocrinol. Metab. 2017, 102, 3470–3479. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wu, L.; Zhu, X.; Xu, J.; Jin, R.; Li, G.; Wu, F. MiR-29a modulates the angiogenic properties of human endothelial cells. Biochem. Biophys. Res. Commun. 2013, 434, 143–149. [Google Scholar] [CrossRef]
- Davis, E.F.; Newton, L.; Lewandowski, A.J.; Lazdam, M.; Kelly, B.A.; Kyriakou, T.; Leeson, P. Pre-eclampsia and offspring cardiovascular health: Mechanistic insights from experimental studies. Clin. Sci. (Lond.) 2012, 123, 53–72. [Google Scholar] [CrossRef]
- Butalia, S.; Audibert, F.; Cote, A.M.; Firoz, T.; Logan, A.G.; Magee, L.A.; Mundle, W.; Rey, E.; Rabi, D.M.; Daskalopoulou, S.S.; et al. Hypertension Canada’s 2018 Guidelines for the Management of Hypertension in Pregnancy. Can. J. Cardiol. 2018, 34, 526–531. [Google Scholar] [CrossRef]
- Yu, G.Z.; Reilly, S.; Lewandowski, A.J.; Aye, C.Y.L.; Simpson, L.J.; Newton, L.; Davis, E.F.; Zhu, S.J.; Fox, W.R.; Goel, A.; et al. Neonatal Micro-RNA Profile Determines Endothelial Function in Offspring of Hypertensive Pregnancies. Hypertension 2018, 72, 937–945. [Google Scholar] [CrossRef] [PubMed]
- BAVELLONI, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A Current Overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar] [PubMed]
- Kulshreshtha, R.; Ferracin, M.; Wojcik, S.E.; Garzon, R.; Alder, H.; Agosto-Perez, F.J.; Davuluri, R.; Liu, C.-G.; Croce, C.M.; Negrini, M.; et al. A MicroRNA Signature of Hypoxia. Mol. Cell. Biol. 2006, 27, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Zhang, H.; Huang, P.; Luthra, R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010, 29, 4362–4368. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.J.; Souza, A.L.; Clish, C.B.; Puigserver, P. A Hypoxia-Induced Positive Feedback Loop Promotes Hypoxia-Inducible Factor 1 Stability through miR-210 Suppression of Glycerol-3-Phosphate Dehydrogenase 1-Like. Mol. Cell. Biol. 2012, 32, 898. [Google Scholar] [CrossRef]
- Fasanaro, P.; Di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 Modulates Endothelial Cell Response to Hypoxia and Inhibits the Receptor Tyrosine Kinase Ligand Ephrin-A3. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef] [PubMed]
- Cabello, C.M.; Bair, W.B.; Lamore, S.D.; Ley, S.; Alexandra, S.; Azimian, S.; Wondrak, G.T. The cinnamon-derived Michael acceptor cinnamic aldehyde impairs melanoma cell proliferation, invasiveness, and tumor growth. Free Radic. Biol. Med. 2009, 46, 220–231. [Google Scholar]
- Lee, D.-C.; Romero, R.; Kim, J.-S.; Tarca, A.L.; Montenegro, D.; Pineles, B.L.; Kim, E.; Lee, J.; Kim, S.Y.; Draghici, S.; et al. miR-210 Targets Iron-Sulfur Cluster Scaffold Homologue in Human Trophoblast Cell Lines. Am. J. Pathol. 2011, 179, 590–602. [Google Scholar] [CrossRef]
- Muralimanoharan, S.; Maloyan, A.; Mele, J.; Guo, C.; Myatt, L.G.; Myatt, L. MIR-210 modulates mitochondrial respiration in placenta with preeclampsia. Placenta 2012, 33, 816–823. [Google Scholar] [CrossRef]
- Kopriva, S.E.; Chiasson, V.L.; Mitchell, B.M.; Chatterjee, P. TLR3-Induced Placental miR-210 Down-Regulates the STAT6/Interleukin-4 Pathway. PLoS ONE 2013, 8, e67760. [Google Scholar] [CrossRef]
- Luo, R.; Shao, X.; Xu, P.; Liu, Y.; Wang, Y.; Zhao, Y.; Liu, M.; Ji, L.; Li, Y.X.; Chang, C.; et al. MicroRNA-210 contributes to preeclampsia by downregulating potassium channel modulatory factor 1. Hypertension 2014, 64, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Wang, Y.; Xu, P.; Cao, G.; Zhao, Y.; Shao, X.; Li, Y.X.; Chang, C.; Peng, C.; Wang, Y.L. Hypoxia-inducible miR-210 contributes to preeclampsia via targeting thrombospondin type I domain containing 7A. Sci. Rep. 2016, 6, 19588. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, L.; Wang, D.; Xu, Y.; Gao, H.; Tan, W.; Wang, C. Contribution of regulatory T cells to immune tolerance and association of microRNA-210 and Foxp3 in preeclampsia. Mol. Med. Rep. 2019, 19, 1150–1158. [Google Scholar] [CrossRef]
- Rudensky, A.Y. Regulatory T Cells and Foxp3. Immunol. Rev. 2011, 241, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, L.T.; Liang, G.P.; Zhang, P.; Deng, X.J.; Tang, Q.; Zhai, H.Y.; Chang, C.C.; Su, Y.W.; Lu, Q.J. Up-regulation of microRNA-210 induces immune dysfunction via targeting FOXP3 in CD4(+) T cells of psoriasis vulgaris. Clin. Immunol. 2014, 150, 22–30. [Google Scholar] [CrossRef]
- Zhang, Y.; Diao, Z.; Su, L.; Sun, H.; Li, R.; Cui, H.; Hu, Y. MicroRNA-155 contributes to preeclampsia by down-regulating CYR61. Am. J. Obstet. Gynecol. 2010, 202, e461–e466. [Google Scholar] [CrossRef] [PubMed]
- Mo, F.-E.; Muntean, A.G.; Chen, C.-C.; Stolz, D.B.; Watkins, S.C.; Lau, L.F. CYR61 (CCN1) Is Essential for Placental Development and Vascular Integrity. Mol. Cell. Biol. 2002, 22, 8709–8720. [Google Scholar] [CrossRef] [PubMed]
- Holbourn, K.; Ravi Acharya, K.; Perbal, B. The CCN family of proteins: Structure–function relationships. Trends Biochem. Sci. 2008, 33, 561–573. [Google Scholar] [CrossRef]
- Deloia, J.A.; Burlingame, J.M.; Krasnow, J.S. Differential Expression of G1 Cyclins During Human Placentogenesis. Placenta 1997, 18, 9–16. [Google Scholar] [CrossRef]
- Baldin, V.; Marcote, M.J.; Lukas, J.; Draetta, G.; Pagano, M. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 2007, 7, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Yung, H.-w.; Calabrese, S.; Hynx, D.; Hemmings, B.A.; Cetin, I.; Charnock-Jones, D.S.; Burton, G.J. Evidence of Placental Translation Inhibition and Endoplasmic Reticulum Stress in the Etiology of Human Intrauterine Growth Restriction. Am. J. Pathol. 2008, 173, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, J.; Ding, Y. Association of microRNA-155, interleukin 17A, and proteinuria in preeclampsia. Medicine 2017, 96, e6509. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.B.; Nelson, P.S.; Knudsen, B.S.; Parkin, R.K.; Noteboom, J.; Kroh, E.M.; O’Briant, K.C.; Drescher, C.W.; Vessella, R.L.; Gentleman, R.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar]
- Dai, Y.; Diao, Z.; Sun, H.; Li, R.; Qiu, Z.; Hu, Y. MicroRNA-155 is involved in the remodelling of human-trophoblast-derived HTR-8/SVneo cells induced by lipopolysaccharides. Hum. Reprod. 2011, 26, 1882–1891. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.C.; Fusi, L.; Haskard, D.O.; Swiet, M.D.; Page, P. Association of Maternal Endothelial Dysfunction With Preeclampsia. JAMA J. Am. Med. Assoc. 2014, 28, 1607–1612. [Google Scholar]
- Cheng, W.; Liu, T.; Jiang, F.; Liu, C.; Zhao, X.; Gao, Y.; Wang, H.; Liu, Z. microRNA-155 regulates angiotensin II type 1 receptor expression in umbilical vein endothelial cells from severely pre-eclamptic pregnant women. Int. J. Mol. Med. 2011, 27, 393–399. [Google Scholar] [PubMed]
- Shan, H.Y.; Bai, X.J.; Chen, X.M. Angiotensin II induces endothelial cell senescence via the activation of mitogen-activated protein kinases. Cell Biochem. Funct. 2008, 26, 459–466. [Google Scholar] [CrossRef]
- Seligman, S.P.; Buyon, J.P.; Clancy, R.M.; Young, B.K.; Abramson, S.B. The role of nitric oxide in the pathogenesis of preeclampsia. Am. J. Obstet. Gynecol. 1994, 171, 944–948. [Google Scholar] [CrossRef]
- Sun, H.X.; Zeng, D.Y.; Li, R.T.; Pang, R.P.; Yang, H.; Hu, Y.L.; Zhang, Q.; Jiang, Y.; Huang, L.Y.; Tang, Y.B.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Shen, L.; Li, Y.; Li, R.; Diao, Z.; Yany, M.; Wu, M.; Sun, H.; Yan, G.; Hu, Y. Placenta-associated serum exosomal miR-155 derived from patients with preeclampsia inhibits eNOS expression in human umbilical vein endothelial cells. Int. J. Mol. Med. 2018, 41, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Rensen, S.S.M.; Doevendans, P.A.F.M.; Van Eys, G.J.J.M. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 2007, 15, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Boerth, N.J.; Dey, N.B.; Cornwell, T.L.; Lincoln, T.M. Cyclic GMP-Dependent Protein Kinase Regulates Vascular Smooth Muscle Cell Phenotype. J. Vasc. Res. 1997, 34, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, T.M.; Dey, N.B.; Boerth, N.J.; Cornwell, T.L.; Soff, G.A. Nitric oxide—Cyclic GMP pathway regulates vascular smooth muscle cell phenotypic modulation: Implications in vascular diseases. Acta Physiol. Scand. 1998, 164, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, M.; Kim, J.; Park, W.; Kim, S.; Lee, D.K.; Hwang, J.Y.; Choe, J.; Won, M.H.; Ryoo, S.; et al. TNF-α elicits phenotypic and functional alterations of vascular smooth muscle cells by miR-155-5p–dependent down-regulation of cGMP-dependent kinase 1. J. Biol. Chem. 2018, 293, 14812–14822. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Choi, S.; Kim, S.; Kim, J.; Lee, D.K.; Park, W.; Kim, T.; Jung, J.; Hwang, J.Y.; Won, M.H.; et al. NF-κB-responsive miR-155 induces functional impairment of vascular smooth muscle cells by downregulating soluble guanylyl cyclase. Exp. Mol. Med. 2019, 51, 17. [Google Scholar] [CrossRef]
- Lo, Y.M.; Chiu, R.W. Prenatal diagnosis: Progress through plasma nucleic acids. Nat. Rev. Genet. 2007, 8, 71–77. [Google Scholar]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Gunel, T.; Zeybek, Y.G.; Akcakaya, P.; Kalelioglu, I.; Benian, A.; Ermis, H.; Aydinli, K. Serum microRNA expression in pregnancies with preeclampsia. Genet. Mol. Res. 2011, 10, 4034–4040. [Google Scholar] [CrossRef]
- Li, Q.; Long, A.; Jiang, L.; Cai, L.; Xie, L.I.; Gu, J.; Chen, X.; Tan, L. Quantification of preeclampsia-related microRNAs in maternal serum. Biomed. Rep. 2015, 3, 792–796. [Google Scholar] [CrossRef]
- Munaut, C.; Tebache, L.; Blacher, S.; Noel, A.; Nisolle, M.; Chantraine, F. Dysregulated circulating miRNAs in preeclampsia. Biomed. Rep. 2016, 5, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Liu, Z.; Wei, M.; Chen, Y.; Yang, X.; Chen, L.; Xiao, X. MIR-210 and miR-155 as potential diagnostic markers for pre-eclampsia pregnancies. Medicine 2017, 96, e7515. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef] [PubMed]
- Winger, E.E.; Reed, J.L.; Ji, X.; Nicolaides, K. Peripheral blood cell microRNA quantification during the first trimester predicts preeclampsia: Proof of concept. PLoS ONE 2018, 13, e0190654. [Google Scholar] [CrossRef] [PubMed]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014, 18, 371–390. [Google Scholar] [CrossRef] [PubMed]
- El-Khoury, V.; Pierson, S.; Kaoma, T.; Bernardin, F.; Berchem, G. Assessing cellular and circulating miRNA recovery: The impact of the RNA isolation method and the quantity of input material. Sci. Rep. 2016, 6, 19529. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhao, Y.; Liu, M.; Wang, Y.; Wang, H.; Li, Y.X.; Zhu, X.; Yao, Y.; Wang, H.; Qiao, J.; et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 2014, 63, 1276–1284. [Google Scholar] [CrossRef]
- Adel, S.; Mansour, A.; Louka, M.; Matboli, M.; Elmekkawi, S.F.; Swelam, N. Evaluation of MicroRNA-210 and Protein tyrosine phosphatase, non-receptor type 2 in Pre-eclampsia. Gene 2017, 596, 105–109. [Google Scholar] [CrossRef]
- Manaster, I.; Goldman-Wohl, D.; Greenfield, C.; Nachmani, D.; Tsukerman, P.; Hamani, Y.; Yagel, S.; Mandelboim, O. MiRNA-Mediated Control of HLA-G Expression and Function. PLoS ONE 2012, 7, e33395. [Google Scholar] [CrossRef]
- Su, M.-T.; Tsai, P.-Y.; Tsai, H.-L.; Chen, Y.-C.; Kuo, P.-L. miR-346 and miR-582-3p-regulated EG-VEGF expression and trophoblast invasion via matrix metalloproteinases 2 and 9. BioFactors 2017, 43, 210–219. [Google Scholar] [CrossRef]
- Tan, Z.; Randall, G.; Fan, J.; Camoretti-Mercado, B.; Brockman-Schneider, R.; Pan, L.; Solway, J.; Gern, J.E.; Lemanske, R.F.; Nicolae, D.; et al. Allele-Specific Targeting of microRNAs to HLA-G and Risk of Asthma. Am. J. Hum. Genet. 2007, 81, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-H.; Sun, H.-M.; Zheng, R.-Z.; Li, Y.-C.; Zhang, Q.; Cheng, P.; Tang, Z.-H.; Huang, F. Meta-analysis of microRNA-183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene 2013, 527, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Lasabova, Z.; Vazan, M.; Zibolenova, J.; Svecova, I. Overexpression of miR-21 and miR-122 in preeclamptic placentas. Neuro Endocrinol. Lett. 2015, 36, 695–699. [Google Scholar] [PubMed]
- Gao, S.; Wang, Y.; Han, S.; Zhang, Q. Up-regulated microRNA-300 in maternal whole peripheral blood and placenta associated with pregnancy-induced hypertension and preeclampsia. Int. J. Clin. Exp. Pathol. 2017, 10, 4232–4242. [Google Scholar]
- Turco, M.Y.; Gardner, L.; Kay, R.G.; Hamilton, R.S.; Prater, M.; Hollinshead, M.S.; McWhinnie, A.; Esposito, L.; Fernando, R.; Skelton, H.; et al. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 2018, 564, 263–267. [Google Scholar] [CrossRef]
- Coutifaris, C.; Kao, L.C.; Sehdev, H.M.; Chin, U.; Babalola, G.O.; Blaschuk, O.W.; Strauss, J.F. E-cadherin expression during the differentiation of human trophoblasts. Development (Camb. Engl.) 1991, 113, 767–777. [Google Scholar]
- Chakraborty, D.; Cui, W.; Rosario, G.X.; Scott, R.L.; Dhakal, P.; Renaud, S.J.; Tachibana, M.; Rumi, M.A.; Mason, C.W.; Krieg, A.J.; et al. HIF-KDM3A-MMP12 regulatory circuit ensures trophoblast plasticity and placental adaptations to hypoxia. Proc. Natl. Acad. Sci. USA 2016, 113, E7212–E7221. [Google Scholar] [CrossRef]
- Alahari, S.; Post, M.; Rolfo, A.; Weksberg, R.; Caniggia, I. Compromised JMJD6 Histone Demethylase Activity Affects VHL Gene Repression in Preeclampsia. J. Clin. Endocrinol. Metab. 2018, 103, 1545–1557. [Google Scholar] [CrossRef]
- Xie, D.; Zhu, J.; Liu, Q.; Li, J.; Song, M.; Wang, K.; Zhou, Q.; Jia, Y.; Li, T. Dysregulation of HDAC9 Represses Trophoblast Cell Migration and Invasion Through TIMP3 Activation in Preeclampsia. Am. J. Hypertens. 2019, 32, 515–523. [Google Scholar] [CrossRef]
- Zadora, J.; Singh, M.; Herse, F.; Przybyl, L.; Haase, N.; Golic, M.; Yung, H.W.; Huppertz, B.; Cartwright, J.E.; Whitley, G.; et al. Disturbed Placental Imprinting in Preeclampsia Leads to Altered Expression of DLX5, a Human-Specific Early Trophoblast Marker. Circulation 2017, 136, 1824–1839. [Google Scholar] [CrossRef]
- Van Dijk, M.; Mulders, J.; Poutsma, A.; Konst, A.A.; Lachmeijer, A.M.; Dekker, G.A.; Blankenstein, M.A.; Oudejans, C.B. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat. Genet. 2005, 37, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, M.; van Bezu, J.; van Abel, D.; Dunk, C.; Blankenstein, M.A.; Oudejans, C.B.; Lye, S.J. The STOX1 genotype associated with pre-eclampsia leads to a reduction of trophoblast invasion by alpha-T-catenin upregulation. Hum. Mol. Genet. 2010, 19, 2658–2667. [Google Scholar] [CrossRef] [PubMed]
- Rigourd, V.; Chauvet, C.; Chelbi, S.T.; Rebourcet, R.; Mondon, F.; Letourneur, F.; Mignot, T.M.; Barbaux, S.; Vaiman, D. STOX1 overexpression in choriocarcinoma cells mimics transcriptional alterations observed in preeclamptic placentas. PLoS ONE 2008, 3, e3905. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A. Genomic imprinting, development and disease--is pre-eclampsia caused by a maternally imprinted gene? Reprod. Fertil. Dev. 1998, 10, 23–29. [Google Scholar] [CrossRef] [PubMed]
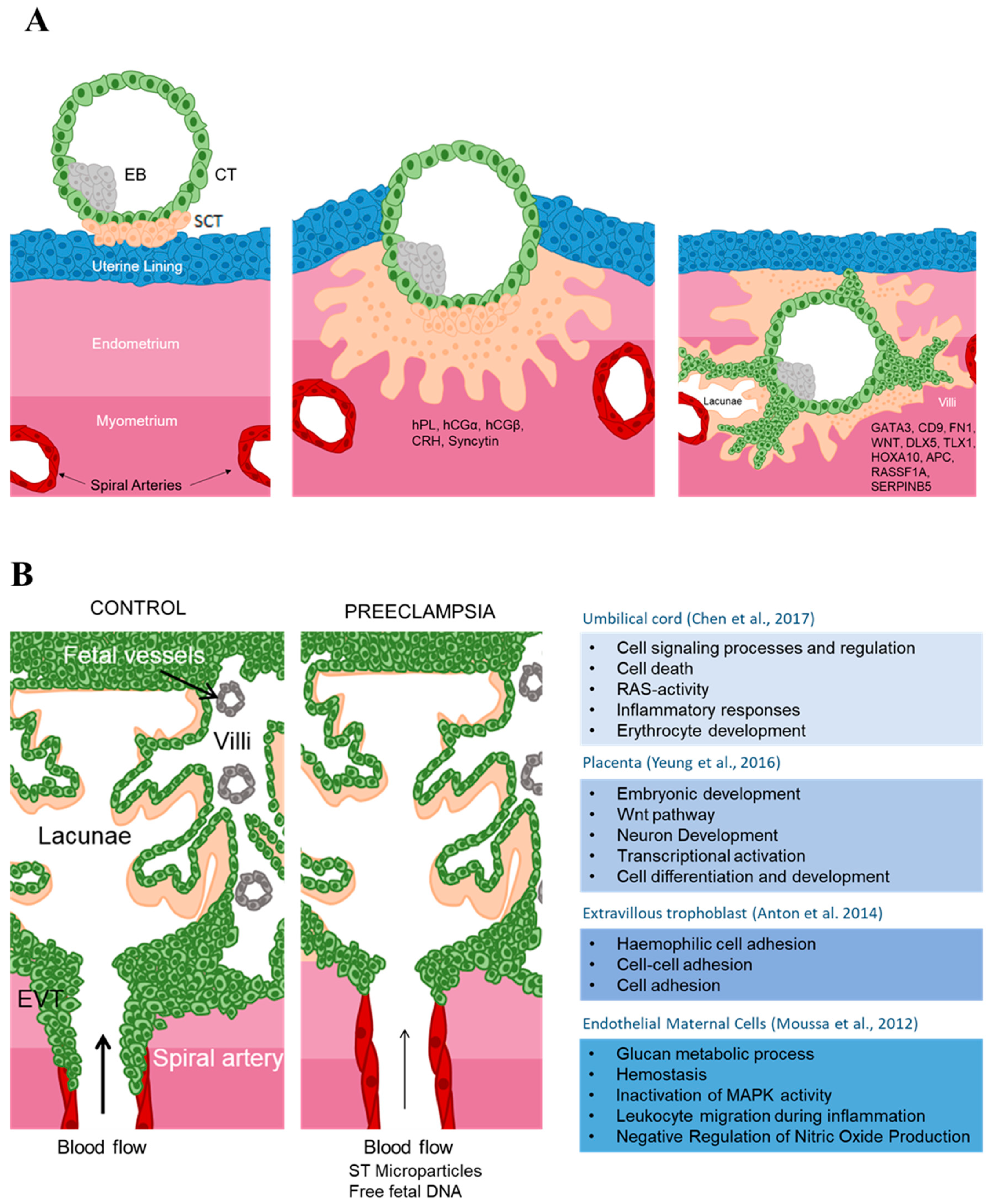


| Epigenetic Mechanism | Target | Cell Type | Biological Relevance | Reference |
|---|---|---|---|---|
| H3K9/27me3 | MMP-2, MMP-9 | Human placenta | Related to trophoblasts motility and invasion | [23] |
| H3K4 acetylation + H3K9 methylation | Maspin | Human placenta | Negatively correlated with human trophoblasts motility and invasion | [24,25] |
| Acetylated H3 | Pregnancy-Specific Glycoproteins | JEG-3 | Inhibition of HDACs in JEG-3 cells up-regulated PSG protein and mRNA expression levels | [26] |
| HDAC3 | GCMa | Cell Line | HDAC3 associates with the proximal GCMa-binding site (pGBS) in the syncytin promoter and inhibits its expression | [27] |
| Acetylation of H2A and H2B | Murine TSCs | Decreases the EMT and invasiveness of murine TSCs while maintaining their stemness phenotype | [28] | |
| H3K4Me2; H4K20me3 | Genome Wide | SCTs | H3K4Me2 co-localizes with active RNAP II in the majority of STB nuclei | [29] |
| H3K27me3 | Genome Wide | vCT | H3K27me3 highly represented in vCT | [30] |
| lncRNA TUG1 | RND3 | HTR-8/SVneo, JEG-3 | TUG1 epigenetically silences RND3 transcription by interacting with EZH2 involved in cellular proliferation, migration and invasion in trophoblasts | [31] |
| lncRNA RPAIN | C1q | HTR8/SVneo | Inhibition of proliferation and invasion. Inhibits C1q expression | [32] |
| lncRNA MALAT1 | JEG-3 | Regulates proliferation, migration, invasion and apoptosis | [33] | |
| lncRNA MEG3 | HTR8/SVneo and JEG-3 | Regulates migration and apoptosis | [34] | |
| lncRNA MIR503HG | JEG-3 | Regulates migration and invasion | [35] | |
| lncRNA LINC00629 | JEG-3 | Regulates migration and invasion | [35] | |
| lncRNA SPRY4-IT1 | HuR | HTR8/SVneo | Regulates migration and apoptosis/interferes with the β-catenin Wnt signaling | [36,37] |
| lncRNA H19 | Binds small RNAs and proteins | vCT, JAR | Regulates proliferation and apoptosis | [38] |
| miR-141-3p and miR-200a-3p | Transthyretin (TTR) | syncytitialized BeWo | Inhibits TTR expression by directly binding to the 3’UTR of TTR. Regulate thyroxin uptake by the SCT | [39] |
| miR-34 | Plasminogen activator inhibitor-1 (PAI-1), SERPINA3 | JAR | Regulates invasion | [40,41] |
| miR-155 | Cyclin D1 | HTR-8/SVneo | attenuates trophoblast proliferation | [42] |
| miR-17_92, miR-106a_363, miR-106b_25 | GCM1 | attenuate differentiation of trophoblasts | [43] | |
| miR-675 | NOMO1, Igf1R | JEG3 cells | restricts trophoblast proliferation | [44] |
| C19MC miR cluster | HTR8/SVneo | impaired migration | [45] | |
| methylation of gene body | DAXX | Human placenta | Loss of methylation during both vCT syncytialization to SCT and EVTs differentiation to invasive EVTs | [46] |
| methylation of gene promoter | APC | Human placenta and choriocarcinoma cells | trophoblast invasiveness | [47] |
| hypomethylated promoter | MASPIN | Human placenta | inhibits EVTs migration and invasion | [24,25,48] |
| Hypermethylated promoter | RASSF1A | Human placenta; JAR; JEG3 | Possible role in cytotrophoblast development through its effects on ID2 | [49] |
| Genome wide methylation | PMDs (Partially Methylated Domains) | human placenta: Chorionic Villi | genes involved in immune response, Epithelial-mesenchymal transition and inflammation | [50,51,52] |
| Genome wide methylation | Genome Wide | human SCTs compared to vCTs | hypomethylated SCTs compared to vCTS | [53] |
| Genome wide methylation | Genome Wide | BeWo and BeWo + Forskolin | DNA methylation status of numerous genes regulated at the expression level were altered by forskolin-induced fusion | [54] |
| Methylation | HOX genes: TLX1, HOXA10, DLX5 | Human placenta | Increased methylation across gestation correlates with decreased expression. Involved in SCTs differentiation | [46] |
| Genome wide methylation | Genome Wide | Side-population trophoblasts, vCTs and EVTs | Each cell population has a distinctive methylome | [55,56] |
| Methylation | Cdx2; Eomes; Plet1; TcFap2c | Mice trophoblast stem cells (TSCs) | methylation regulates the expression of genes involved in the establishment of the TSCs | [57,58,59] |
| Methylation | Genome Wide | Blastocyst | hypomethylation of the trophectoderm compared to the inner cell mass | [60] |
| Sample | Method | GEO ID | Findings | Reference |
|---|---|---|---|---|
| First-trimester and term placenta and maternal blood | Illumina HM450 | 2944 hypermethylated CpG sites in the first and 5218 in third trimester placenta. | [62] | |
| First-trimester placenta and maternal blood | MeDIP-Seq and Illumina HM450 | 3759 CpG sites in 2188 regions were differentially methylated | [63] | |
| Placenta (first, second and third trimester) | Illumina HM450 and MethylC-Seq & RNA-Seq | GSE39777 | Identification of partially methylated domains (PMDs) and differences between placenta and other tissues | [51] |
| Placenta (first, second and third trimester) | Illumina HM27 | Increase in overall genome methylation observed from first to third trimester. | [64] | |
| Term placenta | MeDIP + custom microarray | Tissue-specific differentially methylated regions in the placenta | [65] | |
| Various human trophoblast populations | Illumina HiSeq 2000 | GSE109682 | Human trophoblasts are different from somatic cells in terms of global CpG methylation | [56] |
| Methylation profiles of E18.5 term placenta of WT and Hltf−/− mouse | Illumina HiSeq 2000 (Mus musculus) | GSE114145 | Hltf-gene deletion alters the epigenetic landscape of the placenta. | [66] |
| Fetal placental tissue of both sexes in GR+/+ vs. GR+/− mice | Illumina HiSeq 2000 | GSE123188 | GR mutation in mice changes the epigenome of placental tissue in a sex-specific manner | [67] |
| Human placentas | Illumina HM450 | GSE108567 | Adjusting for batch effects in DNA methylation | [68] |
| Epigenetic mechanism of mouse embryo development | Illumina HiSeq 2500 (Mus musculus) | GSE104243 | H3K27me3 and DNA methylation in extraembryonic and embryonic lineages | [69] |
| Samples from different normal human tissues | Illumina HM450 | GSE103413 | Identifying candidate imprinted genes | Database, unpublished |
| Bisulphite and oxidative bisulphite converted placental DNA | Illumina HM450 | GSE93429 | Hydroxymethylcytosine and methylcytosine profiles in the human placenta | [70] |
| Methylation in first and third trimester placental samples | Illumina Genome Analyzer Iix | GSE98752 | Complex Association between DNA Methylation and Gene Expression | [71] |
| DNA Methylation in Human Fetal Tissues and Human IPSC | Illumina HM450 | GSE76641 | DNA methylation and transcriptional trajectories in human development. | [72] |
| DNA methylation of fetal membranes, trophoblasts and villi 2nd trimester | Illumina HM450 | GSE98938 | Genome-scale fluctuations in the cytotrophoblast epigenome | Database, unpublished |
| Developing mouse placenta | Illumina HiSeq 2000 | GSE84350 | DNA Methylation Divergence and Tissue Specialization in the Developing Mouse Placenta | [73] |
| Villous cytotrophoblasts samples | Illumina HM450 | GSE93208 | DNA methylation profiling of first trimester villous cytotrophoblasts | [52] |
| Placental tissue collected at term. | Illumina HM450 | GSE71719 | DNA methylation and hydroxymethylation assessment. | [74] |
| DNA from chorionic villus from the 1st trimester and maternal blood cell samples | Illumina HiSeq 2000 (Homo sapiens) | GSE58826 | DNA Methylation Predictors of Gene Expression in the 1st Trimester Chorionic Villus | Database, unpublished |
| Methylation patterns of human placenta, blood neutrophils and somatic tissue | Illumina HiSeq 2000 (Homo sapiens) | GSE59988 | The human placenta exhibits a dichotomized DNA methylation pattern compared to somatic tissues | [75] |
| mRNA and DNA methylation profiling of Dnmt3a/3b-null trophoblasts | Illumina HiSeq 2000 (Mus musculus) | GSE66049 | Maternal DNA methylation in early trophoblast development | [76] |
| Imprinted differentially methylated regions in hu-man villous trophoblast and blood samples | Illumina MiSeq (Homo sapiens) | GSE76273 | Polymorphic imprinted methylation in the human placenta | [77] |
| Placental villous explant culture in different growth conditions | Illumina HM450 | GSE60885 | Genome-wide DNA methylation identifies trophoblast invasion-related genes. | [78] |
| Trophoblast methylation in NLRP7 knockdown | Illumina HM450 | GSE45727 | NLRP7 alters CpG methylation | [79] |
| Bisulphite converted DNA | Illumina HumanMethylation27 BeadChip | GSE36829 | Epigenome analysis of placenta samples from newborns | Database, unpublished |
| First trimester, second trimester and full-term placentas | Illumina HumanMethylation27 BeadChip | GSE31781 | Widespread changes in promoter methylation profile in human placentas. | [80] |
| Chorionic villus and maternal blood cell samples | Illumina HumanMethylation27 BeadChip | GSE23311 | DNA Methylation Analysis in Human Chorionic Villus and Maternal Blood Cells | [81] |
| Cell Type | Gene | Methylation State in PE | Possible Target | Reference |
|---|---|---|---|---|
| Placenta and maternal plasma | SERPINB5 | Hypomethylated | Trophoblast Invasion | [177] |
| First-trimester maternal white blood cell and placenta samples | ABCA1 | Hypomethylated | Cholesterol transporter in macrophages | [178,179] |
| First-trimester maternal white blood cell, placenta samples, umbilical cord blood | GNAS | Hypomethylated | Diabetes, hypertension and metabolic diseases | [178,179] |
| First-trimester maternal white blood cell and placenta samples | TAPBP | Hypomethylated | Peptide loading in the Histocompatibility complex | [178] |
| First-trimester maternal white blood cell and placenta samples | DYNLL1 | Hypomethylated | Phosphate metabolic processing | [178] |
| First-trimester maternal white blood cell and placenta samples | ORPD1 | Hypomethylated | Opioid Receptor | [178] |
| Placenta | TIMP3 | Hypomethylated | Metalloprotease Inhibitor | [180] |
| Placenta | P2RX4 | Hypomethylated | Apoptosis and Inflammation | [170] |
| Placenta | PAPPA2 | Hypomethylated | Insuline-like growth factor regulator | [170] |
| Placenta | DLX5 | Hypomethylated | Trophoblast proliferation and differentiation | [181] |
| Placenta | KRT15 | Hypomethylated | Cytoskeleton | [182] |
| Placenta | SERPINA3 | Hypomethylated | Inhibition of inflammation, pathogen degradation and tissue remodeling | [183] |
| Placenta | FN1 | Hypomethylated | Cell adhesion, trophoblast proliferation, differentiation and apoptosis | [182] |
| Placenta | TEAD3 | Hypomethylated | Cell homeostasis, Inflammation, Coagulation, complement activation | [184] |
| Placenta | JUNB | Hypomethylated | TNF signaling pathway | [182] |
| Placenta | PKM2 | Hypomethylated | Cellular metabolism | [182] |
| Placenta | NDRG1 | Hypomethylated | Trophoblast invasion | [182] |
| Placenta | BHLHE40 | Hypomethylated | Inhibition of trophoblast differentiation | [171] |
| Placenta | INHBA | Hypomethylated | Inhibition of trophoblast differentiation | [171] |
| Placenta | CYP11A1 | Hypomethylated | Trophoblast autophagy and steroidogenic pathway | [184] |
| Placenta | HSD3B1 | Hypomethylated | Steroidogenic pathway | [184] |
| Placenta | TEAD3 | Hypomethylated | Steroidogenic pathway | [184] |
| Placenta | CYP19 | Hypomethylated | Steroidogenic pathway | [184] |
| Placenta | CRH | Hypomethylated | Cortisol bioavailability in the placenta | [184] |
| Placenta | TFPI-2 | Hypomethylated | Block in endothelial dysfunction | [185] |
| Placenta | VEGF | Hypomethylated | Angiogenesis | [186] |
| Umbilical cord blood, placenta samples | IGF2 | Hypomethylated | Embryonic development and fetal growth | [179,187] |
| Placenta and Peripheral Blood | GNA12 | Hypomethylated | Blood pressure | [188] |
| Placenta | CAPG | Hypomethylated | Macrophage function | [189] |
| Placenta | GLI2 | Hypomethylated | Embryo development | [189] |
| Placenta | KRT13 | Hypomethylated | Cytoskeleton | [189] |
| Placenta | LEP | Hypomethylated | Cell homeostasis and metabolism | [190] |
| Placenta | LP1 | Hypomethylated | Lipid metabolism | [191] |
| Placenta | CEBPα | Hypomethylated | Transcription stimulation of LEP promoter | [191] |
| Placenta | SH3PXD2A | Hypomethylated | Trophoblast invasion and podosome formation | [191] |
| Placenta | NCAM1 | Hypomethylated | Trophoblast-trophoblast interactions and adhesion | [174] |
| Cord blood samples | HSD11B2 | Hypomethylated | Cortisol transmission from the mother to the fetus | [192] |
| Placenta | WNT2 | Hypermethylated | Placentation and cell signaling | [158,193] |
| Placenta | SPESP1 | Hypermethylated | Fertilization | [158] |
| Placenta | NOX5 | Hypermethylated | Reactive Oxygen Species signaling | [158] |
| Placenta | ALCAM | Hypermethylated | Cell Adhesion | [158] |
| Placenta | IGF-1 | Hypermethylated | Placentation, trophoblast function, fetal growth. | [194] |
| Placenta | SOX7 | Hypermethylated | Embryonic development and cell fate | [155] |
| Placenta | CDX1 | Hypermethylated | Trophoblast invasion restriction | [155] |
| Placenta | CXCL1 | Hypermethylated | Chemokine inducer of angiogenesis | [155] |
| Placenta | ADORA2B | Hypermethylated | Placenta impairment and fetal growth restriction | [155] |
| Placenta | FAM3B | Hypermethylated | Cytokine activity | [182] |
| Placenta | SYNE1 | Hypermethylated | Nuclear organization and structural integrity | [182] |
| Placenta | AGAP1 | Hypermethylated | Cellular development, assembly and function | [182] |
| Placenta | CRHBP | Hypermethylated | Cortisol bioavailability in the placenta | [190] |
| Placenta and maternal blood | STAT5A | Hypermethylated | Transcription activation | [195] |
| Placenta and maternal plasma | RASSF1A | Hypermethylated | Tumor suppressor gene | [177] |
| Placenta | PTPRN2 | Hypermethylated | Phosphate metabolic processing | [173] |
| Placenta | GATA4 | Hypermethylated | Placenta Growth | [173] |
| YWHAQ | Hypermethylated | Cellular response to reduce oxygen levels | [196] | |
| Placenta | TNF | Hypermethylated | MMP-9 stimulation, Immune system activation, cell survival, migration and differentiation | [174] |
| Placenta | COL5A1 | Hypermethylated | Extracellular matrix | [174] |
| Placenta | CDH11 | Hypermethylated | Trophoblast anchoring to the decidua, syncytiotrophoblast differentiation | [174] |
| Placenta | HLA-G | Hypermethylated | Maternal Immune tolerance and immune rejection | [197] |
| microRNA | PE Placenta | PE Plasma | Function | Gene targets | AUC | References |
|---|---|---|---|---|---|---|
| miR-214 | DOWN | [308] | ||||
| miR-152 | DOWN | [300] | ||||
| miR-218 | DOWN | [308] | ||||
| miR-590 | DOWN | [308] | ||||
| miR-18a | DOWN | DOWN | Promoting trophoblast migration | SMAD2 | [225,308] | |
| miR-19a | DOWN | [308] | ||||
| miR-19b1 | DOWN | TGFβ-signaling | SMAD factors | [225] | ||
| miR-379 | DOWN | [308] | ||||
| miR-411 | DOWN | [308] | ||||
| miR-195 | DOWN | [308] | ||||
| miR-223 | DOWN | [308] | ||||
| miR-363 | DOWN | [308] | ||||
| miR-542-3p | DOWN | [308] | ||||
| miR-144 | DOWN | Ischemia, hypoxia | [225] | |||
| miR-15b | DOWN | Angiotensin-renin system | [225] | |||
| miR-181a | UP | UP | [225,308] | |||
| miR-584 | UP | [308] | ||||
| miR-30a-3p | UP | [308] | ||||
| miR-151 | UP | [308] | ||||
| miR-31 | UP | [308] | ||||
| miR-210 | UP | UP | PTPN2 | 0.7 < AUC < 0.9 | [225,255,300,302,303,305,308,309] | |
| miR-17-3p | UP | [308] | ||||
| miR-193b | UP | [308] | ||||
| miR-638 | UP | [308] | ||||
| miR-525 | UP | [308] | ||||
| miR-515-3p | UP | [308] | ||||
| miR-519e | UP | [308] | ||||
| miR-517-5p | UP | UP | AUC = 0.7 | [304] | ||
| miR-518b | UP | UP | [225,304,308] | |||
| miR-524 | UP | [308] | ||||
| miR-296 | UP | [308] | ||||
| miR-362 | UP | [308] | ||||
| miR-574-5p | UP | AUC > 0.7 | [302] | |||
| miR-1233-3p | UP | AUC > 0.6 | [302] | |||
| miR-155 | UP | AUC > 0.7 | [225,303] | |||
| miR-1267 | UP | AUC > 0.8 | [305] | |||
| miR-148a | UP | Immune response | HLA-G | AUC > 0.9 | [305,310] | |
| miR-196a | UP | AUC = 1 | [305] | |||
| miR-33a | UP | AUC = 1 | [305] | |||
| miR-575 | UP | AUC > 0.9 | [305] | |||
| miR-582 | UP | Trophoblast invasion, migration | VEGF | 1 | [305,311] | |
| miR-152 | UP | UP | Immune response | HLA-G | AUC > 0.9 | [256,301,312] |
| miR-183 | UP | UP | Cell differentiation, apoptosis, invasion | AUC > 0.9 | [255,301,313] | |
| miR-215 | UP | [225] | ||||
| miR-650 | UP | [225] | ||||
| miR-21 | UP | UP | Apoptosis | [225,314] | ||
| miR-29a | UP | [225] | ||||
| miR-300 | UP | Trophoblast differentiation | ETS-1 | [315] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apicella, C.; Ruano, C.S.M.; Méhats, C.; Miralles, F.; Vaiman, D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int. J. Mol. Sci. 2019, 20, 2837. https://doi.org/10.3390/ijms20112837
Apicella C, Ruano CSM, Méhats C, Miralles F, Vaiman D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. International Journal of Molecular Sciences. 2019; 20(11):2837. https://doi.org/10.3390/ijms20112837
Chicago/Turabian StyleApicella, Clara, Camino S. M. Ruano, Céline Méhats, Francisco Miralles, and Daniel Vaiman. 2019. "The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia" International Journal of Molecular Sciences 20, no. 11: 2837. https://doi.org/10.3390/ijms20112837
APA StyleApicella, C., Ruano, C. S. M., Méhats, C., Miralles, F., & Vaiman, D. (2019). The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. International Journal of Molecular Sciences, 20(11), 2837. https://doi.org/10.3390/ijms20112837






