Serum Proteome Alterations in Human Cystathionine ?-Synthase Deficiency and Ischemic Stroke Subtypes
Abstract
:1. Introduction
2. Results
2.1. Serum Proteins Affected by CBS Deficiency
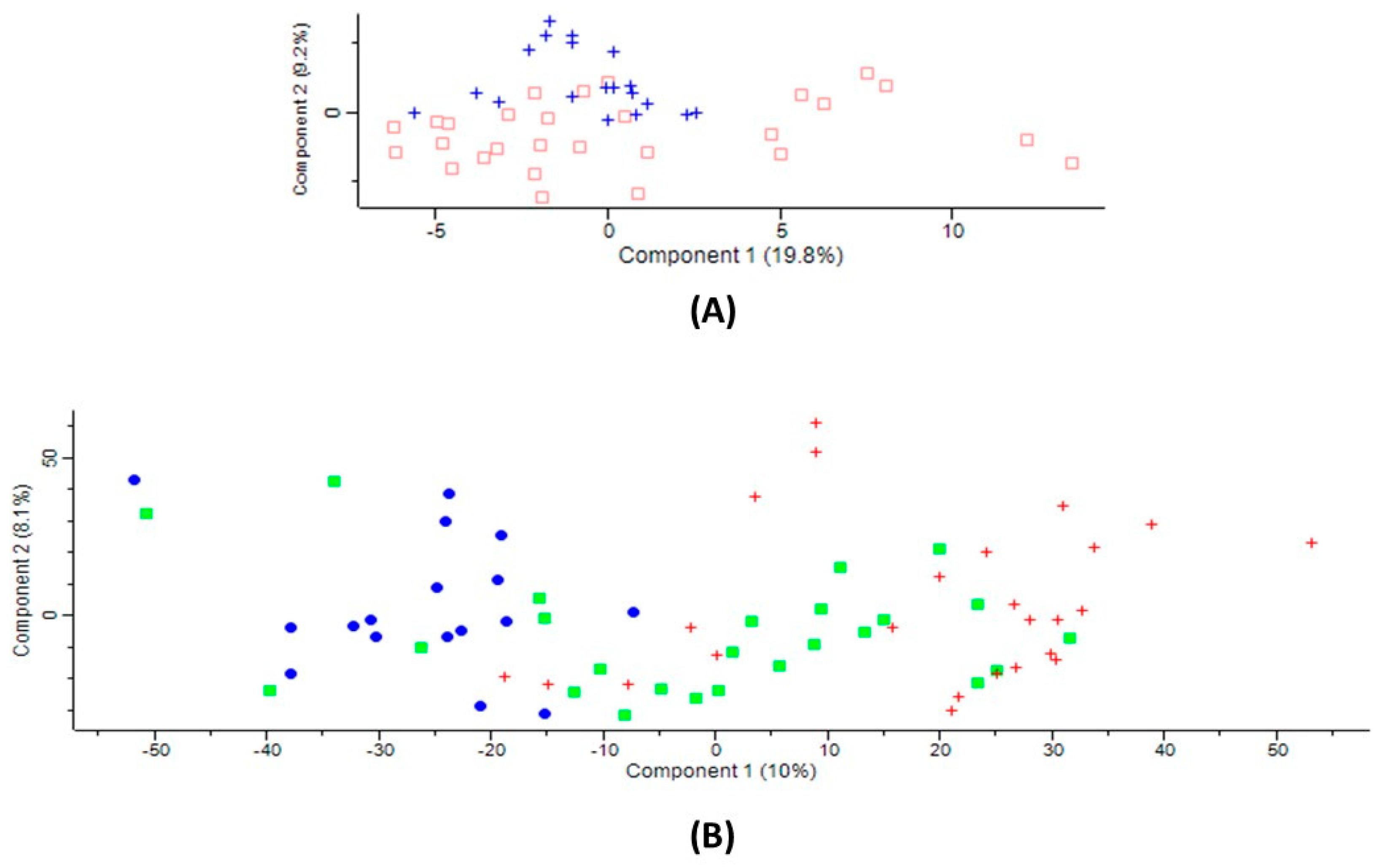
2.2. Serum Proteins Affected by Ischemic Stroke Subtype
2.3. Ischemic Stroke Subtype-Specific Proteins
2.4. Overlap between Proteins Affected by CBS Deficiency and Ischemic Stroke Subtype
2.5. Validation of Label-Free Mass Spectrometry Analyses by ELISA
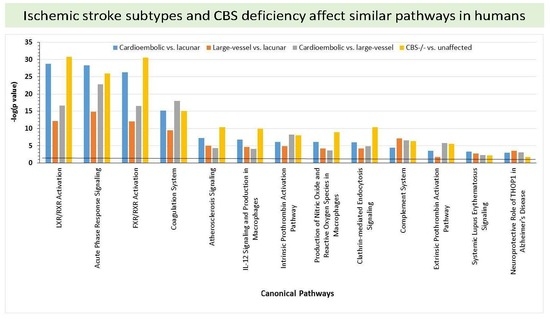
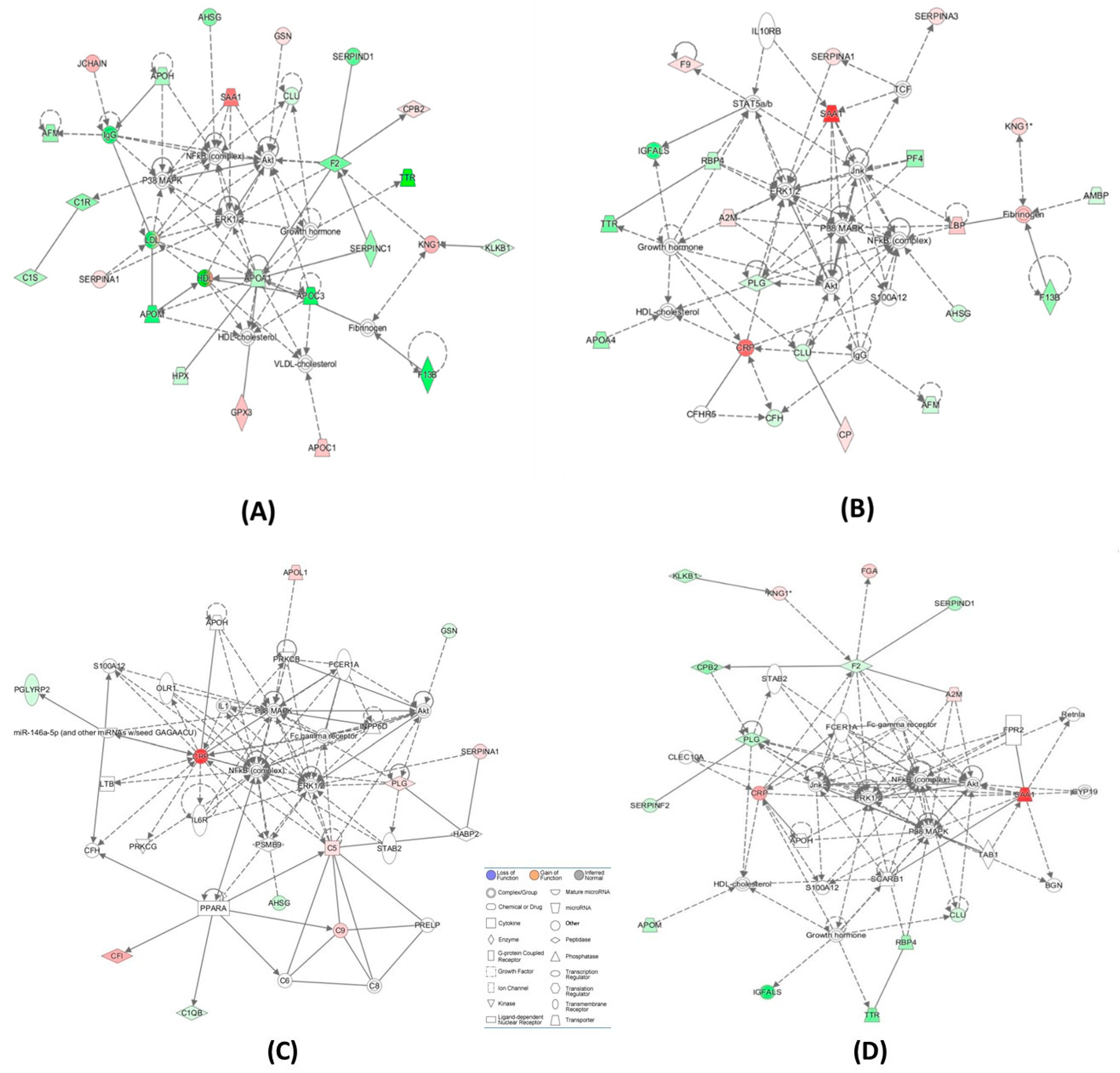
2.6. Bioinformatic Analyses
2.7. Involvement of Homocysteine and Anti-N-Hcy-Protein Autoantibodies
3. Discussion
4. Materials and Methods
4.1. CBS-Deficient Patients
4.2. Ischemic Stroke Patients
4.3. Serum Samples
4.4. Digestion with Trypsin
4.5. Label-Free Mass Spectrometry
4.6. Data Analysis
4.7. Statistical Analyses
4.8. Pathway and Network Analyses
4.9. ELISA Assays
4.10. Anti-N-Hcy-Protein Antibody Assays
4.11. Homocysteine Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics-2019 Update: A Report from the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.L.; Norrving, B.; Mensah, G.A. Global Burden of Stroke. Circ. Res. 2017, 120, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.D. Cardioembolic stroke: everything has changed. Stroke Vasc. Neurol. 2018, 3, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.F.; Sarracino, D.A.; Prakash, A.; Athanas, M.; Krastins, B.; Rezai, T.; Sutton, J.N.; Peterman, S.; Gvozdyak, O.; Chou, S.; et al. Discrimination of ischemic and hemorrhagic strokes using a multiplexed, mass spectrometry-based assay for serum apolipoproteins coupled to multi-marker ROC algorithm. Proteom. Clin. Appl. 2012, 6, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Mudd, S.H.; Levy, H.L.; Kraus, J.P. Disorders of transsulfuration. In The Metabolic and Molecular Bases of Inherited Disease, 8th ed.; Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Childs, B., Kinzler, K.W., Vogelstein, B., Eds.; Mc Graw-Hill: New York, NY, USA, 2001; Volume 2, pp. 2007–2056. [Google Scholar]
- Kelly, P.J.; Furie, K.L.; Kistler, J.P.; Barron, M.; Picard, E.H.; Mandell, R.; Shih, V.E. Stroke in young patients with hyperhomocysteinemia due to cystathionine beta-synthase deficiency. Neurology 2003, 60, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Chwatko, G.; Boers, G.H.; Strauss, K.A.; Shih, D.M.; Jakubowski, H. Mutations in methylenetetrahydrofolate reductase or cystathionine beta-synthase gene, or a high-methionine diet, increase homocysteine thiolactone levels in humans and mice. FASEB J. 2007, 21, 1707–1713. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H.; Boers, G.H.; Strauss, K.A. Mutations in cystathionine beta-synthase or methylenetetrahydrofolate reductase gene increase N-homocysteinylated protein levels in humans. FASEB J. 2008, 22, 4071–4076. [Google Scholar] [CrossRef] [PubMed]
- Gurda, D.; Handschuh, L.; Kotkowiak, W.; Jakubowski, H. Homocysteine thiolactone and N-homocysteinylated protein induce pro-atherogenic changes in gene expression in human vascular endothelial cells. Amino Acids 2015, 47, 1319–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol. Rev. 2019, 99, 555–604. [Google Scholar] [CrossRef]
- Undas, A.; Perla, J.; Lacinski, M.; Trzeciak, W.; Kazmierski, R.; Jakubowski, H. Autoantibodies against N-homocysteinylated proteins in humans: implications for atherosclerosis. Stroke 2004, 35, 1299–1304. [Google Scholar] [CrossRef]
- Sikora, M.; Marczak, L.; Twardowski, T.; Stobiecki, M.; Jakubowski, H. Direct monitoring of albumin lysine-525 N-homocysteinylation in human serum by liquid chromatography/mass spectrometry. Anal. Biochem. 2010, 405, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Eikelboom, J.W.; Hankey, G.J.; Anand, S.S.; Lofthouse, E.; Staples, N.; Baker, R.I. Association between high homocyst(e)ine and ischemic stroke due to large- and small-artery disease but not other etiologic subtypes of ischemic stroke. Stroke 2000, 31, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, A.; Brattstrom, L.; Norrving, B.; Hultberg, B.; Andersson, A.; Johansson, B.B. Plasma homocysteine in the acute and convalescent phases after stroke. Stroke 1995, 26, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Coull, B.M.; Malinow, M.R.; Beamer, N.; Sexton, G.; Nordt, F.; de Garmo, P. Elevated plasma homocyst(e)ine concentration as a possible independent risk factor for stroke. Stroke 1990, 21, 572–576. [Google Scholar] [CrossRef] [PubMed]
- Bots, M.L.; Launer, L.J.; Lindemans, J.; Hoes, A.W.; Hofman, A.; Witteman, J.C.; Koudstaal, P.J.; Grobbee, D.E. Homocysteine and short-term risk of myocardial infarction and stroke in the elderly: the Rotterdam Study. Arch. Intern. Med. 1999, 159, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.Y.; Ampil, E.R.; Chen, C.P.; Auchus, A.P. The relationship between homocysteine, cognition and stroke subtypes in acute stroke. J. Neurol. Sci. 2006, 250, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Weng, Y.; Zheng, L.; Li, H.; Gong, Q.; Fu, Y.; Zhao, J. Polymorphism of the complement 5 gene is associated with large artery atherosclerosis stroke in Chinese patients. Arq. Neuropsiquiatr. 2016, 74, 881–886. [Google Scholar] [CrossRef] [Green Version]
- Jickling, G.C.; Sharp, F.R. Biomarker panels in ischemic stroke. Stroke 2015, 46, 915–920. [Google Scholar] [CrossRef]
- Alvarez-Perez, F.J.; Castelo-Branco, M.; Alvarez-Sabin, J. Usefulness of measurement of fibrinogen, D-dimer, D-dimer/fibrinogen ratio, C reactive protein and erythrocyte sedimentation rate to assess the pathophysiology and mechanism of ischaemic stroke. J. Neurol. Neurosurg. Psychiatry 2011, 82, 986–992. [Google Scholar] [CrossRef] [Green Version]
- Turaj, W.; Slowik, A.; Pulyk, R.; Adamski, M.; Szczudlik, A. Comparison of plasma concentrations of fibrinogen in patients with ischemic stroke due to large vessel disease and small vessel disease. Neurol. Neurochir. Pol. 2006, 40, 297–301. [Google Scholar]
- Malik, R.; Dau, T.; Gonik, M.; Sivakumar, A.; Deredge, D.J.; Edeleva, E.V.; Götzfried, J.; van der Laan, S.W.; Pasterkamp, G.; Beaufort, N.; et al. Common coding variant in SERPINA1 increases the risk for large artery stroke. Proc. Natl. Acad. Sci. USA 2017, 114, 3613–3618. [Google Scholar] [CrossRef] [PubMed]
- Meschia, J.F. Alpha-1 antitrypsin dysfunction and large artery stroke. Proc. Natl. Acad. Sci. USA 2017, 114, 3555–3557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, A.; Chen, C.P.; Sze, S.K. Discovery of prognostic biomarker candidates of lacunar infarction by quantitative proteomics of microvesicles enriched plasma. PLoS ONE 2014, 9, e94663. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Zhou, L.; Guo, T.; Wang, N.; Hao, H.; Zhou, Y.; Yu, D. Plasma proteomics reveals coagulation, inflammation, and metabolic shifts in H-type hypertension patients with and without acute ischemic stroke. Oncotarget 2017, 8, 100384–100395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fullerton, H.J.; deVeber, G.A.; Hills, N.K.; Dowling, M.M.; Fox, C.K.; Mackay, M.T.; Kirton, A.; Yager, J.Y.; Bernard, T.J.; Hod, E.A.; et al. Inflammatory Biomarkers in Childhood Arterial Ischemic Stroke: Correlates of Stroke Cause and Recurrence. Stroke 2016, 47, 2221–2228. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Zhan, X.; Su, X.; Guo, L.; Lv, L.; Su, B. Proteomic analysis of serum proteins in acute ischemic stroke patients treated with acupuncture. Exp. Biol. Med. (Maywood) 2011, 236, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Ambrosius, W.; Michalak, S.; Kazmierski, R.; Andrzejewska, N.; Kozubski, W. Predictive value of serum transthyretin for outcome in acute ischemic stroke. PLoS ONE 2017, 12, e0179806. [Google Scholar] [CrossRef] [PubMed]
- Allard, L.; Lescuyer, P.; Burgess, J.; Leung, K.Y.; Ward, M.; Walter, N.; Burkhard, P.R.; Corthals, G.; Hochstrasser, D.F.; Sanchez, J.C. ApoC-I and ApoC-III as potential plasmatic markers to distinguish between ischemic and hemorrhagic stroke. Proteomics 2004, 4, 2242–2251. [Google Scholar] [CrossRef]
- Maclean, K.N.; Gaustadnes, M.; Oliveriusova, J.; Janosik, M.; Kraus, E.; Kozich, V.; Kery, V.; Skovby, F.; Rudiger, N.; Ingerslev, J.; et al. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum. Mutat. 2002, 19, 641–655. [Google Scholar] [CrossRef]
- Orendae, M.; Pronicka, E.; Kubalska, J.; Janosik, M.; Sokolova, J.; Linnebank, M.; Koch, H.G.; Kozich, V. Identification and functional analysis of two novel mutations in the CBS gene in Polish patients with homocystinuria. Hum. Mutat. 2004, 23, 631. [Google Scholar] [CrossRef]
- Sikora, M.; Marczak, L.; Kubalska, J.; Graban, A.; Jakubowski, H. Identification of N-homocysteinylation sites in plasma proteins. Amino Acids 2014, 46, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.P., Jr.; Bendixen, B.H.; Kappelle, L.J.; Biller, J.; Love, B.B.; Gordon, D.L.; Marsh, E.E., 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993, 24, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H.; Perla-Kajan, J.; Finnell, R.H.; Cabrera, R.M.; Wang, H.; Gupta, S.; Kruger, W.D.; Kraus, J.P.; Shih, D.M. Genetic or nutritional disorders in homocysteine or folate metabolism increase protein N-homocysteinylation in mice. FASEB J. 2009, 23, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Xu, Y.K.; Chan, P.; Pattengale, P.K. Simple, Fast, and Simultaneous Detection of Plasma Total Homocysteine, Methylmalonic Acid, Methionine, and 2-Methylcitric Acid Using Liquid Chromatography and Mass Spectrometry (LC/MS/MS). JIMD Rep. 2013, 10, 69–78. [Google Scholar] [PubMed] [Green Version]

| Variable | CBS-Deficient Patients (n = 10) | Healthy Controls (n = 14) | p-Value |
|---|---|---|---|
| Female sex, n (%) | 5 (50) | 7 (50) | NS |
| Mean age, years | 37.3 ± 7.3 | 38.0 ± 12.5 | NS |
| History of stroke, n (%) | 4 (40) | 0 (0) | <0.05 |
| Total cholesterol, mg/dL | 180 ± 48 | 180 ± 37 | NS |
| HDL cholesterol, mg/dL | 58 ± 9 | 67 ± 22 | NS |
| LDL cholesterol, mg/dL | 193 ± 42 | 96 ± 35 | NS |
| Triglyceride, mg/dL | 102 ± 46 | 84 ± 44 | NS |
| Methionine, µM | 567 ± 16 | 14.9 ± 4.5 | <0.001 |
| tHcy, µM | 71.2 ± 55.6 | 9.7 ± 5.9 | <0.001 |
| Anti-N-Hcy-protein antibodies, A492 | 0.33 ± 0.16 | 0.12 ± 0.10 | 0.005 |
| Variable | All Patients (n = 68) | Ischemic Stroke Subtype Patients | ANOVA p-Value | ||
|---|---|---|---|---|---|
| Large-Vessel (n = 28) | Cardioembolic (n = 16) | Lacunar (n = 24) | |||
| Female sex, % | 41.2 | 32.1 | 56.2 | 41.7 | |
| Age, years | 66.4 ± 12.7 | 63.8 ± 11.0 | 78.3 ± 15.0 | 62.0 ± 8.0 | <0.001 |
| Atrial fibrillation, % | 34 | 6 | 91 | 0 | |
| Hypertension, % | 72 | 67 | 67 | 83 | |
| Cholesterol, mg/dL | 207.6 ± 57.4 | 218.0 ± 64.7 | 173.0 ± 64.5 | 211.5 ± 40.5 | 0.005 |
| HDL cholesterol, mg/dL | 52.0 ± 14.0 | 55.9 ± 14.75 | 51.0 ± 15.0 | 49.3 ± 13.0 | NS |
| LDL cholesterol, mg/dL | 131.6 ± 48.7 | 138.0 ± 59.0 | 114.5 ± 53.3 | 136.0 ± 33.3 | NS |
| Triglycerides, mg/dL | 116.0 ± 59.4 | 120.0 ± 51.0 | 77.8 ± 25.8 | 133.0 ± 72.0 | 0.001 |
| Creatinine, µmol/L | 94.0 ± 34.0 | 89.0 ± 24.7 | 100.6 ± 38.4 | 94.3 ± 34.6 | NS |
| Glucose, mmol/L | 5.85 ± 1.37 | 5.70 ± 1.26 | 5.96 ± 1.54 | 5.86 ± 1.36 | NS |
| Alanine aminotransferase, U/L | 24.87 ± 18.7 | 21.7 ± 8.6 | 26.8 ± 27.6 | 26.2 ± 13.2 | NS |
| Aspartate aminotransferase, U/l | 27.4 ± 16.4 | 25.9 ± 10.0 | 30.2 ± 23.3 | 26.2 ± 11.7 | NS |
| Thyroid-stimulating hormone, mU/L | 1.64 ± 1.47 | 1.28 ± 0.86 | 1.68 ± 1.08 | 1.94 ± 2.23 | NS |
| Free triiodothyronine, pmol/L | 5.08 ± 3.43 | 6.94 ± 5.4 | 3.76 ± 1.09 | 4.57 ± 0.72 | 0.007 |
| Leukocytes, × 109/L | 8.5 ± 2.75 | 9.8 ± 3.4 | 7.3 ± 1.9 | 7.9 ± 2.0 | 0.01 |
| tHcy, μM | 3.3 ± 1.6 | 3.3 ± 1.2 | 3.5 ± 1.6 | 3.0 ± 2.0 | NS |
| Anti-N-Hcy-protein antibodies, A492 | 0.13 ± 0.08 | 0.12 ± 0.08 | 0.14 ± 0.11 | 0.10 ± 0.07 | NS |
| Gene Name | Protein Name | Cardioembolic vs. Large-Vessel Stroke | Cardioembolic vs. Lacunar Stroke | Large-Vessel vs. Lacunar Stroke | ANOVA p Value | CBS−/− vs. Control | Molecular Function/Biological Process | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold Change | p Value | Fold Change | p Value | Fold Change | p Value | Fold Change | T-test p Value | ||||
| AFM | Afamin | 0.84 | 0.011 | 0.85 | 0.005 | 0.0148 | 0.83 | 0.007 | Vitamin transport | ||
| ORM1 | α-1-acid glycoprotein1 | 1.35 | 0.001 | 1.20 | 0.010 | 0.0018 | Acute inflammatory response/acute-phase response | ||||
| ORM2 | α-1-acid glycoprotein2 | 0.81 | 0.042 | Acute-phase response | |||||||
| SERPINA3 | α-1-antichymotrypsin | 1.21 | 0.046 | 1.42 | <1 × 10−4 | 1.18 | 0.021 | 0.0002 | Acute inflammatory response/acute-phase response | ||
| SERPINA1 | α-1-antitrypsin | 1.17 | 0.007 | 1.18 | 0.011 | 0.0106 | 1.14 | 0.045 | Acute phase response; Blood coagulation | ||
| SERPINF2 | α-2-antiplasmin | 0.85 | 0.027 | 0.0305 | 0.80 | 1.2 × 10−5 | Complement/coagulation cascades | ||||
| AHSG | α-2-HS-glycoprotein | 0.85 | 0.001 | 0.81 | 0.0001 | 0.0001 | 0.80 | 5.3 × 10−5 | Acute phase response | ||
| SERPINC1 | Antithrombin-III | 0.84 | 4.5 × 10−5 | Complement/coagulation cascades | |||||||
| A2M | α-2-macroglobulin | 1.29 | 0.024 | 1.29 | 0.006 | Blood coagulation | |||||
| APOA1 | Apolipoprotein A-I | 0.87 | 0.038 | Fat digestion/absorption | |||||||
| APOA4 | Apolipoprotein A-IV | 0.71 | 0.006 | Cholesterol transport | |||||||
| APOC1 | Apolipoprotein C-I | 0.67 | 0.004 | 0.52 | <1 × 10−4 | 0.77 | 0.035 | 4.5E-5 | 1.40 | 0.032 | Cholesterol efflux/lipid and lipoprotein metabolic process |
| APOC3 | Apolipoprotein C-III | 0.68 | 0.014 | Cholesterol transport | |||||||
| APOL1 | Apolipoprotein L1 | 1.23 | 0.042 | Cholesterol metabolism | |||||||
| APOM | Apolipoprotein M | 0.81 | 0.012 | 0.0375 | 0.70 | 0.001 | Cholesterol transport, antioxidant activity | ||||
| APOH | Beta-2-glycoprotein 1T | 0.86 | 0.015 | Blood coagulation | |||||||
| C4BPA | C4b-binding protein α-chain | 0.44 | 0.032 | Complement activation/immune response | |||||||
| CPB2 | Carboxypeptidase B2 | 0.74 | 0.018 | 0.0431 | 1.12 | 0.044 | Complement/coagulation cascades; Protein digestion | ||||
| CP | Ceruloplasmin | 1.21 | 0.001 | 1.12 | 0.007 | 0.0020 | Cellular iron ion homeostasis | ||||
| CLU | Clusterin | 0.88 | 0.014 | 0.88 | 0.004 | 0.0072 | 0.93 | 0.019 | Negative regulation of amyloid-beta formation | ||
| F9;factor IX | Coagulation factor IX | 1.45 | 0.004 | 1.29 | 0.018 | 0.0010 | Blood coagulation | ||||
| F13B | Coagulation factor XIII B-chain | 0.74 | 0.010 | 0.71 | 0.005 | 0.0010 | 0.69 | 0.0005 | Blood coagulation | ||
| C1QA | Complement C1q subunit A | 1.66 | 0.042 | Complement and coagulation cascades | |||||||
| C1QB | Complement C1q subunit B | 1.12 | 0.038 | 0.90 | 0.047 | 0.0433 | Complement/coagulation cascades | ||||
| C5 | Complement C5 | 1.14 | 0.003 | 0.0136 | Complement/coagulation cascades | ||||||
| C1R | Complement C1r subcomponent | 0.82 | 5 × 10−5 | Complement/coagulation cascades | |||||||
| C1S | Complement C1s subcomponent | 0.86 | 0.004 | Complement/coagulation cascades | |||||||
| C9 | Complement component C9 | 1.36 | 1 × 10−4 | 1.25 | 0.002 | 0.0002 | 1.36 | 0.004 | Complement/coagulation cascades | ||
| CFI | Complement factor I | 0.85 | 0.002 | Complement/coagulation cascades | |||||||
| CRP | C-reactive protein | 2.44 | 0.046 | 5.89 | 1 × 10−5 | 2.41 | 0.011 | 2.5 × 10−7 | Acute inflammatory response/acute-phase response | ||
| HEL-213 | Epididymis luminal protein 213 | 1.55 | 0.003 | ||||||||
| FGA | Fibrinogen α-chain; | 1.53 | 0.006 | 2.01 | 1 × 10−6 | 1.32 | 0.031 | 1.6 × 10−5 | Blood coagulation | ||
| FBLN1 | Fibulin-1 | 1.99 | 0.0005 | 0.0326 | 2.13 | 0.001 | Blood coagulation/fibrin clot formation | ||||
| FCN3 | Ficolin-3 | 0.72 | 0.007 | 0.0211 | 0.76 | 0.003 | Complement activation | ||||
| GSN | Gelsolin | 0.87 | 0.004 | 0.0160 | 1.11 | 0.015 | Actin filament capping/Amyloid fibril formation | ||||
| GPX3 | Glutathione peroxidase 3 | 1.26 | 0.018 | 1.41 | 0.005 | Cellular response to oxidative stress | |||||
| HPR | Haptoglobin-related | 0.61 | 0.002 | 0.64 | 0.001 | 0.0018 | Receptor-mediated endocytosis | ||||
| HPX | Hemopexin | 0.90 | 0.004 | Cellular iron ion homeostasis | |||||||
| SERPIND1 | Heparin cofactor 2 | 0.80 | 0.027 | 0.71 | 4 × 10−4 | 0.0023 | 0.77 | 0.005 | Complement/coagulation cascades | ||
| H2AFJ | Histone H2A | 0.49 | 0.027 | Chromatin silencing | |||||||
| HIST1H4A | Histone H4 | 0.35 | 0.001 | 0.43 | 0.003 | 1.2 × 10−5 | Telomere organization | ||||
| IGHV3-7 | Ig heavy chain V-III region GAL | 1.64 | 0.019 | Immune response | |||||||
| IGKV1D-12 | IgK chain V-I region Wes | 0.77 | 0.032 | Immune response | |||||||
| IGH@ | IGH@ protein | 0.71 | 0.006 | Immune response | |||||||
| IGK@ | IGK@ protein | 0.88 | 0.019 | 1.31 | 0.002 | Immune response | |||||
| IGHD | Immunoglobulin heavy constant delta | 3.39 | 0.009 | Immune response | |||||||
| IGHV3-72 | Ig heavy variable 3-72 | 1.48 | 0.025 | Immune response | |||||||
| IGJ; JCHAIN | Ig J-chain | 1.51 | 0.006 | Immune response | |||||||
| IGKV2D-24 | IgK variable | 0.70 | 0.024 | Immune response | |||||||
| IGFALS | Insulin-like growth fac-tor-binding complex acid-labile subunit | 0.59 | 0.003 | 0.58 | 0.002 | 0.0005 | Cell adhesion | ||||
| ITIH2 | Inter-α-trypsin inhibi-tor heavy chain H2 | 0.92 | 0.022 | Amine metabolic process | |||||||
| ITIH3 | Inter-α-trypsin inhibi-tor heavy chain H3 | 1.46 | 0.001 | 1.72 | 1 × 10−5 | 1.1 × 10−5 | Amine metabolic process | ||||
| ITIH4 | Inter-α-trypsin inhi-bitor heavy chain H4 | 1.12 | 0.001 | Amine metabolic process | |||||||
| N/A | cDNA FLJ53075, high-ly similar to KNG1 | 1.55 | 0.001 | ||||||||
| SERPINA4 | Kallistatin | 0.70 | 0.012 | 0.59 | <1 × 10−4 | 0.84 | 0.018 | 4.8 × 10−5 | Platelet degranulation | ||
| KNG1 | Kininogen-1 | 0.92 | 0.029 | 0.92 | 0.009 | 0.0002 | 0.86 | 0.000 | Blood coagulation/inflammatory response | ||
| LBP | Lipopolysaccharide-binding protein | 1.90 | 0.002 | Acute phase response | |||||||
| LUM | Lumican | 1.27 | 0.017 | 0.78 | 0.002 | 0.0037 | Collagen binding | ||||
| PGLYRP2 | N-acetylmuramoyl-L-alanine amidase | 0.87 | 0.001 | 0.91 | 0.023 | 0.0055 | Immune resonse/inflammatory response | ||||
| KLKB1 | Plasma kallikrein | 0.81 | 0.016 | 0.91 | 0.043 | Complement/coagulation cascades | |||||
| PLG | Plasminogen | 0.82 | 0.0002 | 0.89 | 0.004 | 1.09 | 0.039 | 1 × 10−4 | Blood coagulation, fibrynolysis | ||
| PF4 | Platelet factor 4 | 0.72 | 0.001 | 0.0264 | Platelet degranulation, inflammatory response | ||||||
| PZP | Pregnancy zone protein | 3.87 | 0.0003 | 0.38 | 0.004 | 0.0001 | Female pregnancy | ||||
| AMBP | Protein AMBP; α-1-microglobulin; Trypstatin | 0.90 | 0.005 | 0.0497 | Cell adhesion; Heme metabolic process | ||||||
| F2 | Prothrombin | 0.90 | 0.008 | 0.0142 | 0.79 | 8 × 10−6 | Complement/coagulation cascades; Neuroactive ligand-receptor interaction; Regulation of actin cytoskeleton | ||||
| RBP4 | Retinol-binding protein 4 | 0.75 | 0.010 | 0.79 | 0.012 | 0.0093 | Cardiac muscle tissue development | ||||
| SAA1 | Serum amyloid A-1 | 6.31 | 0.004 | 11.13 | <1 × 10−5 | 0.0052 | 1.97 | 0.020 | Acute-phase response | ||
| APCS | Serum amyloid P | 0.83 | 0.010 | 0.0237 | Immune response | ||||||
| HEL111;TTR | Transthyretin | 0.68 | 0.004 | 0.64 | 0.004 | 0.0060 | 0.57 | 5.4 × 10−9 | Retinol metabolic process, thyroid hormone transport | ||
| HEL-S-51;GC | Vit. D-binding protein | 0.91 | 0.020 | 0.87 | <1 × 10−3 | 0.0007 | 0.85 | 0.0001 | Vitamin D metabolic process | ||
| Cardioembolic vs. Large-Vessel Stroke | Cardioembolic vs. Lacunar Stroke | Large-Vessel vs. Lacunar Stroke | CBS−/− vs. Control |
|---|---|---|---|
| APCS | AMBP | APOL1 | APOA1 |
| APOM | APOA4 | C5 | APOC3 |
| C1QA | FCN3 | GSN | APOH |
| C4BPA | ITIH4 | GPX3 | C1R |
| CPB2 | LBP | H2AFJ | C1S |
| FBLN1 | PF4 | IGK@ | CFI |
| IGKV1D-12 | HEL0213 | ||
| KLKB1 | HPX | ||
| SERPINF2 | IGHV3-7 | ||
| IGHD | |||
| IGHV3-7 | |||
| IGH@ | |||
| IGJ; JCHAIN | |||
| IGKV2D-24 | |||
| ITIH2 | |||
| ORM2 | |||
| SERPINC1 | |||
| cDNA FLJ53075, highly similar to KNG1 |
| Analysis | Molecules in Network | Score | Focus Molecules | Top Diseases and Functions |
|---|---|---|---|---|
| Cardioembolic vs. large-vessel stroke (Figure 3A) | ↑A2M, Akt, APOH, ↓APOM, BGN, CLEC10A, ↓CLU, ↓CPB2,↑CRP, CYP19, ERK1/2, ↓F2, Fc gamma receptor, FCER1A, ↑FGA, FPR2, Growth hormone, HDL-cholesterol, ↓IGFALS, Jnk, KLKB1,↑KNG1, NFkB (complex), P38 MAPK, ↓PLG, ↓RBP4, Retnla, S100A12, ↑SAA1, SCARB1, ↓SERPIND1, ↓SERPINF2, STAB2, TAB1, ↓TTR | 36 | 16 | Hematological System Development and Function, Cell-To-Cell Signaling and Interaction, Organismal Functions |
| Cardioembolic vs. large-vessel stroke | ↓AFM,ALT,↓APCS, ↓APOC1,APOE,C6, ↑C7,C8,C9,C1q,↑C1QA, ↑C1QB, ↓C4BPA, Ccl2, ↑CRP, CRYAB, CTSB, CXCL2, ↓F13B,↑FBLN1, ↑FGA,↓GC, HNF1A, HNF4A, HRG, IL1R1, INSR, ↑ITIH3, LDL-cholesterol, ↑LUM, PSEN2, SCARB1, ↑SERPINA3, STK40, VLDL-cholesterol | 33 | 15 | Metabolic Disease, Developmental Disorder, Hereditary Disorder |
| Cardioembolic vs. lacunar stroke (Figure 3B) | ↑A2M, ↓AFM, ↓AHSG, Akt, ↓AMBP, ↓APOA4, ↓CFH,CFHR5, ↓CLU, ↑CP, ↑CRP, ERK1/2, ↑F9, ↓F13B, Fibrinogen, Growth hormone, HDL-cholesterol, ↓IGFALS, IgG, IL10RB, Jnk, ↑KNG1, ↑LBP, NFkB (complex), P38 MAPK, ↓PF4, ↓PLG, ↓RBP4,S100A12, ↑SAA1, ↑SERPINA1, ↑SERPINA3,STAT5a/b, TCF, ↓TTR | 50 | 21 | Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Neurological Disease |
| Cardioembolic vs. lacunar stroke | ↓AHSG, ↓APOC1, ARG1, C5, C6,↑C7, C8,↑C9, CCND1, CPB2, ↑CRP, CTSB, EHF, ↓FCN3, MASP1, MASP2, Fc-γ receptor, FGA, FGB, FGG, ↓GC, ↓HIST1H4H, HNF1A, HNF4A, IL6, ↑ITIH3, ↑ITIH4, LDL-cholesterol, LGALS3, N-cor, NR1H2, NR5A2, ↑ORM1, PGLYRP2, PRELP, ↑SAA1, ↓SERPIND1, TTR | 32 | 15 | Inflammatory Response, Metabolic Disease, Cell-To-Cell Signaling and Interaction |
| Large-vessel vs. lacunar stroke (Figure 3C) | ↓AHSG, Akt, APOH, ↑APOL1, ↑C5, C6, C8, ↑C9, ↓C1QB, CFH, ↑CFI, ↑CRP, ERK1/2, Fc-γ receptor, FCER1A, ↓GSN, HABP2, IL1, IL6R, INPP5D, LTB, miR-146a-5p (and other miRNAs w/seed GAGAACU), NFkB (complex), OLR1, P38 MAPK, ↓PGLYRP2, ↑PLG, PPARA, PRELP, PRKCB, PRKCG, PSMB9, S100A12, ↑SERPINA1, STAB2 | 24 | 11 | Inflammatory Response, Cellular Movement, Immune Cell Trafficking |
| Large-vessel vs. lacunar stroke | AGER, ↓APOC1, CCND1, CDK4, ↑CP, CRYAB, EHF, F2, ↑F9, Fc-γ receptor, FCER1A, Ferritin, ↑FGA, FGB, FGG, ↑GPX3, ↓H2AFJ, ↓HIST1H4H, INPP5D, ↑KNG1, Ldh (complex), LRPAP1, ↓LUM, MAPK1,N-cor, NR5A2, Nuclear factor 1, ↑ORM1,PLCG2, PSMB9, PSME2, ↑SERPINA3, TNF, TNFSF10, TP53 | 24 | 11 | Cell-To-Cell Signaling and Interaction, Hematological System Development and Function, Inflammatory Response |
| CBS−/− vs. control (Figure 3D) | ↓AFM, ↓AHSG, Akt, ↓APOA1,↑APOC1,↓APOC3, ↓APOH, ↓APOM, ↓C1R, ↓C1S, ↓CLU,↑CPB2,ERK1/2,↓F2, ↓F13B, Fibrinogen, ↑GPX3, Growth hormone, ↑GSN, HDL, HDL-cholesterol, ↓HPX, IgG, ↑JCHAIN, ↓KLKB1, ↑KNG1, LDL, NFkB (complex), P38 MAPK, ↑SAA1, ↑SERPINA1, ↓SERPINC1, ↓SERPIND1, ↓TTR, VLDL-cholesterol | 60 | 24 | Metabolic Disease, Hematological System Development and Function, Lipid Metabolism |
| CBS−/− vs. control | ADAMTS1, ↓AHSG,ALT, ↓APOC3, BGN, BIRC5, C6, C7,↑C9, ↓C1S,CCND1, ↓CFI, ↑CPB2, ↓CPN1, F13A1, ↑FBLN1, ↓FCN3, FGA, FPR2, ↓GC, GCNT3, GPR119, HNF1A, ↓IGHG1, IL1B, ↓ITIH2, MASP1, MASP2, ↓ORM2, PPARA, ↑SAA1, ↓SERPIND1, ↓SERPINF2, TG, TNF | 35 | 16 | Humoral Immune Response, Inflammato-ry Response, Develop-mental Disorder |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sikora, M.; Lewandowska, I.; Kupc, M.; Kubalska, J.; Graban, A.; Marczak, Ł.; Kaźmierski, R.; Jakubowski, H. Serum Proteome Alterations in Human Cystathionine ?-Synthase Deficiency and Ischemic Stroke Subtypes. Int. J. Mol. Sci. 2019, 20, 3096. https://doi.org/10.3390/ijms20123096
Sikora M, Lewandowska I, Kupc M, Kubalska J, Graban A, Marczak Ł, Kaźmierski R, Jakubowski H. Serum Proteome Alterations in Human Cystathionine ?-Synthase Deficiency and Ischemic Stroke Subtypes. International Journal of Molecular Sciences. 2019; 20(12):3096. https://doi.org/10.3390/ijms20123096
Chicago/Turabian StyleSikora, Marta, Izabela Lewandowska, Małgorzata Kupc, Jolanta Kubalska, Ałła Graban, Łukasz Marczak, Radosław Kaźmierski, and Hieronim Jakubowski. 2019. "Serum Proteome Alterations in Human Cystathionine ?-Synthase Deficiency and Ischemic Stroke Subtypes" International Journal of Molecular Sciences 20, no. 12: 3096. https://doi.org/10.3390/ijms20123096
APA StyleSikora, M., Lewandowska, I., Kupc, M., Kubalska, J., Graban, A., Marczak, Ł., Kaźmierski, R., & Jakubowski, H. (2019). Serum Proteome Alterations in Human Cystathionine ?-Synthase Deficiency and Ischemic Stroke Subtypes. International Journal of Molecular Sciences, 20(12), 3096. https://doi.org/10.3390/ijms20123096