Abstract
Brassinosteroid (BR) is an essential hormone in plant growth and development. The BR signaling pathway was extensively studied, in which BRASSINAZOLE RESISTANT 1 (BZR1) functions as a key regulator. Here, we carried out a functional study of the homolog of BZR1 in Medicago truncatula R108, whose expression was induced in nodules upon Sinorhizobium meliloti 1021 inoculation. We identified a loss-of-function mutant mtbzr1-1 and generated 35S:MtBZR1 transgenic lines for further analysis at the genetic level. Both the mutant and the overexpression lines of MtBZR1 showed no obvious phenotypic changes under normal growth conditions. After S. meliloti 1021 inoculation, however, the shoot and root dry mass was reduced in mtbzr1-1 compared with the wild type, caused by partially impaired nodule development. The transcriptomic analysis identified 1319 differentially expressed genes in mtbzr1-1 compared with wild type, many of which are involved in nodule development and secondary metabolite biosynthesis. Our results demonstrate the role of MtBZR1 in nodule development in M. truncatula, shedding light on the potential role of BR in legume–rhizobium symbiosis.
1. Introduction
Medicago truncatula is a model fabacean species for nodulation and nitrogen fixation. During the initiation of symbiotic nodules, for example, the root nodule microsymbionts (collectively termed rhizobia) secrete nodulation (Nod) factors, which are lipochito-oligosaccharides functioning as signal molecules, in response to flavonoids from the host plant. The Nod factors are then perceived by the corresponding receptor kinases in the root tissue, which in turn trigger nodule organogenesis and root hair deformation [1]. After nodule formation, the host plant generates a class of nodule-specific cysteine-rich (NCR) peptides to induce the differentiation of endosymbionts into enlarged and polyploid bacteroids [2]. There are about 600 MtNCR genes in the M. truncatula genome, indicating their important roles in nitrogen fixation [3]. Hormones also play important roles during nodule development [4,5].
Flavonoids are known as secondary metabolites that play important roles in legume–rhizobium symbiosis [6]. Based on the molecular structure, flavonoids consist of flavones, flavonols, isoflavones, flavanones, flavanols, and anthocyanins [7]. The flavonoid biosynthesis pathway starts from downstream of the phenylalanine pathway, in which p-coumaroyl co-enzyme A (CoA) is produced and catalyzed into chalcone by chalcone synthase. In the flavonoid biosynthesis pathway, chalcone is catalyzed into dihydrokaempferol by chalcone isomerase and flavanone 3-hydroxylase, and then by flavonol synthases into flavonols. Flavonoids production is suggested to be highly localized at infection sites in legume [8].
Brassinosteroid (BR) is an essential plant hormone in plant growth and developmental processes, such as cell elongation, photomorphogenesis, seed germination, stomata differentiation and movement, flowering, and plant response to biotic and abiotic stresses [9,10,11]. AtBZR1 (Arabidopsis thaliana BRASSINAZOLE-RESISTANT 1) is a key transcriptional regulator in the BR signaling pathway [12,13,14]. AtBZR1 has a homolog, AtBES1 (At BRASSINOSTEROID INSENSITIVE 1-EMS SUPPRESSOR 1), which shares 88% similarity in the A. thaliana genome [15]. Because of the redundancy with AtBES1, the atbzr1 mutant showed no obvious phenotypic change, while the gain-of-function mutant atbzr1-D showed enhanced BR signaling in A. thaliana. Recently, mtbri1 was reported to show defective nodule development, indicating that BR signaling is involved in legume nodulation [16]. As MtBZR1 is an important transcription factor downstream of MtBRI1, it is intriguing to explore its role in nodule development in M. truncatula.
This study was undertaken with the aim to determine the function of MtBZR1 in M. truncatula R108 nodule development. In order to achieve this, we isolated mutants of MtBZR1 and characterized their phenotypes. Next, we performed transcriptomic analysis of the mtbzr1-1 null mutant, in comparison to the wild-type plants, to identify the deregulated genes. The results indicate that MtBZR1 is involved in the control of nodule development, but not its initiation, in M. truncatula. MtBZR1 was induced by the rhizobium Sinorhizobium meliloti 1021, and mtbzr1-1 displayed partially impaired nodule development compared to the control R108. In addition, transcriptomic analysis found that the genes involved in nodule development and secondary metabolite biosynthesis were under transcriptional reprogramming, consistent with the morphological changes in mtbzr1-1. These results indicate that MtBZR1 plays a role in controlling nodule development after the initiation in M. truncatula.
2. Results
2.1. Search for AtBZR1 Homologs in M. truncatula
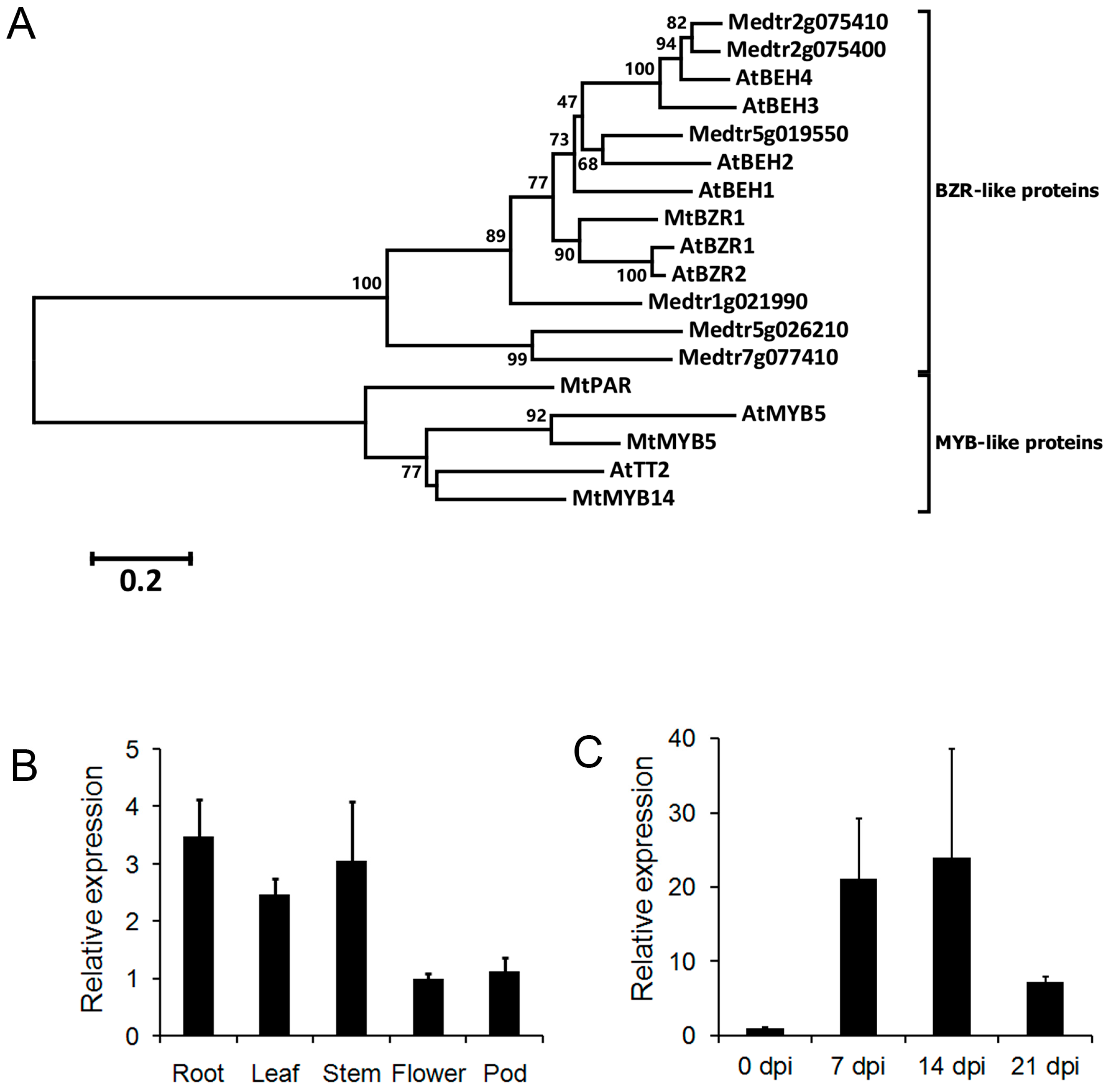
We searched the annotated M. truncatula genome for the homolog of A. thaliana AtBZR1. Medtr3g111680 encodes a 315-amino-acid (aa) protein that shares 64% identify with the A. thaliana AtBZR1 protein. According to the phylogenic analysis of 11 A. thaliana and M. truncatula homologs, Medtr3g111680 is the closest homolog of AtBZR1 in M. truncatula (Figure 1A; Figure S1, Supplementary Materials). It is clustered with two other proteins, Medtr1g021990 and AtBZR2. In addition to the high similarity in the protein sequence, Medtr3g111680 also possesses a BZR domain with conservative sites (Figure S2, Supplementary Materials). We, therefore, denoted Medtr3g111680 as MtBZR1.
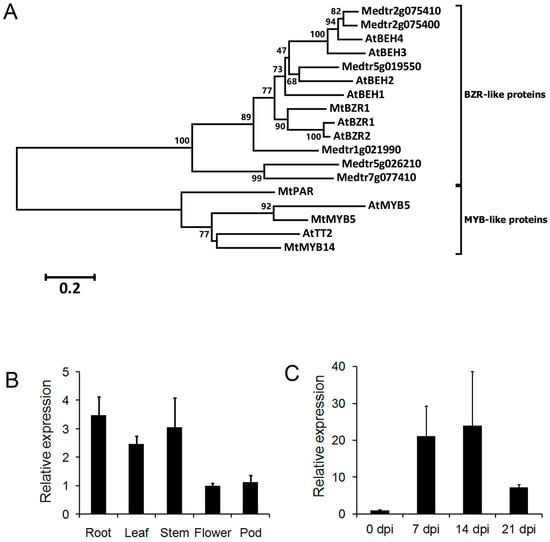
Figure 1.
The phylogenetic analysis and expression of Medicago truncatula brassinazole resistant 1 (MtBZR1). (A) A neighbor-joining tree of BRASSINAZOLE-RESISTANT 1 (BZR1)-like proteins in A. thaliana and M. truncatula. AtMYB5 and A. thaliana TRANSPARENT TESTA 2 (AtTT2) acted as an outgroup sequence. Multiple alignment and phylogenetic analysis were run with the MEGA6 program with 1000 bootstraps. Scores higher than 50 are labeled by the branches. The protein sequences from A. thaliana start with “At”; those from M. truncatula start with “Mt”. (B) Expression of MtBZR1 in M. truncatula tissues. The expression in the flower was set as 1.0. (C) Expression of MtBZR1 in nodules after inoculation with rhizobium (dpi, days post inoculation). The expression at zero dpi was set as 1.0. In (B,C), MtUbiquitin gene was used as an internal control. Bars represent standard errors from three biological replicates.
2.2. Induction of MtBZR1 upon S. meliloti 1021 Inoculation
To characterize its role in plant development, an expression assay of MtBZR1 by quantitative real-time PCR (qRT-PCR) was carried out in various tissue types. As shown in Figure 1B, MtBZR1 expression was highest in the root tissue, and comparable in stem and leaf, and relatively low in flower and pod. As nodulation is a specific characteristic of legume, we then performed another expression assay in nodules of the M. truncatula R108. As shown in Figure 1C, MtBZR1 was induced more than 20-fold in nodules at seven and 14 days post inoculation (dpi) by S. meliloti 1021 strain, and then reduced to seven-fold at 21 dpi, compared with that at zero dpi in R108. These results suggest that MtBZR1 may be involved in nodule development in M. truncatula.
2.3. Identification of the mtbzr1-1 Null Mutant
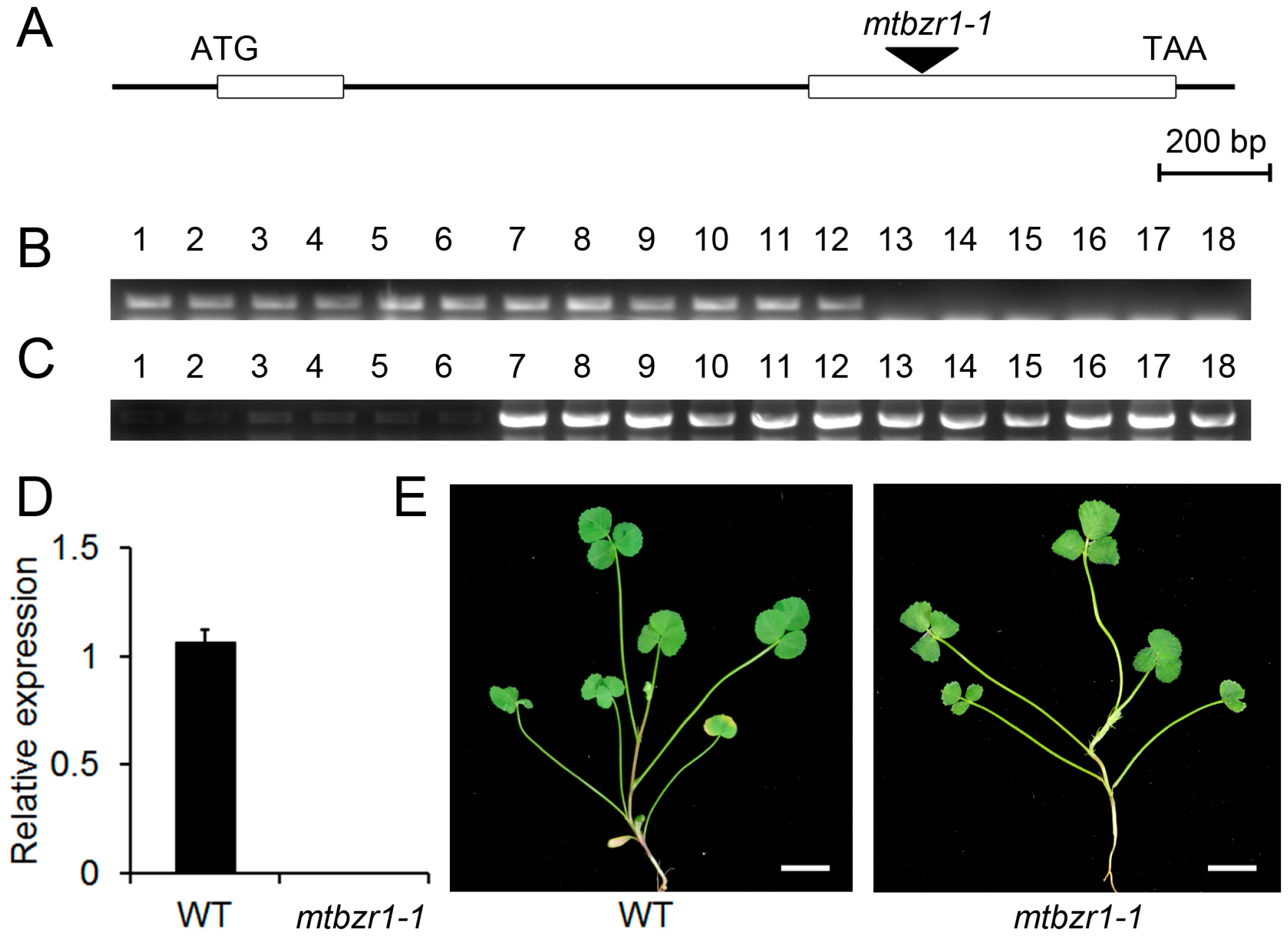
To illustrate the biological function of MtBZR1, we adopted a reverse genetic approach by searching the Noble Research Institute Mutant collection for the loss-of-function mutants. One mutant line (mtbzr1-1) that contains a single insertion in MtBZR1′s second exon was found and used for the following genetic analysis (Figure 2A). The insertion was confirmed by genotyping using the gene-specific primers and a Tnt1 primer (Figure 2B,C), and homozygous plants were identified. As shown in Figure 2D, we detected no expression of MtBZR1 in the mtbzr1-1 mutant plant. There was no obvious difference in developmental phenotypes between mtbzr1-1 and wild-type plants (Figure 2E).
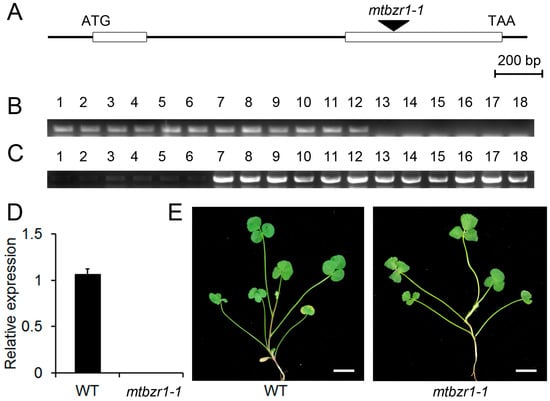
Figure 2.
The gene structure of MtBZR1 and identification of the mtbzr1-1 mutant. (A) The gene structure of MtBZR1. The open boxes represent two exons and the Tnt1 insertion in the null mutant mtbzr1-1 is indicated by the arrow. (B) The PCR of the primers crosses the Tnt1 insertion bygene-specific primers. Lane 1–6, wild type (WT); lane 7–12, heterozygous mutants; lane 13–18 homozygous mutants. (C) PCR using a gene-specific primer and a Tnt1 primer. (D) The expression of MtBZR1 in the wild type (WT) and mtbzr1-1. (E) The shoots of one-month-old WT and mtbzr1-1 plants. Scale bar = 1 cm. In (D), MtUbiquitin gene was used as an internal control. Bars represent standard errors from three biological replicates.
2.4. Loss of Function in MtBZR1 Leads to a Decrease in Plant Dry Mass in S. meliloti 1021 Nodulation
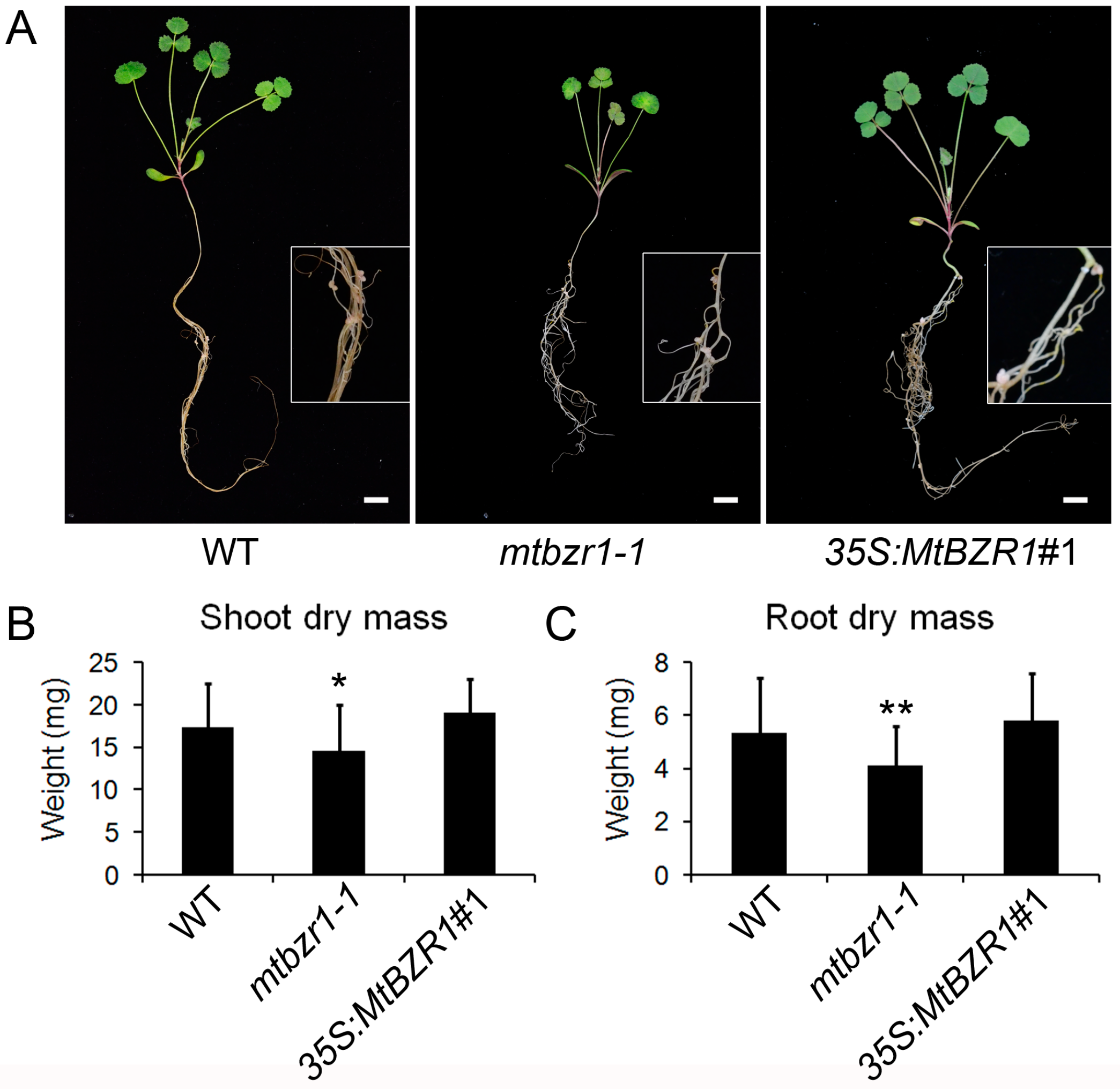
The induction of MtBZR1 in nodulation implies its role in nodule development. Thus, we compared the growth between mtbzr1-1 mutant and wild type after S. meliloti 1021 inoculation. As shown in Figure 3A, the inoculated mtbzr1-1 mutant exhibited a smaller plant size compared with the wild-type R108. To confirm this observation, we measured the dry mass of the shoot and root tissues in mtbzr1-1 and wild-type R108 plants 21 days after inoculation. The dry mass of both shoot and root in mtbzr1-1 was significantly decreased, compared with that of the wild-type R108 control (Figure 3B,C).
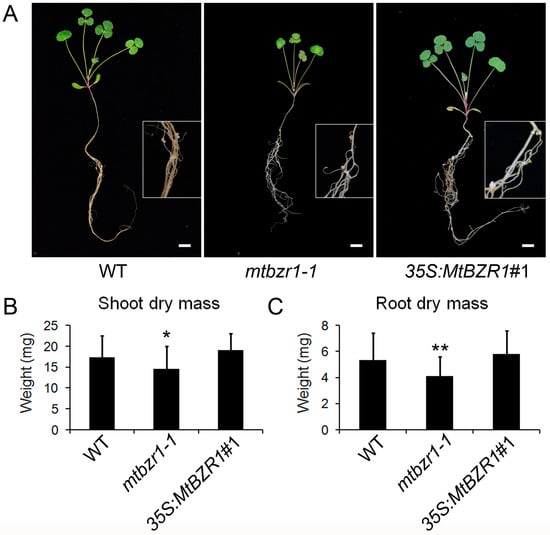
Figure 3.
Reduced dry mass in mtbzr1-1 in nodulation. (A) Photos of one-month-old wild-type (WT), mtbzr1-1, and 35S:MtBZR1#1 transgenic line. Scale bar = 1 cm. Insets are close views of the roots with nodules in WT, mtbzr1-1, and 35S:MtBZR1#1 transgenic plants. Dry mass of shoot (B) and root (C) of WT, mtbzr1-1, 35S:MtBZR1#1 transgenic line 21 days after rhizobial inoculation. Values are the means ± SD (n = 25); T = 2; * p < 0.05, ** p < 0.01.
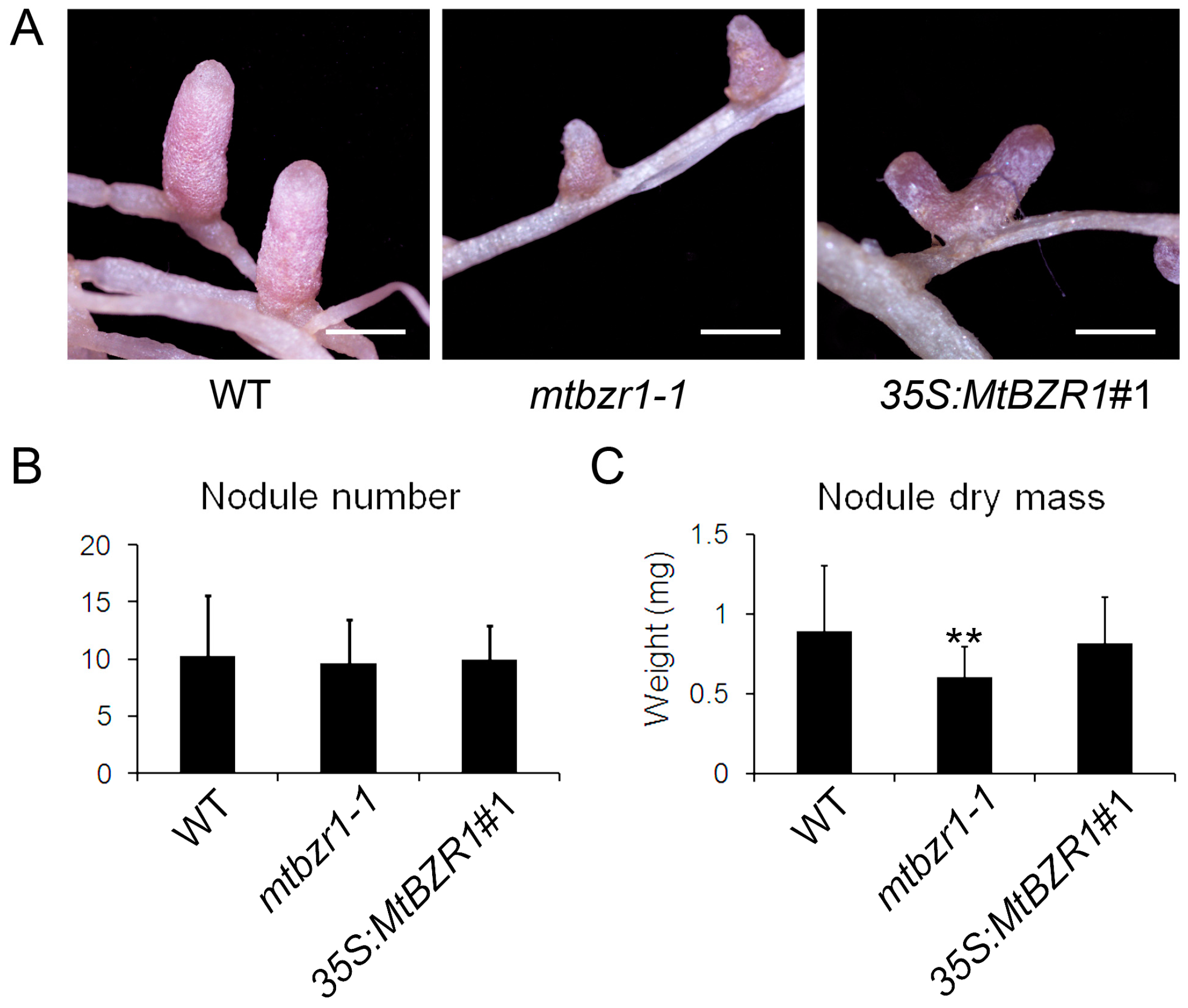
To explore the reason why mtbzr1-1 growth was retarded, we compared the nodule development in the mutant with the wild-type plants. As shown in Figure 4A, the nodules on mtbzr1-1 roots were smaller than those from the wild type. The measurement of nodule dry mass confirmed the observation that the nodules from mtbzr1-1 were significantly smaller/lighter than those from the wild type, although the number of nodules was comparable between them (Figure 4B,C). The genetic assay of mtbzr1-1 demonstrates that the loss of function in MtBZR1 leads to partially impaired nodule development in M. truncatula.
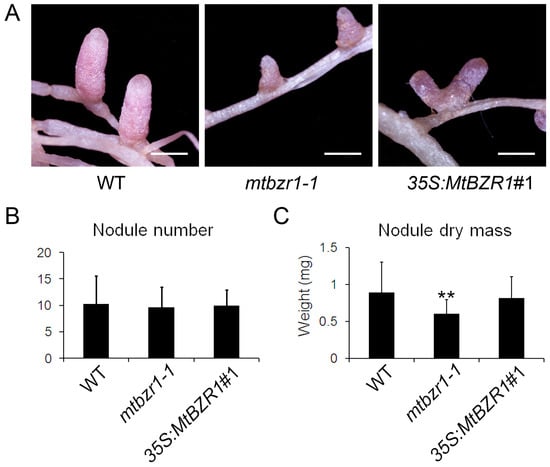
Figure 4.
Nodule development in wild type (WT), mtbzr1-1, and 35S:MtBZR1#1 transgenic line. (A) Photos of nodules from one-month-old WT, mtbzr1-1, and 35S:MtBZR1#1 transgenic line. Scale bar = 1 mm. Nodule number (B) and dry mass (C) in WT, mtbzr1-1, and 35S:MtBZR1#1 transgenic line 21 days after rhizobial inoculation. Values are the means ± SD (n = 25); T = 2; ** p < 0.01.
To further investigate the role of MtBZR1, the full length of the MtBZR1 coding sequence under control of the CaMV 35S promoter was introduced into the wild-type background. A total of six independent transgenic lines were generated and two lines with the highest MtBZR1 expression were used for further analysis (Figure S3A, Supplementary Materials). The 35S:MtBZR1 plants showed no obvious phenotypic changes with normal compound leaves from wild-type plants (Figure S3B, Supplementary Materials). The recombinant protein fusioned with a green fluorescence protein (GFP) showed the detection of the GFP signal of MtBZR1-GFP (Figure S3C, Supplementary Materials).
When we carried out rhizobial inoculation of the S. meliloti 1021 strain on the 35S:MtBZR1 lines, as shown in Figure 3A, the 35S:MtBZR1#1 plant showed a similar stature as the wild-type R108 plant, and also had a similar shoot and root dry mass as wild-type R108 (Figure 3B,C). The phenotype and dry mass of nodules were also comparable between the 35S:MtBZR1#1 plant and the wild-type control (Figure 4A–C).
2.5. Transcriptomic Profiles of Nodules in mtbzr1-1
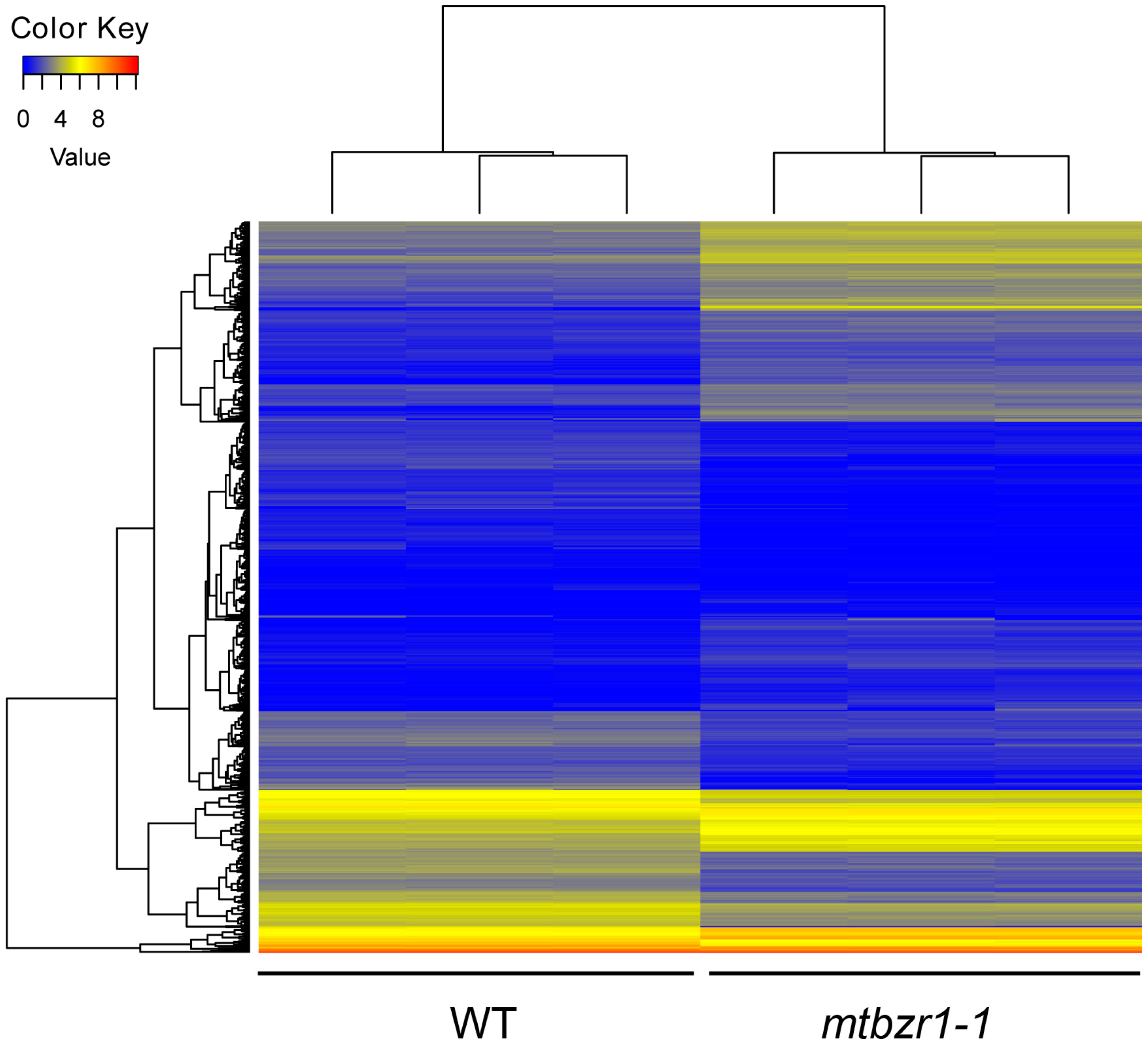
To understand the involvement of MtBZR1 in nodule development, we performed a transcriptomic assay of mtbzr1-1. The nodules were harvested from three biological replicates of mtbzr1-1 and wild-type plants 21 days after rhizobial inoculation by S. meliloti 1021 strain. For each replicate, RNA sequencing generated more than 20 million raw reads, which were filtered and aligned against the annotated reference genome for gene quantification. The correlation efficiency between biological replicates was as high as 99.9%, indicating a good repeatability (Figure S4A, Supplementary Materials). The differentially expressed genes (DEGs) were identified between the mtbzr1-1 and wild-type sample by DEGseq. Genes with more than two-fold expression changes and a false discovery rate (FDR) smaller than 0.001 were identified as DEGs. A total of 1319 DEGs were found in mtbzr1-1, among which 618 were elevated and 701 were suppressed (Figure 5; Figure S4B and Table S2, Supplementary Materials).
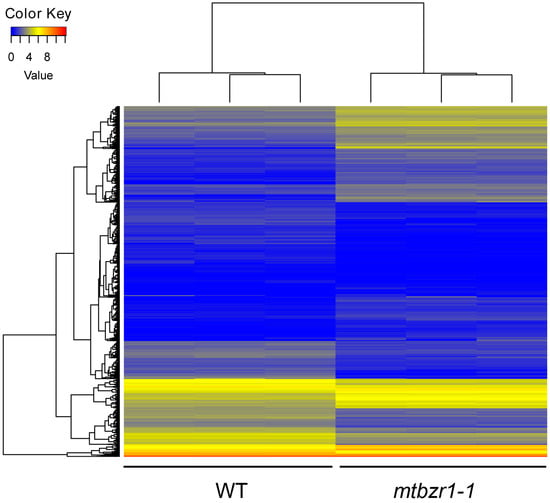
Figure 5.
The heatmap of FPKM (fragments per kilobase of transcript per million reads mapped) values of 1319 differential expressed genes in three biological replicates of nodules from mtbzr1-1 and wild-type plants.
To characterize these differentially expressed genes (DEGs) in mtbzr1-1, we carried out enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Among the most enriched GO terms in mtbzr1-1′s DEGs were nodule morphogenesis (GO:0009878), development involved in symbiotic interaction (GO:0044111), multi-organism process (GO:0009877), nodulation (GO:0044403), interspecies interaction between organisms (GO:0044419), symbiosis, encompassing mutualism through parasitism (GO:0044403), and anatomical structure (GO:0048856) (Table 1). These GO categories consisted of the same 43 Nodule Cysteine-Rich (NCR) genes, most of which were under suppression in mtbzr1-1 compared with wild type (Table 2). The expressions of MtNCR301 (Medtr3g015940), MtNCR237 (Medtr7g065025), MtNCR327 (Medtr7g071315), MtNCR145 (Medtr4g060610), MtNCR332 (Medtr4g065455), and MtNCR224 (Medtr6g406350) were validated by qRT-PCR (Figure S5, Supplementary Materials). The large-scale suppression of these NCRs in mtbzr1-1 suggests their important roles in nodules, and their deregulation may result in impaired nodule development caused by loss of function of MtBZR1.

Table 1.
Enriched Gene Ontology (GO) terms in differentially expressed genes (DEGs). ID—identifier.

Table 2.
The expression of genes involved in GO:0009878. FDR—false discovery rate.
2.6. Differential Expression of Genes Involved in Flavonoid Biosynthesis in mtbzr1-1
From the mtbzr1-1 transcriptome, we identified six significantly enriched secondary metabolite biosynthesis pathways (p < 0.01), including isoflavonoid biosynthesis (ko00943), monoterpenoid biosynthesis (ko00902), sesquiterpenoid and triterpenoid biosynthesis (ko00909), flavonoid biosynthesis (ko00941), phenylpropanoid biosynthesis (ko00940), and stilbenoid, diarylheptanoid, and gingerol biosynthesis (ko000945) (Table 3). In the phenylpropanoid biosynthesis pathway, 28 genes were suppressed and 15 were upregulated (Figure S6, Supplementary Materials); in the flavonoid biosynthesis pathway, 16 genes were suppressed and only four genes were upregulated (Figure S7 and Table S3, Supplementary Materials); in the isoflavonoid biosynthesis pathway, 17 genes were suppressed and only three were upregulated (Figure S8 and Table S4, Supplementary Materials). A validation experiment with qRT-PCR showed that two flavonol synthase (FLS) genes in the flavonoid biosynthesis pathway and four isoflavone 4′-O-methyltransferase (IOMT) genes in the isoflavonoid biosynthesis pathway were all suppressed in mtbzr1-1, consistent with RNA sequencing results (Figure S9, Supplementary Materials). Downregulation of the majority of DEGs involved in the flavonoid and isoflavonoid pathways in mtbzr1-1 indicates the suppression of their biosynthesis and, thus, possibly reduced production.

Table 3.
Enriched KEGG pathways in DEGs.
3. Discussion
In this study, we characterized MtBZR1, a transcriptional regulator of BR signaling in M. truncatula, via genetic and molecular approaches. MtBZR1 expression is elevated in young nodules (seven and 14 dpi) in comparison to roots. Furthermore, the dry mass of shoots and roots was reduced in mtbzr1-1 compared with wild type. Considering the smaller nodules in mtbzr1-1 and suppression of genes involved in nodulation, our results demonstrate that MtBZR1 plays an important role in legume–rhizobia symbiosis.
NCRs play important roles in nodulation in fabaceans [3]. We found that 41 of 43 differential expressed NCR genes were under suppression in mtbzr1-1 compared with wild type. The NCR genes are important for the infection, and most of these genes are expressed exclusively in nodules and through the processes of nodule development [17,18]. In our study, the suppression of these NCR genes is consistent with nodule dry mass reduction of the S. meliloti 1021 strain in mtbzr1-1. Within the promoter regions of NCRs, conserved motifs such as auxin response factor binding sites were reported [3]. Auxin response factor (ARF) binding sites (TGTCTC) are found to be located within 1000-bp promoter regions after searching for conserved cis-elements and motifs in NCR gene promoters [19]. The conserved ARF binding motifs, which are also found in the promoters of NCR downregulated in mtbzr1-1 in this study, suggest the important role of auxin during the symbiosis process between M. truncatula and S. meliloti 1021. In the pathway of auxin signal transduction, the auxin receptor TIR1 (TRANSPORT INHIBITOR RESPONSE 1) and the series of downstream ARF-related transcription factors are target genes of AtBZR1 in A. thaliana [20,21]. Hence, the nodulation-deficient phenotype of mtbzr1-1 is probably due to MtBZR1′s regulation of the elements downstream of other signaling pathways.
Flavonoids function as signaling molecules in legume–rhizobia recognition. Specific flavonoids are produced to activate Nod production of rhizobia’s compatible symbionts in conditions with low nitrogen. In accordance with this, the chalcone synthase knockdown in M. truncatula disrupts the initiation of nodulation, although the deformation of root hair remains [22]. A diverse group of metabolites derived from the flavonoid biosynthesis pathway accumulate in host roots and nodules. Despite this, only a small number of them clearly demonstrated their role in rhizobial infection. One of these flavonoids is methoxychalcone, which significantly enhances inducing activity on the nodulation genes [8]. It is produced from coumaroyl-CoA by chalcone synthases, whose coding genes are downregulated in the mutant mtbzr1-1, and then by chalcone-O-methyltransferases. The flavonoids are related to nodulation gene expression in nodules. For example, the infection zones of nodules contain a high level of flavonoids and the nodulation gene expression levels are high [6]. The dysregulation of flavonoid production would impair nodule initiation development in the mtbzr1-1 mutant. Our data suggest that the suppression of NCR genes and flavonoid metabolism probably lead to the impairment of the development of nodules in the cross-talk pathway in mtbzr1-1.
The mutant of the BR 5α-reductase gene in the BR synthesis pathway in pea displayed defective root development and exhibited reduced nodule organogenesis [5]. Furthermore, mtbzr1-1 developed normal shoots and roots probably due to gene redundancy with its homologs in the genome, similarly to atbzr1 in A. thaliana. Interestingly, mtbzr1-1 showed defects in nodule development, indicating its different role between shoot development, root development, and nodulation. In the KEGG analysis, the MtFLSs were downregulated in mtbzr1-1. Their homolog in A. thaliana, At5g05600, functions as a flavonol synthase and a 2-oxoglutarate/Fe (II)-dependent oxygenase that hydroxylates jasmonic acid (JA) [23]. Consist with this, we found that genes involved in linoleic acid metabolism (ko00591) were enriched with DEGs. Linoleic acids are precursors for JA, which inhibits the calcium spiking induced by Nod factors [23,24]. As an important transcription factor in BR signaling, MtBZR1 might function in hormone cross-talk in nodule development.
In previous studies, the strain S. meliloti 1021 was proven inefficient in nitrogen fixation on A17 and R108 plants [25,26], and its inefficiency might affect the quantitative differences in phenotypes investigated in experiments reported here. However, the mtbzr1-1 mutant displayed significantly less nodule mass although the numbers of nodules were comparable with the wild-type plants. Despite the possible defective differentiation of bacteroids, the differences in nodule phenotypes between mtbzr1-1 and wild type were caused by the loss of function of MtBZR1. These results indicate that MtBZR1 is involved in the nodule development after its initiation. Inoculation with an efficient strain, e.g., S. meliloti 102F34, would likely result in more pronounced variation in nodule phenotype, which will facilitate screening for new mutants in the future.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The plant materials used in this work were the Medicago truncatula ecotype R108. The mtbzr1-1 mutant was requested from the Noble Research Institute Tnt1 Mutant collection [27]. The mtbzr1-1 mutants were identified using gene-specific and Tnt1-specific primers (listed in Table S1, Supplementary Materials). The seeds of wild type, mtbzr1-1, and 35S:MtBZR1 were scarified mechanically and vernalized at 4 °C on moist filter paper for one week before transfer to soil. The plants were grown in a growth chamber at 22 °C under a 16-h light/8-h dark cycle with 150 μmol/m2/s light intensity and 70–80% relative humidity.
4.2. Root Nodule Induction
For rhizobial inoculation, wild-type, mtbzr1-1, and 35S:MtBZR1 seedlings were transferred into plastic trays filled with pearlite/sand (in a 3:1 ratio). The plants were grown in the growth chamber and watered every three days with nutrient solution without nitrogen as previously reported [28]. The rhizobium S. meliloti 1021 strain was cultured in TY (tryptone, yeast extract, and sodium chloride) medium (supplemented with 10 mg∙mL−1 tetracycline, 200 mg∙mL−1 streptomycin, and 6 mmol∙L−1 calcium chloride) at 28 °C in a shaker to reach an OD600 value of 1.0 [28]. Five-day-old seedling of wild type, mtbzr1-1, and 35S:MtBZR1 were inoculated with 5 mL of rhizobial suspension diluted to OD600 = 0.1. Nodule numbers and dry masses were determined 21 days after inoculation.
4.3. Identification of MtBZR1 Gene and Phylogenetic Analysis
To identify the homologs of A. thaliana BZR1 in M. truncatula, we used the amino-acid sequences of the two AtBZRs (AtBZR1 and AtBZR2) and four AtBZR1 homologs (AtBEH1 to AtBEH4) from A. thaliana for a BLASTP search in the M. truncatula (Available online: http://bioinfo3.noble.org/doblast/). A total of seven homologous sequences, Medtr3g111680, Medtr5g019550, Medtr1g021990, Medtr2g075410, Medtr2g075400, Medtr5g026210, and Medtr7g077410, were obtained. To study the phylogenetic relationships, the six amino-acid sequences from A. thaliana and seven from M. truncatula were aligned in the ClustalW program (Available online: https://www.genome.jp/tools-bin/clustalw). The MYB transcription factors related to AtMYB5, AtTT2, MtMYB5 (Medtr3g083540), and MtMYB14 (Medtr4g125520) acted as the outgroup sequences. Then, the phylogenic tree was constructed by the neighbor-joining, maximum-parsimony, or minimum-likelihood method with 1000 bootstrap replicates in MEGA6 [29].
4.4. Plasmid Construction and Plant Transformation
The coding sequence (CDS) of MtBZR1 was amplified from a complementary DNA (cDNA) sample prepared from wild-type root RNA with MtBZR1F and MtBZR1R primers (listed in Table S1, Supplementary Materials). The PCR product was purified and cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA, USA) and MtBZR1′s CDS was confirmed by Sanger sequencing. The resulting pENTR-MtBZR1 plasmid was used to construct 35S:MtBZR1-GFP for plant transformation by recombination with a destination vector pEarleyGate103 (Invitrogen, Carlsbad, CA, USA) [30]. For M. truncatula transformation, the 35S:MtBZR1-GFP plasmid was introduced into Agrobacterium strain EHA105 and used to transform the wild-type plant by the leaf disc method as previously described [31]. The transgenic plants were genotyped with the 35S primer and reverse primer of the gene. The positive ones were accessed for the expression level of MtBZR1 with its qRT primers for a step further.
The 35S:MtBZR1-GFP construct or the 35S:GFP control plasmid were introduced into Agrobacterium tumefaciens strain EHA105 and used for an infiltration assay of Nicotiana benthamiana leaves as previously reported [32]. Leaves from four-week-old N. benthamiana plants were infiltrated with A. tumefaciens at an OD600 value of 0.4 containing 35S:MtBZR1-GFP or 35S:GFP, respectively, and incubated in the dark at 25 °C for 16 h. Leaf discs infiltrated with A. tumefaciens were observed using a Zeiss LSM880 confocal microscope at 488-nm wavelength for the GFP fluorescence (Zeiss, Oberkochen, Germany).
4.5. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
To analyze the expression level of MtBZR1 in different organs, samples of root, stem, leaf, flower, and pod were harvested from two-month-old wild-type plants. For time course analysis of MtBZR1 expression after rhizobial inoculation, nodule samples from wild-type plants were harvested at seven, 14, and 21 days post-inoculation (dpi). To analyze the expression level of MtBZR1 in mtbzr1-1 and 35S:MtBZR1 plants, leaves were harvested from one-month-old wild-type, mtbzr1-1, and 35S:MtBZR1 plants. For expression analysis of NCR genes in mtbzr1-1 nodules, nodule samples from wild type and mtbzr1-1 were harvested at 21 dpi. Each biological replicate included 100 mg of nodules collected from 25 plants. To minimize the differences caused by nodule age, we pooled 100 mg of nodules from those plants for RNA extraction. All samples were frozen immediately in liquid nitrogen. RNA was extracted using RNeasy Mini Kit (Qiagen, Beijing, China), and treated with DNase I (Qiagen, Beijing, China). The quantity and quality of extracted RNA were measured by a Nanodrop 2000 Spectrophotometer (NanoDrop Technologies, Waltham, MA, USA). One microgram of total RNA was transcribed into cDNA using ThermoScript RT-PCR system (ThermoFisher, Waltham, MA, USA). After dilution (in a 1:50 ratio), the cDNA samples were used for qRT-PCR on a CFX Connect Real-Time PCR Detection system (Bio-Rad, Hercules, CA, USA) using SYBR Green PCR Master Mix (Roche, Basel, Switzerland) in three biological replicates. An MtUbiquitin gene, Medtr3g110110 [33], was used as an internal control, and relative gene expression was calculated according to the ∆∆CT method [34]. The primer sequences used for qRT-PCR were designed using Primer Express 3.0 software (https://primer-express.updatestar.com/) and are listed in Table S1 (Supplementary Materials).
4.6. Transcriptomic Analysis
For the transcriptomic analysis of mtbzr1-1, three biological replicates of nodule tissue were harvested from wild-type or mtbzr1-1 plants 21 days after rhizobial inoculation. RNA samples were sequenced on a BGISEQ-500 platform as eukaryote samples following the manufacturer’s instructions at BGI Genomics Institute (BGI-Shenzhen, Shenzhen, China). About 22 million raw reads were generated for each sample. Adapter sequences, low-quality reads, and reads containing more than 5% unknown nucleotides were first filtered out by trimmonmatic (v0.37) [35]. Clean reads were then aligned to the M. truncatula reference transcriptome (version 4.0) using Bowtie [36]. Gene expression (Table S5, Supplementary Materials) was quantified using RSEM (RNA-Seq by Expectation Maximization) [37], and differentially expressed genes with fold change >2 and false discovery rate (FDR) <0.001 were identified using R package DEGseq [38]. The enrichment of Gene Ontology (GO) terms and KEGG pathways was assessed using a hypergeometric test with p-values adjusted by the FDR method.
4.7. Statistical Analysis
Data processing and statistical analysis were performed using Microsoft® Office Excel 2010 (Available online: http://www.microsoft.com/). A Student’s t-test was used to evaluate if the differences were significant in the analysis of gene expression level, nodule numbers, and dry masses between samples.
5. Conclusions
In this study, we characterized MtBZR1, the homolog of A. thaliana BZR1 in M. truncatula, and found evidence that MtBZR1 plays an essential role in nodule development. The transcriptomic analysis of the loss-of-function mtbzr1-1 mutant suggests that MtBZR1 regulates NCR and secondary metabolite biosynthesis possibly via hormone signaling cross-talk. This work demonstrates the biological function of MtBZR1 and a potential role of BR in nodule development, which implies sophisticated hormone cross-talk during nodulation in M. truncatula.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/12/2941/s1. Figure S1. The maximum-parsimony and maximum-likelihood trees of BZR1-like proteins in A. thaliana and M. truncatula. (A) The maximum-parsimony tree of BZR1-like proteins. (B) The maximum-likelihood tree of BZR1-like proteins. Figure S2. Multiple alignment of MtBZR1, BZR1, and BZR1-like proteins from A. thaliana and M. truncatula. The BZR domain is indicated by a red line. Identical and similar amino acids are indicated as asterisks and colons. Figure S3. The expression of MtBZR1 in 35S:MtBZR1 transgenic lines. (A) The expression of MtBZR1 in 35S:MtBZR1 transgenic lines. (B) The leaves from WT and 35S:MtBZR#1 transgenic line. (C) GFP signal was detected in tobacco leaf disc two days after infiltration with Agrobacterium carrying a 35S:MtBZR1-GFP construct. The 35S:GFP construct was used as a negative control. Scale bar = 0.5 cm in (B) and 100 µm in (C). Figure S4. Repeatability and differential analysis of RNA-sequencing data. (A) The heatmap of Pearson’s coefficient between sequencing samples. (B) The volcano plot of differential analysis between mtbzr1-1 and wild-type plants. The induced and suppressed genes are indicated as red and green points, respectively. Figure S5. The expressions of MtNCRs, MtNCR301, MtNCR237, MtNCR327, MtNCR145, MtNCR332, and MtNCR224 in mtbzr1-1 and wild-type plants. MtUbiquitin was used as an internal control. Bars represent standard errors from three biological replicates. Figure S6. The phenylpropanoid biosynthesis pathway with induced and suppressed genes indicated in red and green frames, respectively. Figure S7. The flavonoid biosynthesis pathway with induced and suppressed genes indicated in red and green frames, respectively. Figure S8. The isoflavonoid biosynthesis pathway with induced and suppressed genes indicated in red and green frames, respectively. Figure S9. The expressions of MtFLSs and MtIOMTs in mtbzr1-1 and wild-type plants. MtUbiquitin was used as an internal control. Bars represent standard errors from three biological replicates. Table S1. Primers used in this study. Table S2. The 1319 differentially expressed genes in mtbzr1-1. Table S3. The expression of genes involved in the flavonoid biosynthesis pathway. Table S4. The expression of genes involved in the isoflavonoid biosynthesis pathway. Table S5. The expression of all genes in WT and mtbzr1-1.
Author Contributions
C.C., Y.Z., H.W., and L.H. performed the experiments; H.W. and Y.X. contributed the research materials and analysis tools; Y.Z. and C.Z. designed the experiments and wrote the paper.
Funding
This work was supported by grants from the Ministry of Science and Technology of China (2015CB943500), the Major Program of Shandong Province Natural Science Foundation (ZR2018ZC0334), the National Natural Science Foundation of China (projects 31671507 and 31371235), and the 1000-Talents Plan from China for Young Researchers.
Acknowledgments
We thank Kirankumar Mysore (Noble Research Institute) and Jiangqi Wen (Noble Research Institute) for providing the Tnt1 mutants. We also thank Taiyong Quan from Common Research Technology Platform of Life Science in Shandong University (Qingdao) for help with the confocal microscopy work.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ARF | AUXIN RESPONSE FACTOR |
| BES1 | BRASSINOSTEROID INSENSITIVE 1-EMS SUPPRESSOR 1 |
| BR | brassinosteroid |
| BZR | BRASSINAZOLE RESISTANT 1 |
| CDS | coding sequence |
| DEG | differentially expressed gene |
| DPI | days post inoculation |
| FDR | false discovery rate |
| FLS | flavonol synthase |
| FPKM | fragments per kilobase of transcript per million reads mapped |
| GFP | green fluorescence protein |
| GO | gene ontology |
| IOMT | isoflavone O-methyltransferase |
| JA | jasmonic acid |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LCR | low-molecular-weight cysteine-rich |
| NCR | nodule-specific cysteine-rich |
| qRT-PCR | quantitative real-time PCR |
| RSEM | RNA-seq by expectation maximization |
| TIR1 | TRANSPORT INHIBITOR RESPONSE 1 |
| TT2 | TRANSPARENT TESTA 2 |
| TY | tryptone, yeast extract, and sodium chloride |
| WT | wild type |
References
- Rey, T.; Nars, A.; Bonhomme, M.; Bottin, A.; Huguet, S.; Balzergue, S.; Jardinaud, M.F.; Bono, J.J.; Cullimore, J.; Dumas, B.; et al. Nfp, a lysm protein controlling nod factor perception, also intervenes in medicago truncatula resistance to pathogens. New Phytol. 2013, 198, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, Q.; Fedorova, E.; Liu, J.; Qin, Q.; Zheng, Q.; Price, P.A.; Pan, H.; Wang, D.; Griffitts, J.S.; et al. Microsymbiont discrimination mediated by a host-secreted peptide in medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6848–6853. [Google Scholar] [CrossRef] [PubMed]
- Guefrachi, I.; Nagymihaly, M.; Pislariu, C.I.; Van de Velde, W.; Ratet, P.; Mars, M.; Udvardi, M.K.; Kondorosi, E.; Mergaert, P.; Alunni, B. Extreme specificity of ncr gene expression in medicago truncatula. BMC Genom. 2014, 15, 712. [Google Scholar] [CrossRef] [PubMed]
- Foo, E.; McAdam, E.L.; Weller, J.L.; Reid, J.B. Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J. Exp. Bot. 2016, 67, 2413–2424. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Ross, J.J.; Reid, J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant. Physiol. 2005, 138, 2396–2405. [Google Scholar] [CrossRef] [PubMed]
- Gifford, I.; Battenberg, K.; Vaniya, A.; Wilson, A.; Tian, L.; Fiehn, O.; Berry, A.M. Distinctive patterns of flavonoid biosynthesis in roots and nodules of datisca glomerata and medicago spp. Revealed by metabolomic and gene expression profiles. Front. Plant Sci. 2018, 9, 1463. [Google Scholar] [CrossRef] [PubMed]
- Besseau, S.; Hoffmann, L.; Geoffroy, P.; Lapierre, C.; Pollet, B.; Legrand, M. Flavonoid accumulation in arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant. Cell 2007, 19, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Murray, J.D. The role of flavonoids in nodulation host-range specificity: An update. Plants 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Clouse, S.D. Brassinosteroid signal transduction: From receptor kinase activation to transcriptional networks regulating plant development. Plant. Cell 2011, 23, 1219–1230. [Google Scholar] [CrossRef]
- Clouse, S.D.; Langford, M.; McMorris, T.C. A brassinosteroid-insensitive mutant in arabidopsis thaliana exhibits multiple defects in growth and development. Plant. Physiol. 1996, 111, 671–678. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.Y.; Cao, D.M.; Tang, W.; He, K.; Zhu, J.Y.; He, J.X.; Bai, M.Y.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The gsk3-like kinase bin2 phosphorylates and destabilizes bzr1, a positive regulator of the brassinosteroid signaling pathway in arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. Pp2a activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating bzr1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized bzr1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. Bes1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Cheng, X.; Gou, X.; Yin, H.; Mysore, K.S.; Li, J.; Wen, J. Functional characterisation of brassinosteroid receptor mtbri1 in medicago truncatula. Sci. Rep. 2017, 7, 9327. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, S.; Liu, J.; Terecskei, K.; Abraham, E.; Gombar, A.; Domonkos, A.; Szucs, A.; Kormoczi, P.; Wang, T.; et al. Host-secreted antimicrobial peptide enforces symbiotic selectivity in medicago truncatula. Proc. Natl. Acad. Sci. USA 2017, 114, 6854–6859. [Google Scholar] [CrossRef]
- Alunni, B.; Kevei, Z.; Redondo-Nieto, M.; Kondorosi, A.; Mergaert, P.; Kondorosi, E. Genomic organization and evolutionary insights on grp and ncr genes, two large nodule-specific gene families in medicago truncatula. Mol. Plant Microbe Interact. MPMI 2007, 20, 1138–1148. [Google Scholar] [CrossRef]
- Nallu, S.; Silverstein, K.A.; Samac, D.A.; Bucciarelli, B.; Vance, C.P.; VandenBosch, K.A. Regulatory patterns of a large family of defensin-like genes expressed in nodules of medicago truncatula. PLoS ONE 2013, 8, e60355. [Google Scholar] [CrossRef]
- Chaiwanon, J.; Wang, Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in arabidopsis roots. Curr. Biol. CB 2015, 25, 1031–1042. [Google Scholar] [CrossRef]
- Wang, W.; Bai, M.Y.; Wang, Z.Y. The brassinosteroid signaling network-a paradigm of signal integration. Curr. Opin. Plant Biol. 2014, 21, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Wasson, A.P.; Pellerone, F.I.; Mathesius, U. Silencing the flavonoid pathway in medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant. Cell 2006, 18, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Vega, J.C.; Cady, B.; Kayanja, G.; Mauriello, A.; Cervantes, N.; Gillespie, A.; Lavia, L.; Trujillo, J.; Alkio, M.; Colon-Carmona, A. Detoxification of polycyclic aromatic hydrocarbons (pahs) in arabidopsis thaliana involves a putative flavonol synthase. J. Hazard. Mater. 2017, 321, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cardoza, V.; Mitchell, D.M.; Bright, L.; Oldroyd, G.; Harris, J.M. Crosstalk between jasmonic acid, ethylene and nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant. J. Cell Mol. Biol. 2006, 46, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Kazmierczak, T.; Nagymihaly, M.; Lamouche, F.; Barriere, Q.; Guefrachi, I.; Alunni, B.; Ouadghiri, M.; Ibijbijen, J.; Kondorosi, E.; Mergaert, P.; et al. Specific host-responsive associations between medicago truncatula accessions and sinorhizobium strains. Mol. Plant Microbe Interact. MPMI 2017, 30, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.J.; O’Hara, G.W.; Tiwari, R.P.; Dilworth, M.J.; Howieson, J.G. The model legume medicago truncatula a17 is poorly matched for n2 fixation with the sequenced microsymbiont sinorhizobium meliloti 1021. New Phytol. 2008, 179, 62–66. [Google Scholar] [CrossRef]
- Tadege, M.; Wen, J.; He, J.; Tu, H.; Kwak, Y.; Eschstruth, A.; Cayrel, A.; Endre, G.; Zhao, P.X.; Chabaud, M. Large-scale insertional mutagenesis using the tnt1 retrotransposon in the model legume medicago truncatula. Plant J. 2008, 54, 335–347. [Google Scholar] [CrossRef]
- Beringer, J.E. R factor transfer in rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Saldanha, A.J. Java treeview—Extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant. J. Cell Mol. Biol. 2006, 45, 616–629. [Google Scholar] [CrossRef]
- Cosson, V.; Durand, P.; d’Erfurth, I.; Kondorosi, A.; Ratet, P. Medicago truncatula transformation using leaf explants. Methods Mol. Biol. 2006, 343, 115–127. [Google Scholar] [PubMed]
- Tian, Y.; Fan, M.; Qin, Z.; Lv, H.; Wang, M.; Zhang, Z.; Zhou, W.; Zhao, N.; Li, X.; Han, C.; et al. Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the brassinazole-resistant1 transcription factor. Nat. Commun. 2018, 9, 1063. [Google Scholar] [CrossRef]
- Cheng, X.; Li, G.; Tang, Y.; Wen, J. Dissection of genetic regulation of compound inflorescence development in Medicago truncatula. Development 2018, 145, 158766. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Hou, C.; Metelli, A.; Qi, L.; Tadege, M.; Mysore, K.S.; Wang, Z.Y. Developmental analysis of a medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant. Cell 2011, 23, 2106–2124. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. Soapnuke: A mapreduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. Degseq: An R package for identifying differentially expressed genes from rna-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).