The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants
Abstract
1. Introduction
2. Results
2.1. Identification of Secreted Proteins
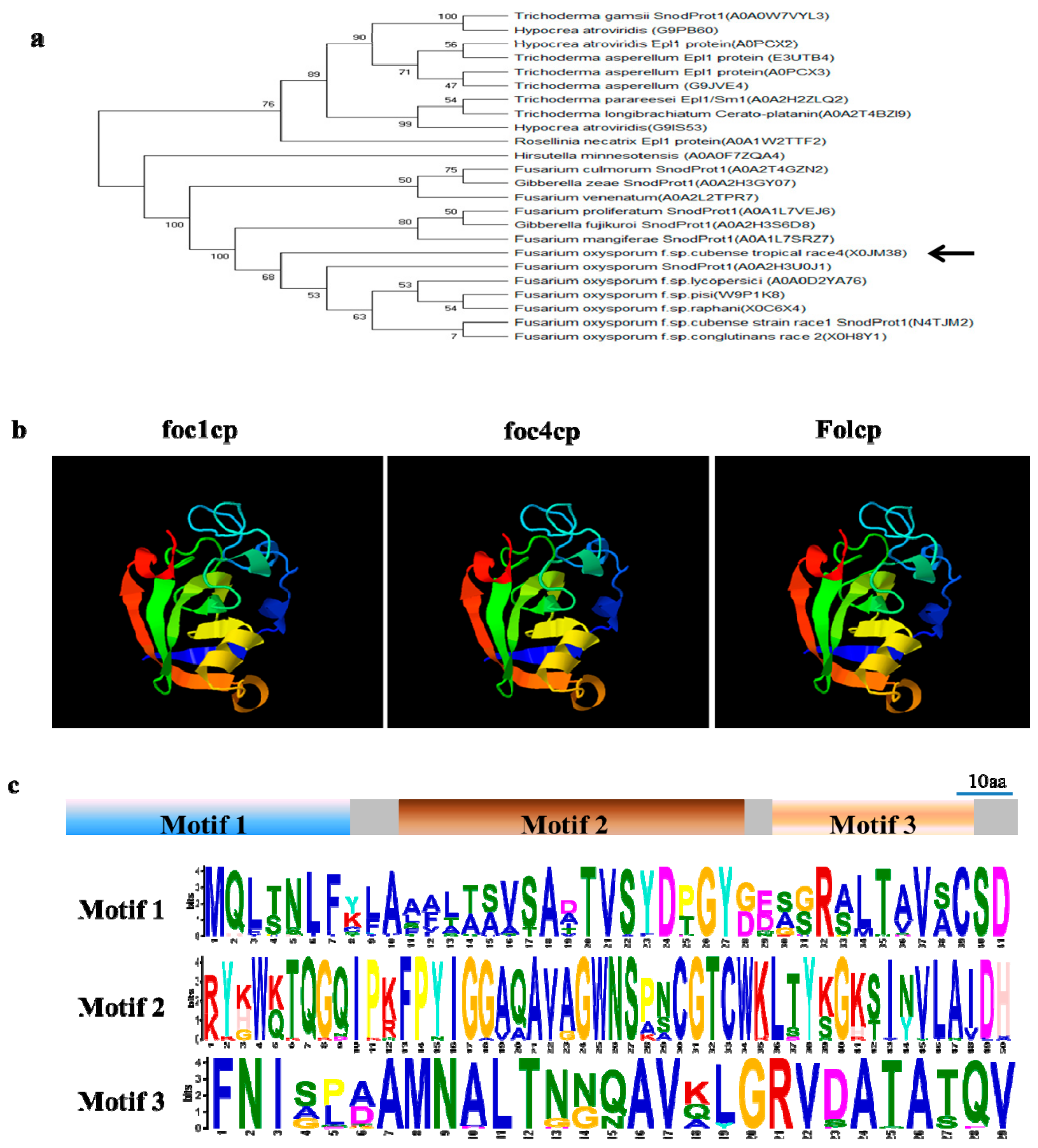
2.2. FocCP1 is A Member of the Cerato-Platanin Family
2.3. Cloning, Expression, and Purification of the Recombinant FocCP1 Protein
2.4. FocCP1 Induced Hypersensitive Response (HR) and H2O2 Accumulation in Tobacco
2.5. Defense Responses in Tobacco Caused by FocCP1
2.6. Deposition of Callose and Phenolic Compounds in Tobacco Induced by FocCP1
2.7. FocCP1 Triggers Plant Disease Resistance
3. Discussion
4. Materials and Methods
4.1. Culture Conditions of Microorganisms and Plants
4.2. Label-Free Analysis of FOC4 Strain
4.3. Bioinformatics Analysis
4.4. Cloning, Expression, and Purification of FocCP1
4.5. Characteristics of FocCP1 Protein
4.6. Detection of Hydrogen Peroxide in Tobacco
4.7. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.8. Quantification of Salicylic Acid (SA) and Jasmonic Acid (JA)
4.9. Assays of Defense Responses in Tobacco
4.10. Bioassay for FocCP1-Induced Disease Resistance in Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FOC(4) | Fusarium oxysporum f. sp. cubense (race 4) |
| CPPs | cerato-platanin proteins |
| FocCP1 | The first cerato-platanin protein from Fusarium oxysporum f. sp. cubense race 4 |
| Pst. 6605 | Pseudomonas syringae pv. tabaci 6605 |
| ROS | reactive oxygen species |
| HR | hypersensitive response |
| SAR | systemic acquired resistance |
| PTI | pathogen-associated molecular pattern triggered immunity |
| ETI | effector-triggered immunity |
| PRRs | pattern recognition receptors |
| Foc1CP | Protein SnodProt1 from Fusarium oxysporum f. sp. cubense race 1 |
| FolCP | Protein SnodProt1 from Fusarium oxysporum f. sp. lycopersici |
References
- Leong, K.S.; Latiffah, Z.; Baharuddin, S. Molecular Characterization of Fusarium oxysporum f. sp. cubense of Banana. Am. J. Appl. Sci. 2009, 6, 1301–1307. [Google Scholar] [CrossRef]
- Stover, R.H. Banana, plantain and abaca diseases. Banan. Plantain Abaca Dis. 1972, 4, 342–349. [Google Scholar]
- Fan, H.; Dong, H.; Xu, C.; Liu, J.; Hu, B.; Ye, J.; Mai, G.; Li, H. Pectin methylesterases contribute the pathogenic differences between races 1 and 4 of Fusarium oxysporum f. sp. cubense. Sci. Rep. 2017, 7, 13140. [Google Scholar] [CrossRef] [PubMed]
- Frías, M.; Brito, N.; González, C. The Botrytis cinerea cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol. Plant Pathol. 2013, 14, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.; Liu, H.; Zhou, T.; Tan, B.; Yuan, J.; Guo, L.; Qiu, D. The purification and characterization of a novel hypersensitive-like response-inducing elicitor from Verticillium dahliae that induces resistance responses in tobacco. Appl. Microbiol. Biotechnol. 2012, 93, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Pazzagli, L.; Cappugi, G.; Manao, G.; Camici, G.; Santini, A.; Scala, A. Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 1999, 274, 24959–24964. [Google Scholar] [CrossRef]
- Bennici, A.; Calamassi, R.; Pazzagli, L.; Comparini, C.; Schiff, S.; Bovelli, R.; Mori, B.; Tani, C.; Scala, A. Cytological and ultrastructural responses of Platanus acerifolia (Ait.) Willd. leaves to cerato-platanin, a protein from Ceratocystis fimbriata f. sp. platani. Phytopathol. Mediterr. 2005, 44, 153–161. [Google Scholar]
- Frías, M.; González, M.; González, C.; Brito, N. BcIEB1, a Botrytis cinerea secreted protein, elicits a defense response in plants. Plant Sci. 2016, 250, 115–124. [Google Scholar] [CrossRef]
- Zaparoli, G.; Cabrera, O.G.; Medrano, F.J.; Tiburcio, R.; Lacerda, G.; Pereira, G.G. Identification of a second family of genes in Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao, encoding necrosis-inducing proteins similar to cerato-platanins. Mycol. Res. 2009, 113, 61–72. [Google Scholar] [CrossRef]
- Jeong, J.S.; Mitchell, T.K.; Dean, R.A. The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. Fems Microbiol. Lett. 2010, 273, 157–165. [Google Scholar] [CrossRef]
- Quarantin, A.; Glasenapp, A.; Schafer, W.; Favaron, F.; Sella, L. Involvement of the Fusarium graminearum cerato-platanin proteins in fungal growth and plant infection. Plant Physiol. Biochem. 2016, 109, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.M.; Idnurm, A.; Howlett, B.J. Characterization of a gene (sp1) encoding a secreted protein from Leptosphaeria maculans, the blackleg pathogen of Brassica napus. Mol. Plant Pathol. 2010, 3, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Djonovic, S.; Pozo, M.J.; Dangott, L.J.; Howell, C.R.; Kenerley, C.M. Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 2006, 19, 838–853. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Kovalchuk, A.; Keriã¶, S.; Asiegbu, F.O. Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 2013, 105, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, Y.; Liang, Y.; Dong, Y.; Yang, X.; Yuan, J.; Qiu, D. The Verticillium dahliae SnodProt1-Like Protein VdCP1 Contributes to Virulence and Triggers the Plant Immune System. Front. Plant Sci. 2017, 8, 1880. [Google Scholar] [CrossRef] [PubMed]
- Marcos, F.; Celedonio, G.; Nélida, B. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar]
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef]
- Pazzagli, L.; Seidl-Seiboth, V.; Barsottini, M.; Vargas, W.A.; Scala, A.; Mukherjee, P.K. Cerato-platanins: Elicitors and effectors. Plant Sci. 2014, 228, 79–87. [Google Scholar] [CrossRef]
- Pazzagli, L.; Zoppi, C.; Carresi, L.; Tiribilli, B.; Sbrana, F.; Schiff, S.; Pertinhez, T.A.; Scala, A.; Cappugi, G. Characterization of ordered aggregates of cerato-platanin and their involvement in fungus-host interactions. BBA Gen. Subj. 2009, 1790, 1334–1344. [Google Scholar] [CrossRef]
- Kishimoto, K.; Kouzai, Y.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. 2010, 64, 343–354. [Google Scholar] [CrossRef]
- Mishina, T.E.; Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2010, 50, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, L.; Faoro, F.; Luti, S.; Baccelli, I.; Martellini, F.; Bernardi, R.; Picciarelli, P.; Scala, A.; Pazzagli, L. Differential timing of defense-related responses induced by cerato-platanin and cerato-populin, two non-catalytic fungal elicitors. Physiol. Plant 2013, 149, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C.; Oldroyd, G.E. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Panstruga, R.; Parker, J.E.; Schulze-Lefert, P. SnapShot: Plant immune response pathways. Cell 2009, 136, 978.e1–978.e3. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Yang, Y.; Zhang, H.; Huang, L.; Li, D.; Song, F. Overexpression of MoSM1, encoding for an immunity-inducing protein from Magnaporthe oryzae, in rice confers broad-spectrum resistance against fungal and bacterial diseases. Sci. Rep. 2017, 7, 41037. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.; Ning, Y.; Ding, B.; Wang, X.; Wang, Z.; Wang, G.L. Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant 2013, 6, 605–620. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nürnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, J.F.; Niu, Y.; Zhang, X.C.; Woody, O.Z.; Xiong, Y.; Djonovic, S.; Millet, Y.; Bush, J.; McConkey, B.J.; et al. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 2015, 521, 213–216. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, C.; Zi, Q.; Qiu, D.; Liu, W.; Zeng, H. A novel elicitor identified from Magnaporthe oryzae triggers defense responses in tobacco and rice. Plant Cell Rep. 2014, 33, 1865–1879. [Google Scholar] [CrossRef]
- González-Rodríguez, V.E.; Liñeiro, E.; Colby, T.; Harzen, A.; Garrido, C.; Cantoral, J.M.; Schmidt, J.; Fernández-Acero, F.J. Proteomic profiling of Botrytis cinerea conidial germination. Arch. Microbiol. 2015, 197, 117. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.L.; Haynes, P.A.; Breci, L.; Francisco, W.A. Analysis of secreted proteins from Aspergillus flavus. Proteomics 2005, 5, 3153–3161. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Quintana, J.; Kovalchuk, A.; Ubhayasekera, W.; Asiegbu, F.O. A cerato-platanin-like protein HaCPL2 from Heterobasidion annosum sensu stricto induces cell death in Nicotiana tabacum and Pinus sylvestris. Fungal. Genet. Biol. 2015, 84, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Brugger, A.; Lamotte, O.; Vandelle, E.; Bourque, S.; Lecourieux, D.; Poinssot, B.; Wendehenne, D.; Pugin, A. Early signaling events induced by elicitors of plant defenses. Mol. Plant Microbe. Interact. 2006, 19, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, H.G.; Li, W.; Wang, X.; Song, F. Ectopic expression of MgSM1, a Cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 2010, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Salas-Marina, M.A.; Isordia-Jasso, M.I.; Islas-Osuna, M.A.; Delgado-Sanchez, P.; Jimenez-Bremont, J.F.; Rodriguez-Kessler, M.; Rosales-Saavedra, M.T.; Herrera-Estrella, A.; Casas-Flores, S. The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front. Plant Sci. 2015, 6, 77. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef]
- Li, S.; Nie, H.; Qiu, D.; Shi, M.; Yuan, Q. A novel protein elicitor PeFOC1 from Fusarium oxysporum triggers defense response and systemic resistance in tobacco. Biochem. Biophys. Res. Commun. 2019, 514, 1074–1080. [Google Scholar] [CrossRef]
- Wang, H.; Yang, X.; Guo, L.; Zeng, H.; Qiu, D. PeBL1, a novel protein elicitor from Brevibacillus laterosporus strain A60, activates defense responses and systemic resistance in Nicotiana benthamiana. Appl. Env. Microbiol. 2015, 81, 2706–2716. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, X.; Qiu, D.; Guo, L.; Zeng, H.; Mao, J.; Gao, Q. PeaT1-induced systemic acquired resistance in tobacco follows salicylic acid-dependent pathway. Mol. Biol. Rep. 2011, 38, 2549–2556. [Google Scholar] [CrossRef]
- Chen, M.; Zeng, H.; Qiu, D.; Guo, L.; Yang, X.; Shi, H.; Zhou, T.; Zhao, J. Purification and characterization of a novel hypersensitive response-inducing elicitor from Magnaporthe oryzae that triggers defense response in rice. PLoS ONE 2012, 7, e37654. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, B.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar]
- Misra, B.B.; Armas, E.D.; Chen, S. Differential metabolomic responses of PAMP-triggered immunity and effector-triggered immunity in Arabidopsis suspension cells. Metabolomics 2016, 12, 1–15. [Google Scholar] [CrossRef]
- Kinkema, M.; Fan, W.; Dong, X. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 2000, 12, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L. EDS1-mediated basal defense and SA-signaling contribute to post-invasion resistance against tobacco powdery mildew in Arabidopsis. Physiol. Mol. Plant Pathol. 2015, 91, 120–130. [Google Scholar] [CrossRef]
- Gong, C.-R.; Li, Y.-M.; Yang, L.-J. Relationship Between LOX Activity, SA and JA Accumulation in Tobacco Leaves Under Water Stress. Agric. Sci. China 2003, 2, 624–628. [Google Scholar]
- Spoel, S.H.; Johnson, J.S.; Xinnian, D. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.; Huang, J.; Meng, F.; Pang, J.; Zhao, Q.; Islam, A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Magnaporthe oryzae chitinase MoChia1 by a rice tetratricopeptide repeat protein allows free chitin to trigger immune responses. Plant Cell 2019. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Jabs, T.; Dietrich, R.A.; Dangl, J.L. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 1996, 273, 1853–1856. [Google Scholar] [CrossRef]
- Frank, V.B.; Julia, B.S.; Ron, M. Unraveling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 2008, 147, 978–984. [Google Scholar]
- Li, X.; Guo, H.; Qi, Y.; Liu, H.; Zhang, X.; Ma, P.; Liang, Z.; Dong, J. Salicylic acid-induced cytosolic acidification increases the accumulation of phenolic acids in Salvia miltiorrhiza cells. Plant Cell Tissue Organ Cult. 2016, 126, 333–341. [Google Scholar] [CrossRef]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauchmani, B.; Ton, J. Callose Deposition: A Multifaceted Plant Defense Response. Mol. Plant Microbe. Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Davis, L.C.; Verpoorte, R. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 2005, 23, 283–333. [Google Scholar] [CrossRef] [PubMed]
- Boller, T.; Felix, G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu. Rev. Plant Biol. 2009, 60, 379. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, D.R.; Fröhlich, T.; Otte, K.A.; Beck, A.; Habermann, F.A.; Wolf, E.; Arnold, G.J. Stage-specific proteome signatures in early bovine embryo development. J. Proteome Res. 2014, 13, 4363–4376. [Google Scholar] [CrossRef] [PubMed]
- Park, G.W.; Hwang, H.; Kim, K.H.; Ju, Y.L.; Lee, H.K.; Ji, Y.P.; Ji, E.S.; Park, S.K.R.; Iii, J.R.Y.; Kwon, K.H. Integrated Proteomic Pipeline Using Multiple Search Engines for a Proteogenomic Study with a Controlled Protein False Discovery Rate. J. Proteome Res. 2016, 15, 4082. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; Von, G.H.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic. Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Timothy, B.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1993, 8, 28–36. [Google Scholar]
- Davis, D.A.; Low, P.S.; Heinstein, P. Purification of a glycoprotein elicitor of phytoalexin formation from Verticillium dahliae. Physiol. Mol. Plant Pathol. 1998, 52, 259–273. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef]
- Pugin, A.; Frachisse, J.M.; Tavernier, E.; Bligny, R.; Gout, E.; Douce, R.; Guern, J. Early Events Induced by the Elicitor Cryptogein in Tobacco Cells: Involvement of a Plasma Membrane NADPH Oxidase and Activation of Glycolysis and the Pentose Phosphate Pathway. Plant Cell 1997, 9, 2077–2091. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.V.; Costa Mdo, N.; de Paula, R.G.; de Azevedo, R.R.; da Silva, F.L.; Noronha, E.F.; Ulhoa, C.J.; Monteiro, V.N.; Cardoza, R.E.; Gutierrez, S.; et al. The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci. Rep. 2015, 5, 17998. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bowling, S.A.; Guo, A.; Cao, H.; Gordon, A.S.; Klessig, D.F.; Dong, X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 1994, 6, 1845–1857. [Google Scholar] [CrossRef]
- Lv, S.; Wang, Z.; Yang, X.; Guo, L.; Qiu, D.; Zeng, H. Transcriptional Profiling of Rice Treated with MoHrip1 Reveal the Function of Protein Elicitor in Enhancement of Disease Resistance and Plant Growth. Front. Plant Sci. 2016, 7, 1818. [Google Scholar] [CrossRef]
- Flokova, K.; Tarkowska, D.; Miersch, O.; Strnad, M.; Wasternack, C.; Novak, O. UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 2014, 105, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Aoi, A.; Satou, C.; Nakaya, M.; Masuta, C.; Nabeta, K. Simultaneous UPLC MS/MS analysis of endogenous jasmonic acid, salicylic acid, and their related compounds. Plant Growth Regul. 2008, 57, 293–301. [Google Scholar] [CrossRef]
- Shivprasad, S.; Pogue, G.P.; Lewandowski, D.J.; Hidalgo, J.; Donson, J.; Grill, L.K.; Dawson, W.O. Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 1999, 255, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2010, 30, 415–429. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Mehta, B.P. Evaluation of Fungicides Against Sigatoka Leaf Spot and Leaf Blight Diseases of Banana (Musa cavedish). J. Mycol. Plant Pathol. 2005, 2, 312–313. [Google Scholar]





| Accessions 1 | Protein Name 2 | Y/T 3 |
|---|---|---|
| Cellulases | ||
| N4UR49 | Putative glucan endo-1,3-beta-glucosidase | Y |
| N4U233 | Beta-glucosidase | T |
| N1S288 | Glucan endo-1,3-beta-glucosidase | Y |
| N1RLP0 | Putative beta-glucosidase | Y |
| N1S1N8 | Glucan endo-1,6-beta-glucosidase | Y |
| Chitinase | ||
| N1RY32 | Chitinase | Y |
| N1RJ75 | Endochitinase | Y |
| Carboxypeptidases | ||
| X0JJ77 | Carboxypeptidase | Y |
| X0K0U8 | Carboxypeptidase | Y |
| N4UD77 | Carboxypeptidase | Y |
| X0KT75 | Glutamate carboxypeptidase | T |
| N4TK61 | Putative metallocarboxypeptidase | Y |
| X0J1W7 | Carboxypeptidase | Y |
| N1RF81 | Putative carboxypeptidase | Y |
| Lipase | ||
| X0J817 | Phospholipase | Y |
| N1RKZ8 | Lysophospholipase | Y |
| N1RV52 | Lipase | Y |
| Aminopeptidases | ||
| X0JV19 | Peptide hydrolase | Y |
| X0JV19 | Peptide hydrolase | Y |
| X0JUF5 | Dipeptidyl aminopeptidase | T |
| N4UIL4 | Peptide hydrolase | Y |
| Glucanase | ||
| X0JFN8 | Endo-1,3(4)-beta-glucanase | Y |
| Transpeptidase | ||
| N4UHE3 | Gamma-glutamyl transpeptidase | Y |
| Cerato-platanin protein | ||
| X0JM38 | Uncharacterized protein | Y |
| Genes | Primer Sequences (5′-3′) | Enzyme Cutting Sites |
|---|---|---|
| FocCP1-F | ATGCAGCTGACCAACCTCTTC | |
| FocCP1-R | TTTGAGACCACAGTTGCTAATAG | |
| pPICZαA-FocCP1-F | CCGGAATTCGCGACTGTCTCCTACG 1 | EcoRI |
| pPICZαA-FocCP1-R | GCTCTAGACCTTTGAGACCACAGTTG 2 | XbaI |
| Genes | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| NtPR1 | CGTTGAGATGTGGGTCGATG | CCTAGCACATCCAACACGAA |
| NtPR5 | CTCATGCTGCCACTTTTGAC | CTCCAAGATTGGCCTGAGTC |
| NtPAL | GTTATGCTCTTAGAACGTCGCCC | CCGTGTAATGCCTTGTTTCTTGA |
| NtHSR203J | TGCCGTCAAAGATGTAGTCG | CAGCATGGCTGACACAAAAG |
| NtEDS1 | GGAGAATGGGAGAAGCAGAA | GAACGCATCATAATACCCGA |
| NtLOX | CTTTAAGAGGAGATGGAACT | TCTAAGCTCATAAGCAATGG |
| Ntactin | ATGCCTATGTGGGTGACGAAG | TCTGTTGGCCTTAGGGTTGAG |
| Disease Stage | Disease Severity Index of Roots/ Bulbs 1 | Disease Severity Index of Leaves 2 |
|---|---|---|
| 0 | pseudostems and bulbs were white and healthy | leaves were green, healthy and bright |
| 1 | vascular bundle of bulb was sporadically brown | leaves were green, healthy and bright |
| 2 | vascular bundle of bulb was sporadically brown, 1/3 of bulb area was yellow | leaves were green, not yellow |
| 3 | vascular bundle of bulb was brown, 1/3–2/3 of bulb area was yellow | a few leaves were a little yellow |
| 4 | vascular bundle of bulb was brown, more than 2/3 of bulb area was yellow | a few leaves were yellow |
| 5 | vascular bundle was brown, 1/2 of the bulb area was brown and decayed | most leaves were yellow |
| 6 | vascular bundle was brown, more than 1/2 of bulb area was brown and decayed | leaves were withered |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Dong, Y.; Li, L.; Zhang, Y.; Yang, X.; Zeng, H.; Shi, M.; Pei, X.; Qiu, D.; Yuan, Q. The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants. Int. J. Mol. Sci. 2019, 20, 2849. https://doi.org/10.3390/ijms20112849
Li S, Dong Y, Li L, Zhang Y, Yang X, Zeng H, Shi M, Pei X, Qiu D, Yuan Q. The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants. International Journal of Molecular Sciences. 2019; 20(11):2849. https://doi.org/10.3390/ijms20112849
Chicago/Turabian StyleLi, Songwei, Yijie Dong, Lin Li, Yi Zhang, Xiufen Yang, Hongmei Zeng, Mingwang Shi, Xinwu Pei, Dewen Qiu, and Qianhua Yuan. 2019. "The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants" International Journal of Molecular Sciences 20, no. 11: 2849. https://doi.org/10.3390/ijms20112849
APA StyleLi, S., Dong, Y., Li, L., Zhang, Y., Yang, X., Zeng, H., Shi, M., Pei, X., Qiu, D., & Yuan, Q. (2019). The Novel Cerato-Platanin-Like Protein FocCP1 from Fusarium oxysporum Triggers an Immune Response in Plants. International Journal of Molecular Sciences, 20(11), 2849. https://doi.org/10.3390/ijms20112849






