Establishment of Efficient Genetic Transformation Systems and Application of CRISPR/Cas9 Genome Editing Technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven
Abstract
:1. Introduction
2. Results
2.1. Effects of Hygromycin (Hyg) and Cefotaxime (Cef)
2.2. Key Transformation Factors
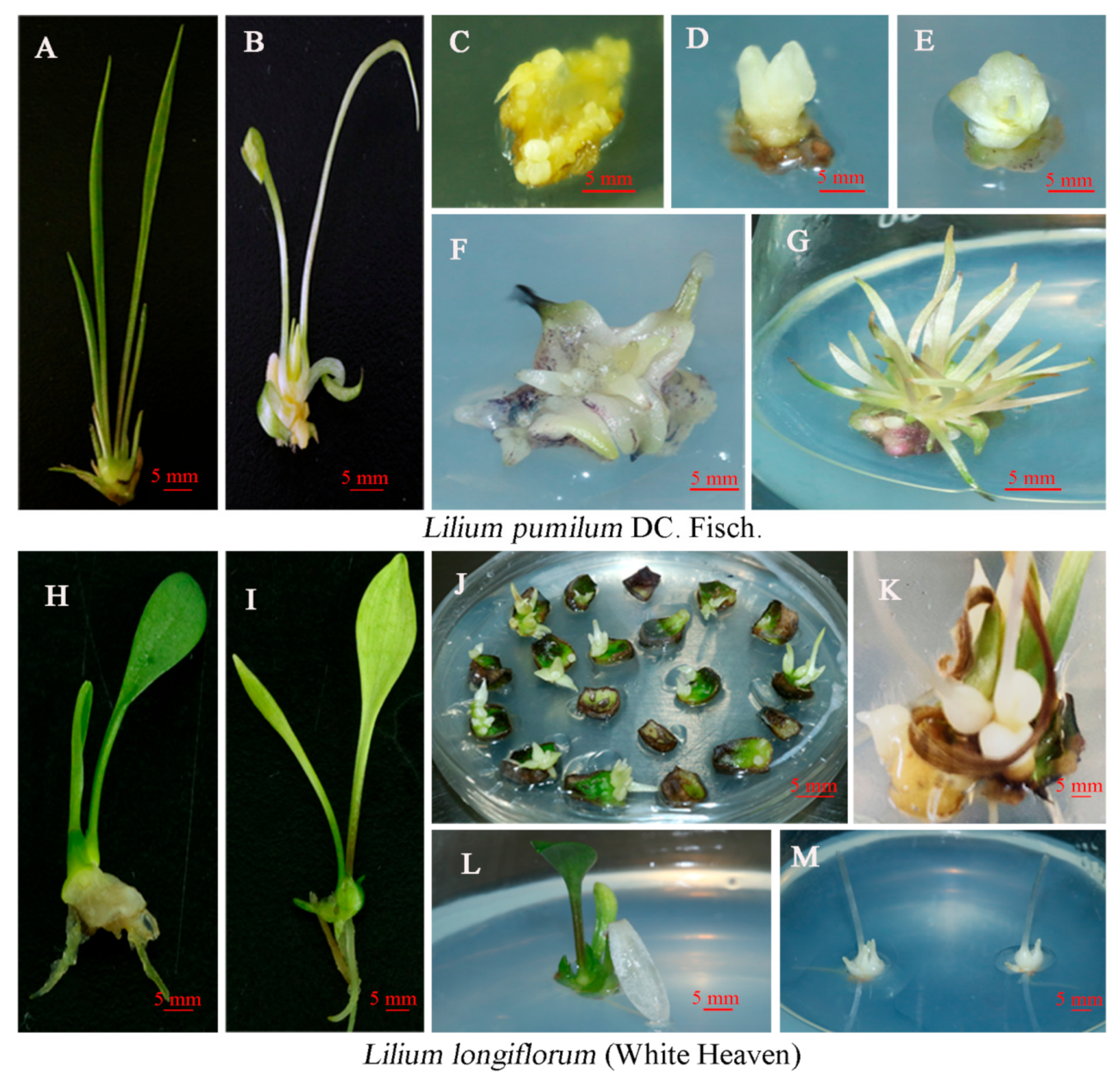
2.3. Regeneration of Transgenic Plants
2.4. β-Glucuronidase (GUS) Histochemical Assay
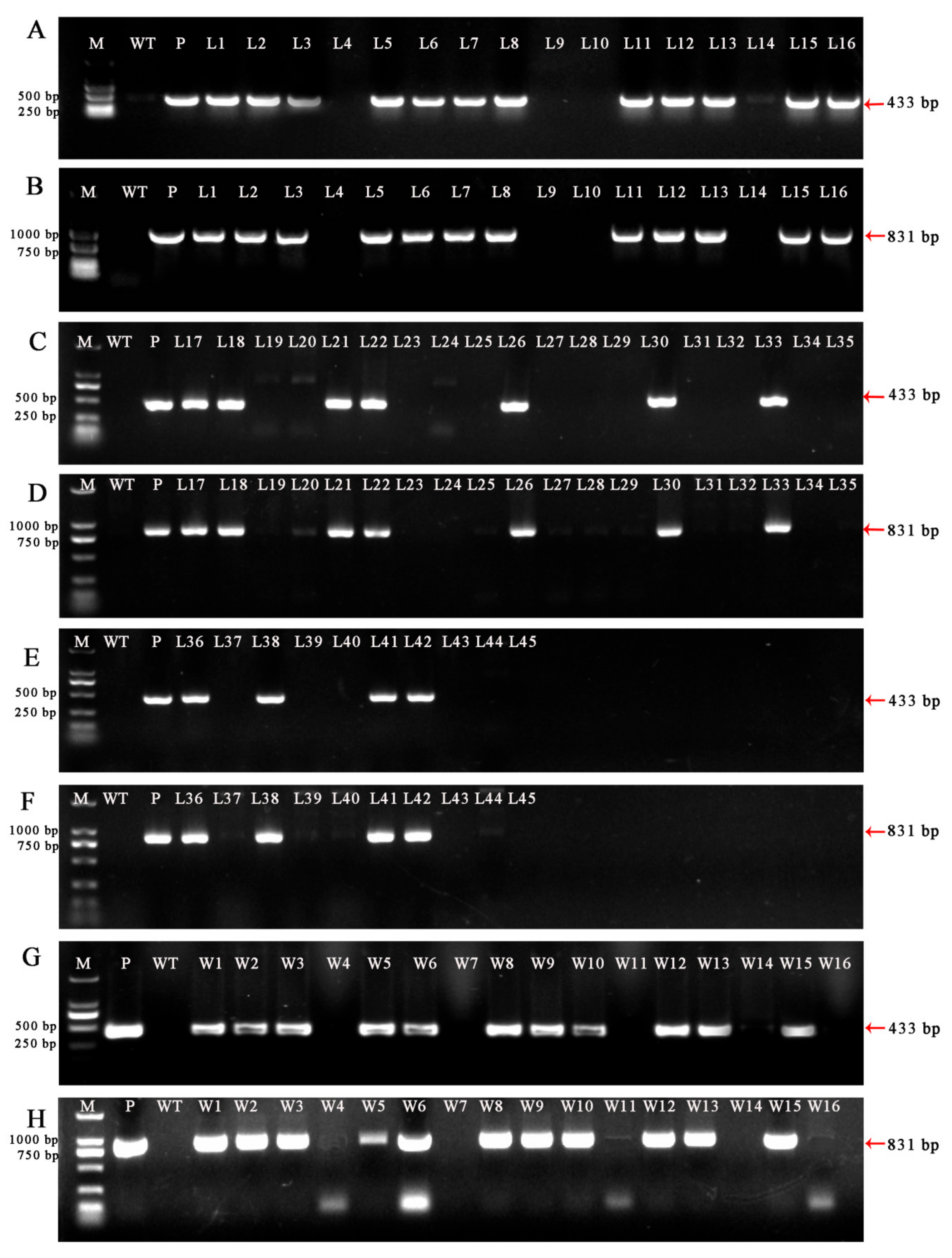
2.5. Polymerase Chain Reaction (PCR) and Southern Blot Analysis
2.6. Phenotype of Transformed Plants with pBUE-LpPDS
2.7. Molecular Analysis of CRISPR/Cas9-Induced Mutations in LpPDS
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Agrobacterium Strain and Plasmid
4.2. Selection Antibiotic Sensitivity Assay
4.3. Preparation of Agrobacterium
4.4. Optimization of Agrobacterium-Mediated Transformation Conditions
4.5. Agrobacterium Transformation
4.6. Identification of Transgenic Plants
4.7. Construction of LpPDS CRISPR/Cas9 Targeting Vector and Transformation in Lilium
4.8. Identification of Transgenic Plants and Detection of Target Edit Types
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MS (Culture medium) | Murashige-Skoog |
| PIC (IAA analogue) | Picloram (Sigma-Aldrich, United States) |
| NAA (Auxin analogue) | Naphthylacetic acid (Sigma-Aldrich, United States) |
| 6-BA (Cytokinin) | N6-benzyladenine (Tiangen, Beijing, China) |
| AS (Chemical material) | Acetosyringone (Tiangen, Beijing, China) |
| Cef (Antibiotic) | Cefotaxime (Tiangen, Beijing, China) |
| Hyg (Antibiotic) | Hygromycin phosphotransferase (Sigma-Aldrich, United States) |
| Kan (Antibiotic) | Kanamycin monosulfate (Tiangen, Beijing, China) |
| Rif (Antibiotic) | Rifampicin (Tiangen, Beijing, China) |
| Bar | Basta resistance gene |
| GUS | β-glucuronidase |
Appendix A
| Media | Composition |
|---|---|
| Lilium pumilum DC. Fisch. | |
| Pre-cultivation I | MS (Murashige-Skoog) + 1.0 mg·L−1 PIC (Picloram) + 0.2 mg·L−1 NAA (Naphthylacetic acid) + 30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Co-cultivation I | MS (NH4NO3-free) + 1.0 mg·L−1 PIC + 0.2 mg·L−1 NAA +100 μmol AS (Acetosyringone) + 60 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Selection I | MS + 1.0 mg·L−1 PIC + 0.2 mg·L−1 NAA + 400 mg·L−1 Cef (Cefotaxime) + 30 mg·L−1 Hyg (Hygromycin) + 30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Selection II | MS + 1.0 mg·L−1 PIC + 0.2 mg·L−1 NAA + 400 mg·L−1 Cef + 10 mg·L−1 Basta +30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Somatic embryo germination medium | MS + 0.5 mg·L−1 6-BA (N6-benzyladenine) + 30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| ‘White Heaven’ | |
| Pre-cultivation II | MS + 1.5 mg·L−1 BA + 0.2 mg·L−1 NAA + 30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Co-cultivation II | MS (NH4NO3-free) + 1.5 mg·L−1 BA + 0.2 mg·L−1 NAA +100 μmol AS + 60 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Selection III | MS (NH4NO3-free) + 1.5 mg·L−1 BA + 0.2 mg·L−1 NAA + 400 mg·L−1 Cef + 40 mg·L−1 Hyg +30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Selection IV | MS (NH4NO3-free) + 1.5 mg·L−1 BA + 0.2 mg·L−1 NAA + 400 mg·L−1 Cef + 20 mg·L−1 Basta + 30 g·L−1 Sucrose + 7.0 g·L−1 Agarose |
| Resuspension solution | ½MS (NH4NO3-free) + 100 μmol AS + 60 g·L−1 Sucrose |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| GUS-F | AGTTCTTTCGGCTTGTTG |
| GUS-R | TTCTACTTTACTGGCTTTGG |
| Hyg-F | TACACAGCCATCGGTCCAGA |
| Hyg-R | CGCAAGGAATCGGTCAATACAC |
| Bar-F | CTGCACCATCGTCAACCACTAC |
| Bar-R | CTGCCAGAAACCCACGTCAT |
| LpPDS-F0 | GACAGGCCTCCATATATTCTTGTTTTAGAGCTAGAAATAGC |
| LpPDS-R0 | ATCATAGCAGGTAGAAGGCCGCTTCTTGGTGCC |
| LpPDS-BsF | AATAATGGTCTCAGGCGACAGGCCTCCATATATTCTT |
| LpPDS-BsR | ATTATTGGTCTCTAAACATCATAGCAGGTAGAAGGC |
| OsU3-FD3 | GACAGGCGTCTTCTACTGGTGCTAC |
| TaU3-RD | CTCACAAATTATCAGCACGCTAGTC |
| pdst-f | CTGAGTTTCGGAGTCGTG |
| pdst-r | CCAGCCAGATAGAACCCT |
References
- Grassotti, A.; Gimelli, F.; Grassotti, A.; Burchi, G. Bulb and cut flower production in the genus Lilium: Current status and the future. Acta Hortic. 2011, 900, 21–36. [Google Scholar] [CrossRef]
- Lazare, S.; Bechar, D.; Fernie, A.R.; Brotman, Y.; Zaccai, M. The proof is in the bulb: Glycerol influences key stages of lily development. Plant J. 2018, 97, 321–340. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhao, Y.Q.; Zhang, M.; Jia, G.X.; Zaccai, M. Functional and evolutionary characterization of the CONSTANS-LIKE Family in Lilium × formolongi. Plant Cell Physiol. 2018, 59, 1874–1888. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.Y.; Lin, L.F.; Lin, M.D.; Hsieh, S.C.; Li, A.Y.; Tsay, Y.S.; Chou, M.L. Overexpression of Lilium formosanum MADS-box (LFMADS) causing floral defects while promoting flowering in Arabidopsis thaliana, whereas only affecting floral transition time in Nicotiana tabacum. Int. J. Mol. Sci. 2018, 19, 2217. [Google Scholar] [CrossRef] [PubMed]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. Biotech 2017, 7, 239. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, T.; Gerszberg, A.; Durańska, P.; Biłas, R.; Hnatuszko-Konka, K. High efficiency transformation of Brassica oleracea var. botrytis plants by Rhizobium rhizogenes. AMB Express 2018, 8, 125. [Google Scholar] [CrossRef]
- Ratjens, S.; Mortensen, S.; Kumpf, A.; Bartsch, M.; Winkelmann, T. Embryogenic callus as target for efficient transformation of Cyclamen persicum enabling gene function studies. Front. Plant Sci. 2018, 9, 1035. [Google Scholar] [CrossRef]
- Cohen, A.; Meredith, C.P. Agrobacterium-mediated transformation of Lilium. Int. Symp. Flower Bulb. 1992, 325, 611–618. [Google Scholar] [CrossRef]
- Mercuri, A.; Benedetti, L.D.; Bruna, S.; Bregliano, R.; Bianchini, C.; Foglia, G.; Schiva, T. Agrobacterium-mediated transformation with rol genes of Lilium Longiflorum thunb. Acta Hortic. 2003, 612, 129–136. [Google Scholar] [CrossRef]
- Hoshi, Y.; Kondo, M.; Mori, S.; Adachi, Y.; Nakano, M.; Kobayashi, H. Production of transgenic lily plants by Agrobacterium-mediated transformation. Plant Cell Rep. 2004, 22, 359–364. [Google Scholar] [CrossRef]
- Wei, C.; Cui, Q.; Lin, Y.; Jia, G.X. Efficient Agrobacterium-mediated transformation of Lilium Oriental ‘Sorbonne’ with genes encoding anthocyanin regulators. Can. J. Plant Sci. 2017, 97, 796–807. [Google Scholar] [CrossRef]
- Azadi, P.; Otang, N.V.; Chin, D.P.; Nakamura, I.; Fujisawa, M.; Harada, H.; Misawa, N.; Mii, M. Metabolic engineering of Lilium × formolongi using multiple genes of the carotenoid biosynthesis pathway. Plant Biol. Rep. 2010, 4, 269–280. [Google Scholar] [CrossRef]
- Qi, Y.; Du, L.; Quan, Y.; Tian, F.; Liu, Y.; Wang, Y. Agrobacterium-mediated transformation of embryogenic cell suspension cultures and plant regeneration in Lilium tenuifolium oriental × trumpet ‘Robina’. Acta Physiol. Plant. 2014, 36, 2047–2057. [Google Scholar] [CrossRef]
- Bassuner, B.M.; Lam, R.; Lukowitz, W.; Yeung, E.C. Auxin and root initiation in somatic embryos of Arabidopsis. Plant Cell Rep. 2007, 26, 1–11. [Google Scholar] [CrossRef]
- Nhut, D.T.; Le, B.V.; Silva, J.A.T.D.; Aswath, C.R. Thin cell layer culture system in Lilium: Regeneration and transformation perspectives. Vitr. Cell. Dev. Biol. Plant 2001, 37, 516–523. [Google Scholar] [CrossRef]
- Karami, O.; Deljou, A.; Kordestani, G.K. Secondary somatic embryogenesis of carnation (Dianthus caryophyllus L.). Plant Cell Tissue Organ Cult. 2008, 92, 273–280. [Google Scholar] [CrossRef]
- Bakhshaie, M.; Babalar, M.; Mirmasoumi, M.; Khalighi, A. Somatic embryogenesis and plant regeneration of Lilium ledebourii (Baker) Boiss., an endangered species. Plant Cell Tissue Organ Cult. 2010, 102, 229–235. [Google Scholar] [CrossRef]
- Ault, J.R.; Siqueira, S.S. Morphogenetic Response of Lilium michiganense to four auxin-type plant growth regulators in vitro. HortScience 2008, 43, 1922–1924. [Google Scholar] [CrossRef]
- Cohen, A.; Lipsky, A.; Arazi, T.; Ion, A.; Stav, R.; Sandler-Ziv, D.; Pintea, C.; Barg, R.; Salts, Y.; Shabtai, S.; et al. Efficient genetic transformation of Lilium Longiflorum and Ornithogalum Dubium by particle acceleration followed by prolonged selection in liquid medium. Acta Hortic. 2004, 651, 131–138. [Google Scholar] [CrossRef]
- Pathi, K.M.; Tula, S.; Tuteja, N. High frequency regeneration via direct somatic embryogenesis and efficient Agrobacterium-mediated genetic transformation of tobacco. Plant Signal Behav. 2013, 8. [Google Scholar] [CrossRef]
- Nyaboga, E.N.; Njiru, J.M.; Tripathi, L. Factors influencing somatic embryogenesis, regeneration, and Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14. Front. Plant Sci. 2015, 6, 411. [Google Scholar] [CrossRef] [PubMed]
- Pikulthong, V.; Teerakathiti, T.; Thamchaipenet, A.; Peyachoknagul, S. Development of somatic embryos for genetic transformation in Curcuma longa L. and Curcuma mangga Valeton & Zijp. Agric. Nat. Resour. 2016, 50, 276–285. [Google Scholar] [CrossRef]
- Kim, M.-J.; An, D.-J.; Moon, K.-B.; Cho, H.-S.; Min, S.-R.; Sohn, J.-H.; Jeon, J.-H.; Kim, H.-S. Highly efficient plant regeneration and Agrobacterium-mediated transformation of Helianthus tuberosus L. Ind. Crop. Prod. 2016, 83, 670–679. [Google Scholar] [CrossRef]
- Su, H.; Jiao, Y.T.; Wang, F.F.; Liu, Y.E.; Niu, W.L.; Liu, G.T.; Xu, Y. Overexpression of VpPR10.1 by an efficient transformation method enhances downy mildew resistance in V. vinifera. Plant Cell Rep. 2018, 37, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Zhang, Z.J.; Kausch, A.P. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 2017, 36, 1477–1491. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Jin, R.; Guo, J.; Zhang, F. Infection of embryonic callus with agrobacterium enables high-speed transformation of maize. Int. J. Mol. Sci. 2019, 279. [Google Scholar] [CrossRef] [PubMed]
- Omar, A.A.; Murata, M.M.; El-Shamy, H.A.; Graham, J.H.; Grosser, J.W. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018, 27, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Eggenberger, A.L.; Banakar, R.; McCaw, M.E.; Zhu, H.; Main, M.; Kang, M.; Gelvin, S.B.; Wang, K. CRISPR/Cas9-mediated targeted T-DNA integration in rice. Plant Mol. Biol. 2019, 99, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Soares, T.D.C.; Silva, C.R.C.D.; Lima, L.M.D.; Severino, L.S.; Santos, R.C.D. Validating a probe from GhSERK1 gene for selection of cotton genotypes with somatic embryogenic capacity. J. Biotechnol. 2018, 270, 44–50. [Google Scholar] [CrossRef]
- Janga, M.R.; Campbell, L.M.; Rathore, K.S. CRISPR/Cas9-mediated targeted mutagenesis in upland cotton (Gossypium hirsutum L.). Plant Mol. Biol. 2017, 94, 349–360. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, N.; Alok, A.; Shivani Kaur, N.; Pandey, P.; Awasthi, P.; Tiwari, S. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genom. 2018, 18, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, S.; Li, D.; Zhang, Q.; Li, L.; Zhong, C.; Liu, Y.; Huang, H. Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol. J. 2018, 16, 1424–1433. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.X.; Mitchell, L.A.; Liu, H.M.; Li, B.Z.; Liu, D.; Agmon, N.; Wu, Y.; Li, X.; Zhou, X.; Li, B.; et al. Rapid and efficient CRISPR/Cas9-based mating-type switching of Saccharomyces cerevisiae. G3 2018, 8, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Tan, Q.; Fan, Q.; Zhu, H.; Hong, Z.; Zhang, Z.; Duanmu, D. Efficient inactivation of symbiotic nitrogen fixation related genes in Lotus japonicus Using CRISPR-Cas9. Front. Plant Sci. 2016, 7, 1333. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Aida, R.; Sasaki, K. Generation of gene-edited chrysanthemum morifolium using multicopy transgenes as targets and markers. Plant Cell Physiol. 2017, 58, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Ma, L.; Peng, Y.; Deng, X.W.; Chen, Z.; Qu, L.-J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 17, 471. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Fan, D.; Liu, T.; Li, C.; Jiao, B.; Li, S.; Hou, Y.; Luo, K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in populus in the first generation. Sci. Rep. 2015, 5, 12217. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, X.; Yang, C.; Li, M.; Guo, Y. Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci. Rep. 2016, 6, 20315. [Google Scholar] [CrossRef]
- Lin, C.S.; Hsu, C.T.; Yang, L.H.; Lee, L.Y.; Fu, J.Y.; Cheng, Q.W.; Wu, F.H.; Hsiao, H.C.; Zhang, Y.; Zhang, R.; et al. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2017, 16, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Hou, Y.; Wang, H.; Ji, R.; Liu, B.; Wen, J.; Niu, L.; Lin, H. Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 2017, 36, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Odipio, J.; Alicai, T.; Ingelbrecht, I.; Nusinow, D.A.; Bart, R.; Taylor, N.J. Efficient CRISPR/Cas9 Genome Editing of Phytoene desaturase in Cassava. Front. Plant Sci. 2017, 8, 1780. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zheng, A.; Jiang, M.; Xue, S.; Yuan, Q.; Jiang, L.; Chen, Q.; Li, M.; Wang, Y.; Zhang, Y.; et al. CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 2018, 8, 16786. [Google Scholar] [CrossRef] [PubMed]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Liu, Y.; Lou, Q.; Xu, W.; Xin, Y.; Bassett, C.; Wang, Y. Characterization of a chalcone synthase (CHS) flower-specific promoter from Lilium orential ‘Sorbonne’. Plant Cell Rep. 2011, 30, 2187–2194. [Google Scholar] [CrossRef]
- Wang, Y.; van Kronenburg, B.; Menzel, T.; Maliepaard, C.; Shen, X.; Krens, F. Regeneration and Agrobacterium-mediated transformation of multiple lily cultivars. Plant Cell Tissue Organ Cult. 2012, 111, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Nunez de Caceres Gonzalez, F.F.; Davey, M.R.; Cancho Sanchez, E.; Wilson, Z.A. Conferred resistance to Botrytis cinerea in Lilium by overexpression of the RCH10 chitinase gene. Plant Cell Rep. 2015, 34, 1201–1209. [Google Scholar] [CrossRef]
- Liu, X.; Gu, J.; Wang, J.; Lu, Y. Lily breeding by using molecular tools and transformation systems. Mol. Biol. Rep. 2014, 41, 6899–6908. [Google Scholar] [CrossRef]
- Sabbadini, S.; Capriotti, L.; Molesini, B.; Pandolfini, T.; Navacchi, O.; Limera, C.; Ricci, A.; Mezzetti, B. Comparison of regeneration capacity and Agrobacterium-mediated cell transformation efficiency of different cultivars and rootstocks of Vitis spp. via organogenesis. Sci. Rep. 2019, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Toki, S.; Hara, N.; Ono, K.; Onodera, H.; Tagiri, A.; Oka, S.; Tanaka, H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006, 47, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, C.; Seng, S.; Khan, M.A.; Sui, J.; Gong, B.; Liu, C.; Wu, C.; Zhong, X.; He, J.; et al. Somatic embryogenesis and Agrobacterium-mediated transformation of Gladiolus hybridus cv. ‘Advance Red’. Plant Cell Tissue Organ Cult. 2015, 120, 717–728. [Google Scholar] [CrossRef]
- Dutt, M.; Madhavaraj, J.; Grosser, J.W. Agrobacterium tumefaciens -mediated genetic transformation and plant regeneration from a complex tetraploid hybrid citrus rootstock. Sci. Hortic. 2010, 123, 454–458. [Google Scholar] [CrossRef]
- Pushyami, B.; Beena, M.R.; Sinha, M.K.; Kirti, P.B. In vitro regeneration and optimization of conditions for Agrobacterium mediated transformation in jute, Corchorus capsularis. J. Plant Biochem. Biotechnol. 2011, 20, 39–46. [Google Scholar] [CrossRef]
- Chen, X.; Equi, R.; Baxter, H.; Berk, K.; Han, J.; Agarwal, S.; Zale, J. A high-throughput transient gene expression system for switchgrass (Panicum virgatum L.) seedlings. Biotechno. Biofuels 2010, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Uchii, S.; Watanabe, S.; Ezura, H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006, 47, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Vergne, P.; Maene, M.; Gabant, G.; Chauvet, A.; Debener, T.; Bendahmane, M. Somatic embryogenesis and transformation of the diploid Rosa chinensis cv Old Blush. Plant Cell Tissue Organ Cult. 2009, 100, 73–81. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Z.T.; Cui, J.; Henny, R.J.; Gray, D.J.; Xie, J.; Chen, J. Efficient somatic embryogenesis and Agrobacterium-mediated transformation of pothos (Epipremnum aureum) ‘Jade’. Plant Cell Tissue Organ Cult. 2013, 114, 237–247. [Google Scholar] [CrossRef]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 532, 293. [Google Scholar] [CrossRef]
- Zhang, J.; Gai, M.; Li, X.; Li, T.; Sun, H. Somatic embryogenesis and direct as well as indirect organogenesis in Lilium pumilum DC. Fisch., an endangered ornamental and medicinal plant. Biosci. Biotech. Biochem. 2016, 80, 1898–1906. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.L.; Dong, L.; Wang, Z.P.; Zhang, H.Y.; Han, C.Y.; Liu, B.; Wang, X.C.; Chen, Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014, 14, 327. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]





| Variety | Concentration of Hyg (mg·L−1) | Rate of Browning Explants (%) | Rate of Proliferation or Induction (%) |
|---|---|---|---|
| Lilium pumilum DC. Fisch. | 0 | 0.83 ± 1.44 e | 99.17 ± 1.44 a |
| 10 | 16.67 ± 1.44 d | 80.83 ± 3.82 b | |
| 20 | 66.67 ± 2.89 c | 68.33 ± 3.82 c | |
| 30 | 83.33 ± 3.82 b | 25.00 ± 2.50 d | |
| 40 | 97.50 ± 2.50 a | 4.17 ± 2.89 e | |
| 50 | 100 a | 0 e | |
| ‘White Heaven’ | 0 | 1.11 ± 0.58 e | 92.33 ± 0.58 a |
| 10 | 24.44 ± 2.08 d | 70.33 ± 2.08 b | |
| 20 | 58.89 ± 3.06 c | 41.11 ± 3.51 c | |
| 30 | 76.67 ± 2.00 b | 20.78 ± 1.53 d | |
| 40 | 92.22 ± 0.58 a | 7.22 ± 0.58 e | |
| 50 | 100 a | 0 e |
| Variety | Concentration of Cef (mg·L−1) | Rate of Browning Explants (%) | Rate of Proliferation or Induction (%) |
|---|---|---|---|
| Lilium pumilum DC. Fisch. | 0 | 0.83 ± 1.44 g | 100 a |
| 100 | 6.67 ± 1.44 f | 96.67 ± 3.82 ab | |
| 200 | 12.50 ± 2.50 e | 91.67 ± 3.82 b | |
| 300 | 20.00 ± 2.50 d | 85.00 ± 2.50 c | |
| 350 | 32.50 ± 2.50 c | 80.00 ± 2.50 c | |
| 400 | 50.00 ± 2.50 b | 71.67 ± 3.82 d | |
| 500 | 74.17 ± 2.89 a | 61.67 ± 3.82 e | |
| ‘White Heaven’ | 0 | 0 f | 100 a |
| 100 | 5.22 ± 0.58 e | 91.11 ± 1.53 b | |
| 200 | 17.56 ± 1.15 d | 83.33 ± 1.00 c | |
| 300 | 24.89 ± 1.00 c | 75.56 ± 0.58 d | |
| 350 | 32.33 ± 1.53 b | 70.00 ± 1.73 d | |
| 400 | 39.56 ± 1.15 b | 57.78 ± 1.53 e | |
| 500 | 47.89 ± 1.53 a | 52.22 ± 0.58 e |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, R.; Wang, Z.; Ren, Y.; Li, H.; Liu, N.; Sun, H. Establishment of Efficient Genetic Transformation Systems and Application of CRISPR/Cas9 Genome Editing Technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. Int. J. Mol. Sci. 2019, 20, 2920. https://doi.org/10.3390/ijms20122920
Yan R, Wang Z, Ren Y, Li H, Liu N, Sun H. Establishment of Efficient Genetic Transformation Systems and Application of CRISPR/Cas9 Genome Editing Technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. International Journal of Molecular Sciences. 2019; 20(12):2920. https://doi.org/10.3390/ijms20122920
Chicago/Turabian StyleYan, Rui, Zhiping Wang, Yamin Ren, Hongyu Li, Na Liu, and Hongmei Sun. 2019. "Establishment of Efficient Genetic Transformation Systems and Application of CRISPR/Cas9 Genome Editing Technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven" International Journal of Molecular Sciences 20, no. 12: 2920. https://doi.org/10.3390/ijms20122920
APA StyleYan, R., Wang, Z., Ren, Y., Li, H., Liu, N., & Sun, H. (2019). Establishment of Efficient Genetic Transformation Systems and Application of CRISPR/Cas9 Genome Editing Technology in Lilium pumilum DC. Fisch. and Lilium longiflorum White Heaven. International Journal of Molecular Sciences, 20(12), 2920. https://doi.org/10.3390/ijms20122920




