Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella
Abstract
1. Introduction
2. Results
2.1. cDNA Cloning and Sequences Analysis of PxTAR1
2.2. Expression Pattern of PxTAR1
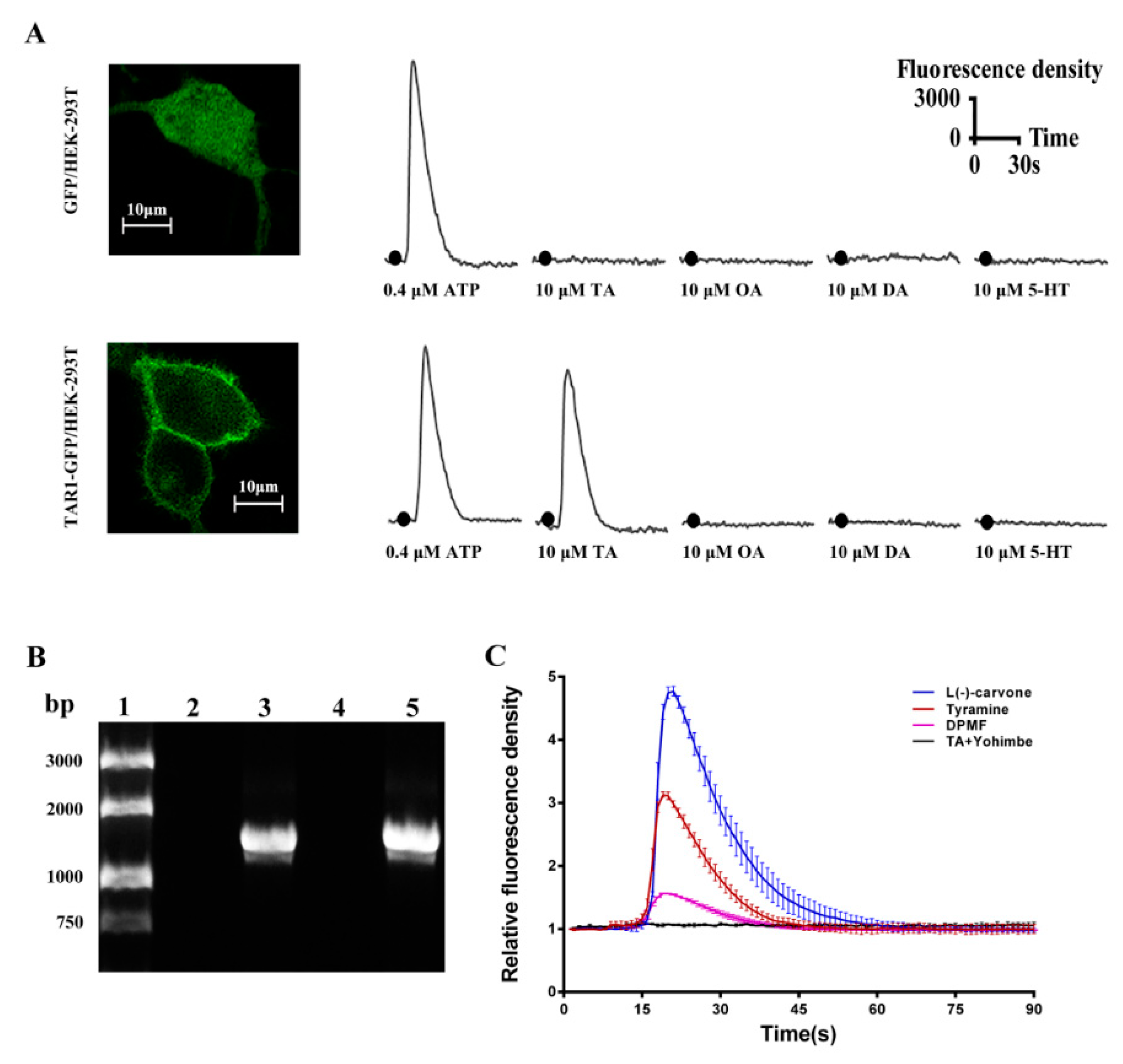
2.3. Functional Characterization of PxTAR1 with the Ca2+ Assay
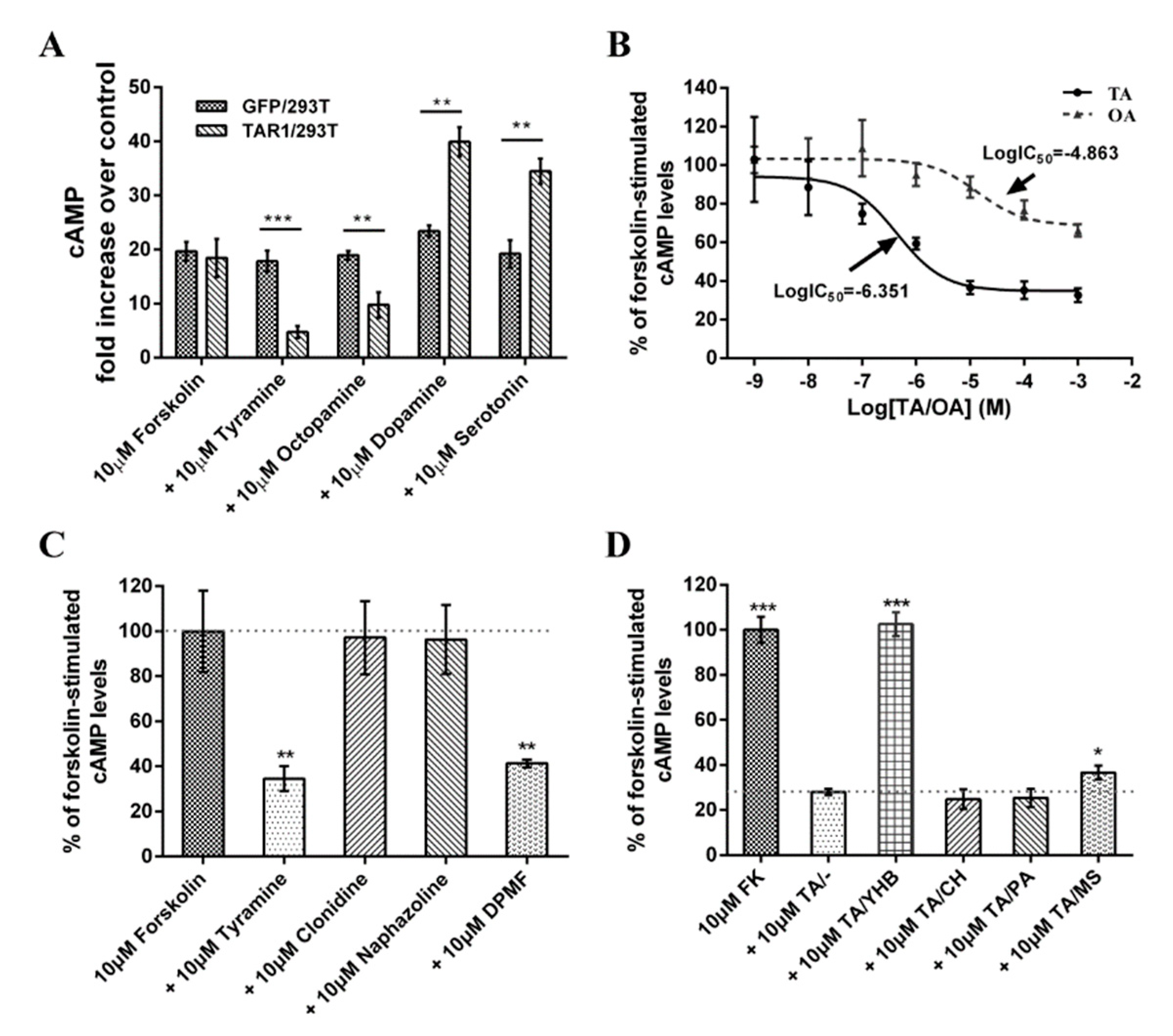
2.4. Pharmacological Characterization of PxTAR1 Based on cAMP Assay
3. Discussion
4. Materials and Methods
4.1. Insect and Chemicals
4.2. Cloning of PxTAR1 cDNAs
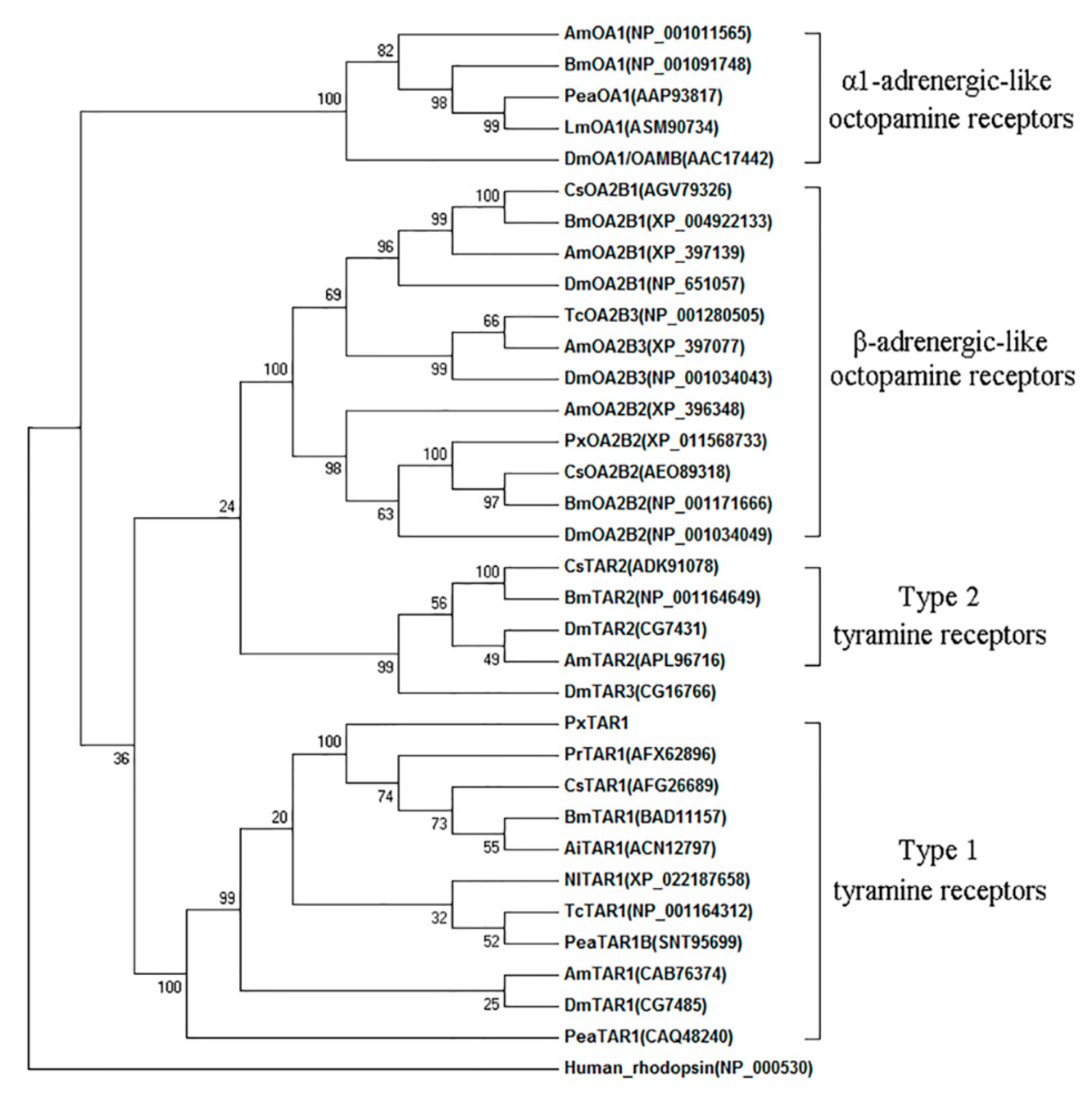
4.3. Sequence Analysis and Phylogenic Tree Construction
4.4. Gene Expression Profile Analysis
4.5. Cell Transfection and Creation of Stable Cell Lines
4.6. Ca2+ and Cyclic AMP (cAMP) Assays
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 5-HT | Serotonin |
| AAs | Amino acids |
| (Ca2+)i | Intracellular Ca2+ concentration |
| (cAMP)i | Intracellular cyclic adenosine monophosphate concentration |
| CH | Chlorpromazine |
| DA | Dopamine |
| DPMF | N2-(2,4-Dimethylphenyl)-N1- methyformamidine |
| FROs | Female reproductive organs |
| GFP | Green fluorescent protein |
| MROs | Male reproductive organs |
| MS | Mianserin |
| OA | Octopamine |
| OARs | Octopamine receptors |
| ORF | Open reading frame |
| PA | Phentolamine |
| TA | Tyramine |
| TARs | Tyramine receptors |
| YHB | Yohimbine |
References
- Blenau, W.; Balfanz, S.; Baumann, A. PeaTAR1B: Characterization of a Second Type 1 Tyramine Receptor of the American Cockroach, Periplaneta americana. Int. J. Mol. Sci. 2017, 18, 2279. [Google Scholar] [CrossRef] [PubMed]
- Roeder, T. Octopamine in invertebrates. Prog. Neurobiol. 1999, 59, 533–561. [Google Scholar] [CrossRef]
- Sinakevitch, I.T.; Daskalova, S.M.; Smith, B.H. The Biogenic Amine Tyramine and its Receptor (AmTyr1) in Olfactory Neuropils in the Honey Bee (Apis mellifera) Brain. Front. Syst. Neurosci. 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.; Lange, A.B. Tyramine as a possible neurotransmitter/neuromodulator at the spermatheca of the African migratory locust, Locusta migratoria. J. Insect Physiol. 2008, 54, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.B. Tyramine: From octopamine precursor to neuroactive chemical in insects. Gen. Comp. Endocrinol. 2009, 162, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Rex, E.; Molitor, S.C.; Hapiak, V.; Xiao, H.; Henderson, M.; Komuniecki, R. Tyramine receptor (SER-2) isoforms are involved in the regulation of pharyngeal pumping and foraging behavior in Caenorhabditis elegans. J. Neurochem. 2004, 91, 1104–1115. [Google Scholar] [CrossRef] [PubMed]
- Fox, L.E.; Soll, D.R.; Wu, C.F. Coordination and modulation of locomotion pattern generators in Drosophila larvae: Effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J. Neurosci. 2006, 26, 1486–1498. [Google Scholar] [CrossRef]
- Vierk, R.; Pflueger, H.J.; Duch, C. Differential effects of octopamine and tyramine on the central pattern generator for Manduca flight. J. Comp. Physiol. Neuroethol. Sens. Neural Behav. Physiol. 2009, 195, 265–277. [Google Scholar] [CrossRef]
- Hirashima, A.; Yamaji, H.; Yoshizawa, T.; Kuwano, E.; Eto, M. Effect of tyramine and stress on sex-pheromone production in the pre- and post-mating silkworm moth, Bombyx mori. J. Insect Physiol. 2007, 53, 1242–1249. [Google Scholar] [CrossRef]
- Blumenthal, E.M. Regulation of chloride permeability by endogenously produced tyramine in the Drosophila Malpighian tubule. Am. J. Physiol. Cell Physiol. 2003, 284, C718–C728. [Google Scholar] [CrossRef]
- Ma, Z.; Guo, X.; Lei, H.; Li, T.; Hao, S.; Kang, L. Octopamine and tyramine respectively regulate attractive and repulsive behavior in locust phase changes. Sci. Rep. 2015, 5, 8036. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, R.; Reim, T.; Sovik, E.; Entler, B.V.; Barron, A.B.; Thamm, M. Learning, gustatory responsiveness and tyramine differences across nurse and forager honeybees. J. Exp. Biol. 2017, 220, 1443–1450. [Google Scholar] [CrossRef] [PubMed]
- Torfs, H.; Van Poyer, W.; Poels, J.; Swinnen, E.; De Loof, A.; Vanden, B.J. Tyramine injections reduce locust viability. Acta Biol. Hung. 2000, 51, 349–354. [Google Scholar] [PubMed]
- Ohta, H.; Ozoe, Y. Molecular Signalling, Pharmacology, and Physiology of Octopamine and Tyramine Receptors as Potential Insect Pest Control Targets. Adv. Insect Physiol. 2014, 46, 73–166. [Google Scholar] [CrossRef]
- Duportets, L.; Barrozo, R.B.; Bozzolan, F.; Gaertner, C.; Anton, S.; Gadenne, C.; Debernard, S. Cloning of an octopamine/tyramine receptor and plasticity of its expression as a function of adult sexual maturation in the male moth Agrotis ipsilon. Insect Mol. Biol. 2010, 19, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.F.; Huang, J.; Ye, G.Y. Molecular cloning and pharmacological characterisation of a tyramine receptor from the rice stem borer, Chilo suppressalis (Walker). Pest Manag. Sci. 2013, 69, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Rotte, C.; Krach, C.; Balfanz, S.; Baumann, A.; Walz, B.; Blenau, W. Molecular characterization and localization of the first tyramine receptor of the American cockroach (Periplaneta americana). Neuroscience 2009, 162, 1120–1133. [Google Scholar] [CrossRef]
- Schützler, N.; Girwert, C.; Hügli, I.; Mohana, G.; Roignan, J.Y.; Ryglewski, S.; Duch, C. Tyramine action on motoneuron excitability and adaptable tyramine/octopamine ratios adjust Drosophila locomotion to nutritional state. Proc. Natl. Acad. Sci. USA 2019, 116, 3805–3810. [Google Scholar] [CrossRef]
- El-Kholy, S.; Stephano, F.; Li, Y.; Bhandari, A.; Fink, C.; Roeder, T. Expression analysis of octopamine and tyramine receptors in Drosophila. Cell Tissue Res. 2015, 361, 669–684. [Google Scholar] [CrossRef]
- Hannan, F.; Hall, L.M. Temporal and spatial expression patterns of two G-protein coupled receptors in Drosophila melanogaster. Invert. Neurosci. 1996, 2, 71–83. [Google Scholar] [CrossRef]
- Saudou, F.; Amlaiky, N.; Plassat, J.L.; Borrelli, E.; Hen, R. Cloning and characterization of a Drosophila tyramine receptor. EMBO J. 1990, 9, 3611–3617. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.C.; He, H.; Davey, R.B. Mutations in a putative octopamine receptor gene in amitraz-resistant cattle ticks. Vet. Parasitol. 2007, 148, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.D.; Temeyer, K.B.; Day, T.A.; Perez de Leon, A.A.; Kimber, M.J.; Coats, J.R. Pharmacological characterization of a tyramine receptor from the southern cattle tick, Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2015, 63, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, N.N.; Klafke, G.; Corley, S.W.; Tidwell, J.; Berry, C.M.; Koh-Tan, H.C. Molecular biology of amitraz resistance in cattle ticks of the genus Rhipicephalus. Front. Biosci. 2018, 23, 796–810. [Google Scholar] [CrossRef]
- Sungirai, M.; Baron, S.; Moyo, D.Z.; De Clercq, P.; Maritz-Olivier, C.; Madder, M. Genotyping acaricide resistance profiles of Rhipicephalus microplus tick populations from communal land areas of Zimbabwe. Ticks Tick Borne Dis. 2018, 9, 2–9. [Google Scholar] [CrossRef]
- Enan, E.E. Molecular response of Drosophila melanogaster tyramine receptor cascade to plant essential oils. Insect Biochem. Mol. Biol. 2005, 35, 309–321. [Google Scholar] [CrossRef]
- Gross, A.D.; Temeyer, K.B.; Day, T.A.; Perez de Leon, A.A.; Kimber, M.J.; Coats, J.R. Interaction of plant essential oil terpenoids with the southern cattle tick tyramine receptor: A potential biopesticide target. Chem. Biol. Interact. 2017, 263, 1–6. [Google Scholar] [CrossRef]
- Li, Z.; Feng, X.; Liu, S.S.; You, M.; Furlong, M.J. Biology, Ecology, and Management of the Diamondback Moth in China. Annu. Rev. Entomol. 2016, 61, 277–296. [Google Scholar] [CrossRef]
- Ohta, H.; Utsumi, T.; Ozoe, Y. Amino acid residues involved in interaction with tyramine in the Bombyx mori tyramine receptor. Insect Mol. Biol. 2004, 13, 531–538. [Google Scholar] [CrossRef]
- Ohta, H.; Khan, M.A.; Nagai, I.; Umemoto, N.; Hamasaki, T.; Ozoe, Y. Responses of the silkworm tyramine receptor to 2-phenylethylamines and 5-phenyloxazoles. Arch. Insect Biochem. Physiol. 2005, 59, 150–160. [Google Scholar] [CrossRef]
- Ohta, H.; Utsumi, T.; Ozoe, Y. B96Bom encodes a Bombyx mori tyramine receptor negatively coupled to adenylate cyclase. Insect Mol. Biol. 2003, 12, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Blenau, W.; Balfanz, S.; Baumann, A. Amtyr1: Characterization of a gene from honeybee (Apis mellifera) brain encoding a functional tyramine receptor. J. Neurochem. 2000, 74, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Brigaud, I.; Grosmaitre, X.; Francois, M.C.; Jacquin-Joly, E. Cloning and expression pattern of a putative octopamine/tyramine receptor in antennae of the noctuid moth Mamestra brassicae. Cell Tissue Res. 2009, 335, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Molaei, G.; Paluzzi, J.P.; Bendena, W.G.; Lange, A.B. Isolation, cloning, and tissue expression of a putative octopamine/tyramine receptor from locust visceral muscle tissues. Arch. Insect Biochem. Physiol. 2005, 59, 132–149. [Google Scholar] [CrossRef] [PubMed]
- Vanden, B.J.; Vulsteke, V.; Huybrechts, R.; De Loof, A. Characterization of a cloned locust tyramine receptor cDNA by functional expression in permanently transformed Drosophila S2 cells. J. Neurochem. 1995, 64, 2387–2395. [Google Scholar] [CrossRef]
- Huang, J.; Liu, W.; Qi, Y.X.; Luo, J.; Montell, C. Neuromodulation of courtship drive through tyramine-responsive neurons in the Drosophila brain. Curr. Biol. 2016, 26, 2246–2256. [Google Scholar] [CrossRef]
- Huang, Q.T.; Ma, H.H.; Deng, X.L.; Zhu, H.; Liu, J.; Zhou, Y.; Zhou, X.M. Pharmacological characterization of a beta-adrenergic-like octopamine receptor in Plutella xylostella. Arch. Insect Biochem. Physiol. 2018, 98, e21466. [Google Scholar] [CrossRef]
- Huang, J.; Ohta, H.; Inoue, N.; Takao, H.; Kita, T.; Ozoe, F.; Ozoe, Y. Molecular cloning and pharmacological characterization of a Bombyx mori tyramine receptor selectively coupled to intracellular calcium mobilization. Insect Biochem. Mol. Biol. 2009, 39, 842–849. [Google Scholar] [CrossRef]
- Kita, T.; Hayashi, T.; Ohtani, T.; Takao, H.; Takasu, H.; Liu, G.; Ohta, H.; Ozoe, F.; Ozoe, Y. Amitraz and its metabolite differentially activate alpha- and beta-adrenergic-like octopamine receptors. Pest Manag. Sci. 2017, 73, 984–990. [Google Scholar] [CrossRef]
- Adnan, M.; Morton, G.; Hadi, S. Analysis of rpoS and bolA gene expression under various stress-induced environments in planktonic and biofilm phase using 2(-ΔΔCT) method. Mol. Cell. Biochem. 2011, 357, 275–282. [Google Scholar] [CrossRef]
- Tischbirek, C.; Birkner, A.; Jia, H.; Sakmann, B.; Konnerth, A. Deep two-photon brain imaging with a red-shifted fluorometric Ca2+ indicator. Proc. Natl. Acad. Sci. USA 2015, 112, 11377–11382. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequences 1 |
|---|---|
| TAR1_HindIII_F | TTTaagcttCTGCCACCATGGGGCAGGCCAACCT |
| TAR1_PstI_R | ATActgcagAGGCTTCATACAGAGCAGCTTC |
| TAR1_XbaI_F | GATtctagaGCCACCATGGGGCAGGCCAACCTC |
| TAR1_SalI_R | AGTgtcgacTCAAGGCTTCATACAGAGCAG |
| TAR1_qF | CTCGGTTGGAACGACTGGC |
| TAR1_qR | CGGTTCTGTCCGCTCGATAT |
| Rpl32_qF | TGCCCAACATTGGTTACGG |
| Rpl32_qR | ACGATGGCCTTGCGCTTC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Huang, Q.; Lai, X.; Liu, J.; Zhu, H.; Zhou, Y.; Deng, X.; Zhou, X. Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella. Int. J. Mol. Sci. 2019, 20, 2953. https://doi.org/10.3390/ijms20122953
Ma H, Huang Q, Lai X, Liu J, Zhu H, Zhou Y, Deng X, Zhou X. Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella. International Journal of Molecular Sciences. 2019; 20(12):2953. https://doi.org/10.3390/ijms20122953
Chicago/Turabian StyleMa, Haihao, Qingting Huang, Xiaoyi Lai, Jia Liu, Hang Zhu, Yong Zhou, Xile Deng, and Xiaomao Zhou. 2019. "Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella" International Journal of Molecular Sciences 20, no. 12: 2953. https://doi.org/10.3390/ijms20122953
APA StyleMa, H., Huang, Q., Lai, X., Liu, J., Zhu, H., Zhou, Y., Deng, X., & Zhou, X. (2019). Pharmacological Properties of the Type 1 Tyramine Receptor in the Diamondback Moth, Plutella xylostella. International Journal of Molecular Sciences, 20(12), 2953. https://doi.org/10.3390/ijms20122953





