Genetic Aberrations Associated with Photodynamic Therapy in Colorectal Cancer Cells
Abstract
1. Introduction
2. Results
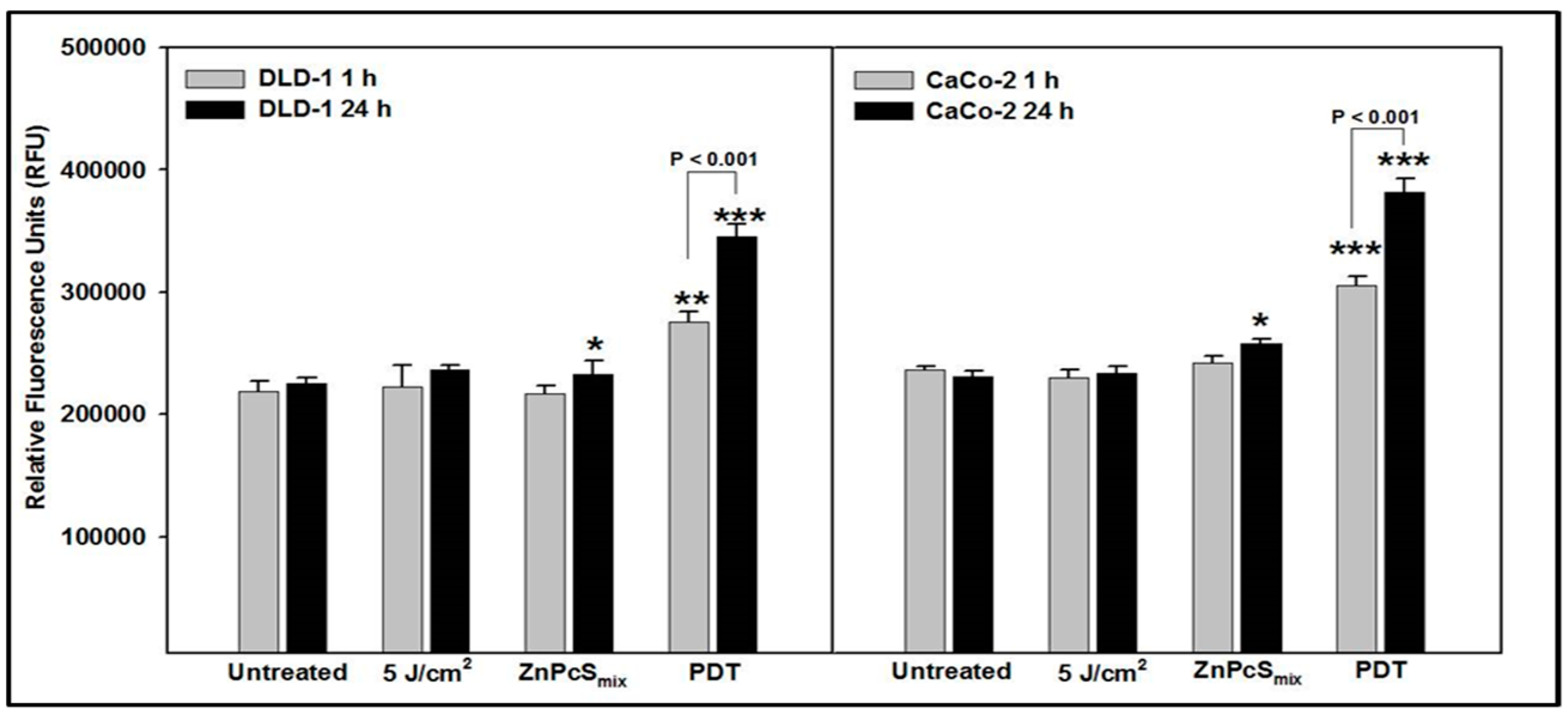
2.1. Cytosolic Acidification
2.2. Mitochondrial Membrane Potential
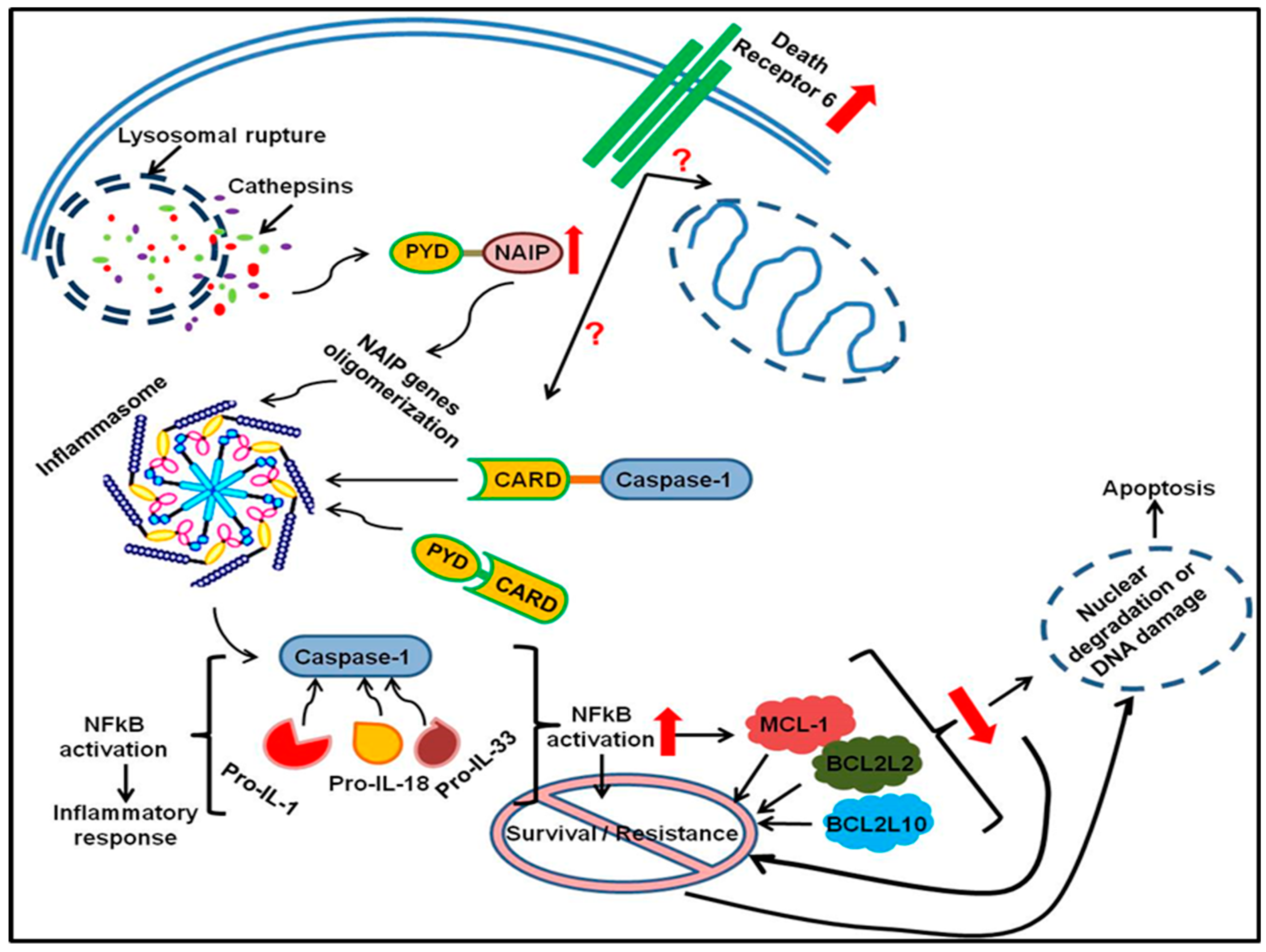
2.3. Cell Death Pathway (Apoptosis Array)
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. PDT Experiments
4.3. Cytosolic Acidification—Hydrogen Peroxide (H2O2) Production
4.4. Mitochondrial Destabilization
4.5. Expression of Apoptotic Genes
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2014, 383, 1490–15023. [Google Scholar] [CrossRef]
- Rodriguez, M.E.; Zhang, P.; Azizuddin, K.; Santos, G.B.D.; Chiu, S.-M.; Xue, L.-Y.; Berlin, J.C.; Peng, X.; Wu, H.; Lam, H.; et al. Structural Factors and Mechanisms Underlying the Improved Photodynamic Cell Killing with Silicon Phthalocyanine Photosensitizers Directed to Lysosomes Versus Mitochondria. Photochem. Photobiol. 2009, 85, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Oleinick, N.L.; Morris, R.L.; Belichenko, I. The Role of Apoptosis in Response to Photodynamic Therapy: What. Where and How. Photochem. Photobiol. 2002, 1, 1–21. [Google Scholar]
- Kessel, D.; Luo, Y. Mitochondrial Photodamage and PDT-Induced Apoptosis. J. Photochem. Photobiol. B Biol. 1998, 42, 89–95. [Google Scholar] [CrossRef]
- Wiseman, H.; Halliwell, B. Damage to DNA by Reactive Oxygen and Nitrogen Species: Role in Inflammatory Disease and Progression to Cancer. J. Biochem. 1996, 313, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, M.; Akashi, M. Catalase Regulates Cell Growth in HL60 Human Promyelocytic Cells: Evidence for Growth Regulation by H2O2. Radiat. Res. 2005, 163, 271–282. [Google Scholar] [CrossRef]
- Repnik, U.; Stoka, V.; Turk, V.; Turk, B. Lysosomes and Lysosomal Cathepsins in Cell Death. Biochem. Biophys. Acta 2012, 1824, 22–33. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Z.; Xing, D. Cell Death via Mitochondrial Apoptotic Pathway due to Activation of Bax by Lysosomal Damage. Free Radic. Biol. Med. 2012, 51, 53–68. [Google Scholar] [CrossRef]
- Ahmad, K.A.; Iskandar, K.B.; Hirpara, J.L.; Clement, M.V.; Pervaiz, S. Hydrogen Peroxide-Mediated Cytosolic Acidification is a Signal for Mitichondrial Translocation of Bax during Drug-Induced Apoptosis of Tumour Cells. Cancer Res. 2004, 64, 7867–7878. [Google Scholar] [CrossRef]
- Gupta, S.; Kass, G.E.N.; Szegezdi, E.; Joseph, B. The Mitochondrial Death Pathway: A Promisisng Therapeutic Target in Disease. J. Cell. Mol. Med. 2009, 13, 1004–1033. [Google Scholar] [CrossRef]
- Boya, P.; Gonzalez-Polo, R.A.; Poncet, D.; Andreau, K.; Vieira, H.L.; Roumier, T.; Perfettini, J.L.; Kroemer, G. Mitochondrial Membrane Permeabilization is a Critical Step of Lysosome-initiated Apoptosis Induced by Hydroxychloroquine. Oncogene 2003, 22, 3927–3936. [Google Scholar] [CrossRef] [PubMed]
- Manoto, S.L.; Sekhejane, P.R.; Houreld, N.N.; Abrahamse, H. Localization and Phototoxic Effect of Zinc Sulfophthalocyanine in Human Colon (DLD-1) and Lung (A549) Carcinoma Cells (in vitro). Photodiagnosis Photodyn. Ther. 2012, 9, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Vaux, D.L.; Strasser, A. The Molecular Biology of Apoptosis. Proc. Natl. Acad. Sci. USA 1996, 93, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Sekhejane, P.R.; Houreld, N.N.; Abrahamse, H. Multiorganelle localization of metallated phthalocyanine photosensitizer in colorectal cancer cells (DLD-1 and CaCo-2) enhances efficacy of photodynamic therapy. Int. J. Photoenergy 2014. [Google Scholar] [CrossRef]
- Persson, H.L. Iron-dependent Lysosomal Destabilization Initiates Silica-induced Apoptosis in Murine Macrophages. Toxicol. Lett. 2005, 159, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.-C.; Appleqvist, H.; Nilsson, C.; Kagedal, K.; Roberg, K.; Ollinger, K. Regulation of Apoptosis-Associated Lysosomal Membrane. Permebilization. Apoptosis 2010, 15, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Wawrzynska, M.; Kalas, W.; Bialy, D.; Ziolo, E.; Arkowski, J.; Mazurek, W.; Strzadala, L. In vitro Photodynamic Therapy with Chlorin e6 Leads to Apoptosis of Human Vascular Smooth Muscle Cells. Arch. Immunol. Ther. Exp. 2010, 58, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.A.; Masud, A.; Kuida, K.; Porter, G.A.; Booth, C.J.; Mehal, W.Z.; Inayat, I.; Flavell, R.A. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science 2006, 311, 847–851. [Google Scholar] [CrossRef]
- Kirkegaard, T.; Jaattela, M. Lysosomal Involvement in Cell Death and Cancer. Biochim. Biophys. Acta 2009, 1973, 746–754. [Google Scholar] [CrossRef]
- Karimpour, S.; Davoodi, J.; Ghahremani, M.-H. Integrity of ATP Binding Sites is Essential for Effective Inhibition of the Extrinsic Apoptosis Pathway by NAIP. Biochem. Biophys. Res. Commun. 2011, 407, 158–162. [Google Scholar] [CrossRef]
- Kumar, S. Caspase Function in Programmed Cell Death. Cell Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Bueg, S.T.; Cheung, H.H.; LaCasse, E.C.; Korneluk, R.G. Modulation of Immune Signalling by Inhibitors of Apoptosis. Trends Immunol. 2012, 33, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Miao, E.A.; Rajan, J.V.; Aderem, A. Caspase-1 Induced Pyroptotic Cell Death. Immunol. Rev. 2012, 243, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.L.; Cookson, B.T. Apoptosis, Pyroptosis and Necrosis: Mechanistic Description of Dead and Dying Eukaryotic Cells. Infect. Immun. 2005, 73, 1907–1916. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Diez, D.; Miranda-Saavedra, D. The IL-10/STAT3-Mediated Anti-Inflammatory and Future Challenges. Brief. Funct. Genom. 2013. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, M.; Rosenthal, R.E.; Mikolajczyk, J.; Stennicke, H.R.; Wiesenthal, T.; Mai, J.; Naito, M.; Salvesen, G.S.; Reed, J.C.; Fiskum, G.; et al. Early processing of Bid and caspase-6, -8, -10, -14 in the canine brain during cardiac arrest and resuscitation. Exp. Neurol. 2004, 189, 261–279. [Google Scholar] [CrossRef]
- Chiu, H.H.L.; Yong, T.M.K.; Wang, J.; Wang, Y.; Vessella, R.L.; Ueda, T.; Wang, Y.-Z.; Sadar, M.D. Induction of Neuronal Apoptosis Inhibitory Protein Expression in Response to Androgen Deprivation in Prostate Cancer. Cancer Lett. 2010, 292, 176–185. [Google Scholar] [CrossRef][Green Version]
- Guar, U.; Aggarwal, B.B. Regulation of Proliferation, Survival and Apoptosis by Members of the TNF Superfamily. Biochem. Pharmacol. 2003, 66, 1403–1408. [Google Scholar] [CrossRef]
- Zeng, L.; Li, T.; Xu, D.C.; Liu, J.; Mao, G.; Cui, M.-Z.; Fu, X.; Xu, X. Death Receptor 6 Induces Apoptosis Not Through Type I or Type II pathways, but via a Unique Mitochondria-Dependent Pathway by Interacting with Bax Protein. J. Biochem. Chem. 2012, 287, 29125–29133. [Google Scholar] [CrossRef]
- Kasof, G.M.; Lu, J.J.; Liu, D.; Speer, B.; Mongan, K.N.; Gomes, B.C.; Lorenzi, M.V. Tumour Necrosis Factor-α Induces the Expression of DR6, a Member of the the TNF Receptor Family, Through Activation of NF-kB. Oncogene 2001, 20, 7965–7975. [Google Scholar] [CrossRef]
- Mahmood, Z.; Shukla, Y. Death Receptors: Targets for Cancer Therapy. Exp. Cell Res. 2010, 316, 877–899. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Bauer, J.H.; Haridas, V.; Wang, S.; Liu, D.; Yu, G.; Vincenz, C.; Aggarwal, B.B.; Ni, J.; Dixit, V.M. Identification and Functional Characterization of DR6, a Novel Death Domain Containing TNF Receptor. FEBS Lett. 1998, 431, 351–356. [Google Scholar] [CrossRef]
- Miyake, K.; Bekisz, J.; Zhao, T.; Clark, C.; Zoon, K.C. Apoptosis-Inducing Factor (AIF) is Targeted in IFN-α2a-Induced Bid Mediated Apoptosis Through Bak Activation in Ovarian Cancer Cells. Biochim. Biophys. Acta 2012, 1823, 1378–1388. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, H.; Zhu, H.; Xu, C.J.; Yuan, J. Cleavage of BID by Caspase 8 Mediates the Mitochondrial Damage in the Fas Pathway of Apoptosis. Cell 1998, 94, 491–501. [Google Scholar] [CrossRef]
- Wang, G.Q.; Gastman, B.R.; Wieckowski, E.; Goldstein, L.A.; Gambotto, A.; Kim, T.H.; Fang, B.; Rabinovitz, A.; Yin, X.M.; Rabinowich, H. A role for Mitochondrial Bak in Apoptotic Response to Anticancer Drugs. J. Biol. Chem. 2001, 276, 34307–34317. [Google Scholar] [CrossRef]
- Beverly, L.J.; Howell, L.A.; Hernandez-Corbacho, M.; Casson, L.; Chipuk, J.E.; Siskind, L.J. BAK Activation is Necessary and Sufficient to Drive Ceramide Synthase-Dependent Ceramide Accumulation Following Inhibition of BCL2-Like Proteins. Biochem. J. 2013, 452, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Beverly, L.J. Regulation of AntiApoptotic BCL2-Proteins by Non-Canonical Interactions: The Next Step Forward or Two Steps back? J. Cell. Biochem. 2012, 113, 3–12. [Google Scholar] [CrossRef]
- Sorenson, C.M. BCL2Family Members and Disease. Biochim. Biophys. Acta 2004, 1644, 169–177. [Google Scholar] [CrossRef]
- Merino, D.; Khaw, S.L.; Glaser, S.P.; Anderson, D.J.; Belmont, L.D.; Wong, C.; Yue, P.; Robati, M.; Phipson, B.; Fairlie, W.D.; et al. Bcl-2, Bcl-x(L), and Bcl-w are not Equivalent Targets of ABT-737 and Navitoclax (ABT-263) in Lymphoid and Leukemic Cells. Blood 2012, 119, 5807–5816. [Google Scholar] [CrossRef]
- Borst, J.; Hendriks, J.; Xiao, Y. CD27 and CD70 in T-Cell and B-Cell Activation. Curr. Opin. Immunol. 2005, 17, 275–281. [Google Scholar] [CrossRef]
- Gravestein, L.A.; Amsen, D.; Boes, M.; Revilla Calvo, C.; Kruisbeek, A.M.; Borst, J. The TNF Receptor Family Member CD27 Signals to Jun N-terminal Kinase via Traf-2. Eur. J. Immunol. 1998, 28, 2208–2216. [Google Scholar] [CrossRef]
- Aggarwal, B.B. Signalling Pathways of the TNF Superfamily: A Double-Edged Sword. Natl. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Wang, W.; Wallach, D. Receptor-Specific Signaling for both the Alternative and the Canonical NF-kB Activation Pathways by NF-kB-Inducing Kinase. Immunity 2004, 21, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Cohen, G.M. Caspases: The Executioners of Apoptosis. Biochem. J. 1997, 326, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. FAS and FASLG Polymorphism and Susceptibility to Idiopathic Azoospermia or Sever Oligozoospermia. Reprod. Biomed. Online 2009, 18, 141–147. [Google Scholar] [CrossRef]
- Liu, Y.; Wen, Q.-J.; Yin, Y.; Lu, X.-T.; Pu, S.-H.; Tian, H.-P.; Lou, Y.-F.; Tang, Y.-N.; Jiang, X.; Lu, G.-S.; et al. FASLG Polymorphism is Associated with Cancer Risk. Eur. J. Cancer 2009, 45, 2574–2578. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hao, Y.L.; Zhou, R.M.; Wang, N.; Qi, B.L. Genetic Polymorphism in the Fas and FasL Genes are Associated with Epithelial Ovarian Cancer Risk and Clinical Outcomes. Gynecol. Oncol. 2013, 128, 584–589. [Google Scholar] [CrossRef]
- Karimi, M.Y.; Kapoor, V.; Sharma, S.C.; Das, S.N. Genetic Polymorphisms in FAS (CD95) and FAS Ligand (CD178) Promoters and Risk of Tobacco-related Oral Carcinoma: Gene-Gene Interactions in High-Risk Indians. Cancer Investig. 2013, 31, 1–6. [Google Scholar] [CrossRef]
- Kornbluth, R.S. An Expanding Role for CD40L and Other Tumour Necrosis Factor Superfamily Ligands in HIV Infection. J. Hematother. Stem Cell Res. 2002, 11, 787–801. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.R.; Shurin, G.V.; Barry, D.A.; Schuh, A.C.; Shurin, M.R.; Robbins, P.D. Comparative Analysis of Antitumor Activity of CD40L, RANKL, and 4-1BBL in vivo Following Intratumoral Administration of Viral Vectors or Transduced Dendritic Cells. J. Gene Med. 2005, 8, 129–137. [Google Scholar] [CrossRef]
- Kwon, B. Regulation of Inflammation by Bidirectional Signaling Through CD137 and its Ligand. Immune Netw. 2012, 12, 176–180. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chakrabarty, S.; Nagata, M.; Yasuda, H.; Wen, L.; Nakayama, M.; Chowdhury, S.A.; Yamada, K.; Jin, Z.; Kotani, R.; Moriyama, H.; et al. Critical Roles of CD30/CD30L Interations in Murine Autoimmune Diabetes. Clin. Exp. Immunol. 2003, 133, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamkhani, A. The Role of CD30 in the Pathogenesis of Haematopoietic Malignancies. Curr. Opin. Pharmacol. 2004, 4, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Srinivasula, S.M.; Wu, G.; Fernandes-Alnemri, T.; Alnemri, E.S.; Shi, Y. Structural Basis of Pro-Caspase 9 Recruitment by the Apoptoti Prorease-Activating Factor 1. Nature 1999, 399, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, A.S.; Proell, M.; Eibl, C.; Page, R.; Schwarzenbacher, R.; Peti, W. Three-Dimensional Structure of NLRP7 Pyrin Domain. J. Biol. Chem. 2010, 258, 27402–27410. [Google Scholar] [CrossRef] [PubMed]
- Reubold, T.F.; Wohlgemuth, S.; Eschenburg, S. Crystal Structure of Full-Length Apaf-1: How the Death Signal is Relayed in the Mitochondrial Pathway of Apoptosis. Structure 2011, 19, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Hwang, I.; Lee, Y.-S.; Park, S.; Lee, W.-K.; Fernandes-Alnemri, T.; Alnemri, E.S.; Kim, Y.-S.; Yu, J.-W. Restoration of ASC Expression Sensitizes Colorectal Cancer Cells to Genotoxic Stress-induced Caspase-independent Cell Death. Cancer Lett. 2013, 331, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Tvrdik, D.; Skalova, H.; Dundr, P.; Povysil, C.; Velenska, Z.; Berkova, A.; Stanek, L.; Petruzelka, L. Apoptosis—Associated Genes and their Role in Predicting Responses to Neoadjuvant Breast Cancer Treatment. Med Sci. Monit. 2012, 18, BR60–BR67. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The Biochemistry of Apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef]
- Chipuk, J.E.; Green, D.R. How do BCL-2 Proteins Induce Mitochondrial Outer Membrane Permeabilization? Trends Cell Biol. 2008, 18, 157–164. [Google Scholar] [CrossRef]
- Kaufmann, T.; Strasser, A.; Jost, P.J. Fas Death Receptor Signalling: Roles of Bid and XIAP. Cell Death Differ. 2012, 19, 42–50. [Google Scholar] [CrossRef]
- Lamers, F.; van der Ploeg, I.; Schild, L.; Ebus, M.E.; Koster, J.; Hansen, B.R.; Koch, T.; Versteeg, R.; Caron, H.B.; Molenaar, J.J. Knockdown of Survivin (BIRC5) Causes Apoptosis in Neuroblastoma via Mitotic Catastrophe. Endocr. Relat. Cancer 2011, 18, 657–688. [Google Scholar] [CrossRef]
- Real, P.J.; Sanz, C.; Gutierrez, O.; Pipaon, C.; Zubiaga, A.M.; Fernandez-Luna, J.L. Transcriptional Activation of the Proapoptotic Bik Gene by E2F Proteins in Cancer Cells. FEBS Lett. 2006, 580, 5905–5909. [Google Scholar] [CrossRef]
- Ogunsipe, A.; Nyokong, T. Photophysical and photochemical studies of sulphonated non-transition metal phthalocyanines in aqueous and non-aqueous media. J. Photochem. Photobiol. A 2005, 173, 211–220. [Google Scholar] [CrossRef]
- Ayuk, S.M.; Houreld, N.N.; Abrahamse, H. Laser irradiation alters the expression profile of genes involved in the extracellular matrix in vitro. Int. J. Photoenergy 2014. [Google Scholar] [CrossRef]



| Gene Symbol | Gene Name | p Value | Fold-Change 2(−∆∆Ct) | Up (↑)/Down (↓) Regulation | Function |
|---|---|---|---|---|---|
| ABL1 | ABL proto-oncogene 1, non-receptor tyrosine kinase | <0.05 | 0.29 | ↓ | Positive Regulation of Apoptosis |
| BAK1 | BCL2 antagonist/killer 1 | <0.05 | 0.11 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| BCL2L10 | BCL2 like 10 | <0.001 | 0.19 | ↓ | Anti-Apoptotic |
| BCL2L2 | BCL2 like 2 | <0.05 | 0.07 | ↓ | Anti-Apoptotic |
| BID | BH3 interacting domain death agonist | <0.05 | 0.29 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| CASP14 | Caspase 14 | <0.001 | 0.19 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase |
| CD27 | CD27 molecule | <0.001 | 0.22 | ↓ | Pro-Apoptotic/Anti-Apoptotic/Negative Regulation of Apoptosis/Caspase inhibitor |
| CD40 | CD40 molecule | <0.001 | 0.22 | ↓ | Positive Regulation of Apoptosis |
| CD40LG | CD40 ligand | <0.05 | 0.25 | ↓ | Anti-Apoptotic |
| CD70 | CD70 molecule | <0.01 | 0.28 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| CIDEA | Cell death inducing DFFA like effector a | <0.01 | 0.17 | ↓ | Negative Regulation of Apoptosis/DNA Damage & Repair |
| IL10 | Interleukin 10 | <0.05 | 0.24 | ↓ | Anti-Apoptotic |
| LTBR | Lymphotoxin beta receptor | <0.001 | 0.10 | ↓ | Positive Regulation of Apoptosis |
| MCL1 | MCL1 apoptosis regulator, BCL2 family member | <0.01 | 0.10 | ↓ | Negative Regulation of Apoptosis |
| NAIP | NLR family apoptosis inhibitory protein | <0.001 | 5.69 | ↑ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| NFkB1 | Nuclear factor kappa B subunit 1 | <0.01 | 3.09 | ↑ | Anti-Apoptotic |
| PYCARD | PYD and CARD domain containing | <0.01 | 0.28 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase Regulator |
| RIPK2 | Receptor interacting serine/threonine kinase 2 | <0.05 | 0.23 | ↓ | Anti-Apoptotic/Positive Regulation of Apoptosis |
| TNF | Tumor necrosis factor | <0.05 | 0.70 | ↓ | Death Domain Receptor/Anti-Apoptotic/Positive Regulation of Apoptosis |
| TNFRSF21 | TNF receptor superfamily member 21 | <0.01 | 284.18 | ↑ | Death Domain Receptor |
| TNFSF8 | TNF superfamily member 8 | <0.001 | 0.18 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| TP73 | Tumor protein p73 | <0.01 | 0.20 | ↓ | DNA Damage & Repair/Negative Regulation of Apoptosis |
| TRADD | TNFRSF1A associated via death domain | <0.05 | 0.09 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Death Domain Receptor |
| Gene Symbol | Gene Name | p Value | Fold-Change 2(−∆∆Ct) | Up (↑)/Down (↓) Regulation | Function |
|---|---|---|---|---|---|
| AKT1 | AKT serine/threonine kinase 1 | <0.001 | 0.11 | ↓ | Anti-Apoptotic/Positive Regulation of Apoptosis |
| APAF1 | Apoptotic peptidase activating factor 1 | <0.01 | 0.21 | ↓ | Caspase Activator |
| BAD | BCL2 associated agonist of cell death | <0.01 | 0.29 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| BAK1 | BCL2 antagonist/killer 1 | <0.05 | 3.07 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| BAX | BCL2 associated X, apoptosis regulator | <0.001 | 0.24 | ↓ | Pro-Apoptotic/Anti-Apoptotic/Positive Regulation of Apoptosis/Caspase Activator |
| BCL2 | BCL2 apoptosis regulator | <0.001 | 0.22 | ↓ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| BCL2L1 | BCL2 like 1 | <0.01 | 0.19 | ↓ | Anti-Apoptotic |
| BCL2L10 | BCL2 like 10 | <0.05 | 2.27 | ↑ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| BCL2L11 | BCL2 like 11 | <0.05 | 2.27 | ↑ | Pro-Apoptotic |
| BIK | BCL2 interacting killer | <0.05 | 2.27 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| BIRC5 | Baculoviral IAP repeat containing 5 | <0.01 | 0.23 | ↓ | Anti-Apoptotic |
| BNIP2 | BCL2 interacting protein 2 | <0.05 | 0.38 | ↓ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| CASP10 | Caspase 10 | <0.05 | 2.27 | ↑ | Positive Regulation of Apoptosis/Caspase |
| CASP14 | Caspase 14 | <0.05 | 2.27 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase |
| CASP2 | Caspase 2 | <0.001 | 0.19 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase |
| CASP6 | Caspase 6 | <0.001 | 0.53 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase |
| CASP8 | Caspase 8 | <0.01 | 0.34 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase |
| CD27 | CD27 molecule | <0.05 | 1.99 | ↑ | Pro-Apoptotic/Anti-Apoptotic/Negative Regulation of Apoptosis/Caspase inhibitor |
| CD40LG | CD40 ligand | <0.01 | 0.40 | ↓ | Anti-Apoptotic |
| CIDEA | Cell death inducing DFFA like effector a | <0.05 | 0.20 | ↓ | Negative Regulation of Apoptosis/DNA Damage & Repair |
| CYCS | Cytochrome c, somatic | <0.01 | 0.34 | ↓ | Pro-Apoptotic |
| DIABLO | Diablo IAP-binding mitochondrial protein | <0.001 | 7.82 | ↑ | Pro-Apoptotic |
| FADD | Fas associated via death domain | <0.05 | 0.18 | ↓ | Positive Regulation of Apoptosis/Death Domain |
| FASLG | Fas ligand | <0.01 | 0.23 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| GADD45A | Growth arrest and DNA damage inducible alpha | <0.03 | 0.58 | ↓ | Pro-Apoptotic |
| IGF1R | Insulin like growth factor 1 receptor | <0.001 | 0.13 | ↓ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| LTBR | Lymphotoxin beta receptor | <0.01 | 0.42 | ↓ | Positive Regulation of Apoptosis |
| MCL1 | MCL1 apoptosis regulator, BCL2 family member | <0.01 | 2.27 | ↑ | Negative Regulation of Apoptosis |
| NAIP | NLR family apoptosis inhibitory protein | <0.05 | 2.27 | ↑ | Anti-Apoptotic/Negative Regulation of Apoptosis |
| PYCARD | PYD and CARD domain containing | <0.05 | 2.73 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis/Caspase Regulator |
| RIPK2 | Receptor interacting serine/threonine kinase 2 | <0.01 | 2.28 | ↑ | Anti-Apoptotic/Positive Regulation of Apoptosis |
| TNF | Tumor necrosis factor | <0.05 | 2.27 | ↑ | Death Domain Receptor/Anti-Apoptotic/Positive Regulation of Apoptosis |
| TNFRSF10A | TNF receptor superfamily member 10a | <0.05 | 2.06 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis/Death Domain Receptor |
| TNFRSF9 | TNF receptor superfamily member 9 | <0.05 | 2.27 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| TNFSF8 | TNF superfamily member 8 | <0.05 | 2.27 | ↑ | Pro-Apoptotic/Positive Regulation of Apoptosis |
| TP53 | Tumor protein p53 | <0.01 | 0.38 | ↓ | DNA Damage & Repair/Positive Regulation of Apoptosis/Caspase Activator |
| TP73 | Tumor protein p73 | <0.05 | 0.22 | ↓ | DNA Damage & Repair/Negative Regulation of Apoptosis |
| TRADD | TNFRSF1A associated via death domain | <0.05 | 0.32 | ↓ | Pro-Apoptotic/Positive Regulation of Apoptosis/Death Domain Receptor |
| Functional Gene Grouping | Genes |
|---|---|
| Death Domain Receptors | CRADD, FADD, TNF, TNFRSF10B (DR5). |
| DNA Damage & Repair | ANL1, CIDEA, CIDEB, TP53, TP73. |
| Extracellular Apoptotic Signals | CFLAR (CASPER), DAPK1, TNFRSF25 (DR3). |
| Other Pro-Apoptotic Genes | BAD, BAK1, BAX, BCL10, BCL2L11, BID, BIK, BNIP3, BNIP3L, CASP1 (ICE), CSP10 (MCH4), CASP14, CASP2, CASP3, CASP4, CASP6, CASP8, CD27 (TNFRSF7), CD70 (TNFSF7), CYCS, DFFA, DIABLO (SMAC), FAS (TNFRSF6), FASLG (TNFSF6), GADD45A, HRK, LTA (TNFB), NOD1 (CARD4), PYCARD (TMS1/ASC), TNFRSF10A, TNFRSF9, TNFSF10 (TRAIL), TNFSF8. TP53BP2, TRADD, TRAF3. |
| Anti-Apoptotic | AKT1, BAG1, BAG3, BAX, BCL2, BCL2A1 (Bfl-1/A1), BCL2L1 (BCL-X), BCL2L10, BCL2L2, BFAR, BIRC3 (cIAP1), BIRC5, BIRC6, BNIP2, BNIP3, BNIP3L, BRAF, CD27 (TNFRSF7), CD40LG (TNFSF5), CFLAR (CASPER, DAPK1, FAS (TNFRSF6, HRK, IGF1R, IL10, MCK1, NAIP (BIRC1), NFKB1, NOL3, RIPK2, TNF, XIAP (BIRC4). |
| Regulation of Apoptosis | Negative regulation:BAG1, BAG3, BCL10, BCL2, BCL2A1 (Bfl-1/A1), BCL2L1 (BCL-X), BCL2L10, BCL2L2, BFAR, BIRC2 (c-IAP2), BIRC3 (c-IAP1), BIRC6, BNIP2, BNIP3, BNIP3L, BRAF, CASP3, CD27 (TNFRSF7), CD40LG (TNFSF5), CFLAR (CASPER), CIDEA, DAPK1, DFFA, FAS (TNFRSF6), IGF1R, MCL1, NAIP (BIRC1), NOL3, TP53, TP73, XIAP (BIRC4). Positive regulation: ABL1, AKT1, BAD, BAK1, BAX, BCL2L11, BID, BIK, BNIP3, BNIP3L, CASP1 (ICE), CASP10 (MCH4), CASP14, CASP2, CASP4, CASP6, CASP8, CD40 (TNFRSF5), CD70 (TNFSF7), CIDEB, CRADD, FADD, FASLG (TNFSF6), HRK, LTA (TNFB), LTBR, NOD1 (CARD4), PYCARD (TMSS1/ASC), RIPK2, TNF, TNFRSF10A, TNFRSF10B (DR5), TNFRSF25 (DR3), TNFRSF9, TNFSF10 (TRAIL), TNFSF8, TP53, TP53BP2, TRADD, TRAF2, TRAF3. |
| Death Domain Receptors | CRADD, DAPK1, FADD, TNFRSF10A, TNFRSF10B (DR5), TNFRSF11B, TNFRSF1A, TNFRSF1B, TNFRSF21, TNFRSF25 (DR3), TRADD. |
| Caspases and Regulators | Caspases:CASP1 (ICE), CASP10 (MCH4), CASP14, CASP2, CASP3, CASP4, CASP5, CASP6, CASP7, CASP8, CASP9, CFLAR (CASPER), CRADD, PYCARD (TMS1/ASC). Caspase activators: AIFM1 (PDCD8), APAF1, BAX, BCL2L10, CASP1 (ICE), CASP9, NOD (CARD4), PYCARD (TMS1/ASC), TNFRSF10A, TNFRSF10B (DR5), TP53. Caspase inhibitors: CD27 (TNFRSF7), XIAP (BIRC4). |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abrahamse, H.; Houreld, N.N. Genetic Aberrations Associated with Photodynamic Therapy in Colorectal Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3254. https://doi.org/10.3390/ijms20133254
Abrahamse H, Houreld NN. Genetic Aberrations Associated with Photodynamic Therapy in Colorectal Cancer Cells. International Journal of Molecular Sciences. 2019; 20(13):3254. https://doi.org/10.3390/ijms20133254
Chicago/Turabian StyleAbrahamse, Heidi, and Nicolette Nadene Houreld. 2019. "Genetic Aberrations Associated with Photodynamic Therapy in Colorectal Cancer Cells" International Journal of Molecular Sciences 20, no. 13: 3254. https://doi.org/10.3390/ijms20133254
APA StyleAbrahamse, H., & Houreld, N. N. (2019). Genetic Aberrations Associated with Photodynamic Therapy in Colorectal Cancer Cells. International Journal of Molecular Sciences, 20(13), 3254. https://doi.org/10.3390/ijms20133254







