Anti-Bacterial Effect of CpG-DNA Involves Enhancement of the Complement Systems
Abstract
:1. Introduction
2. Results
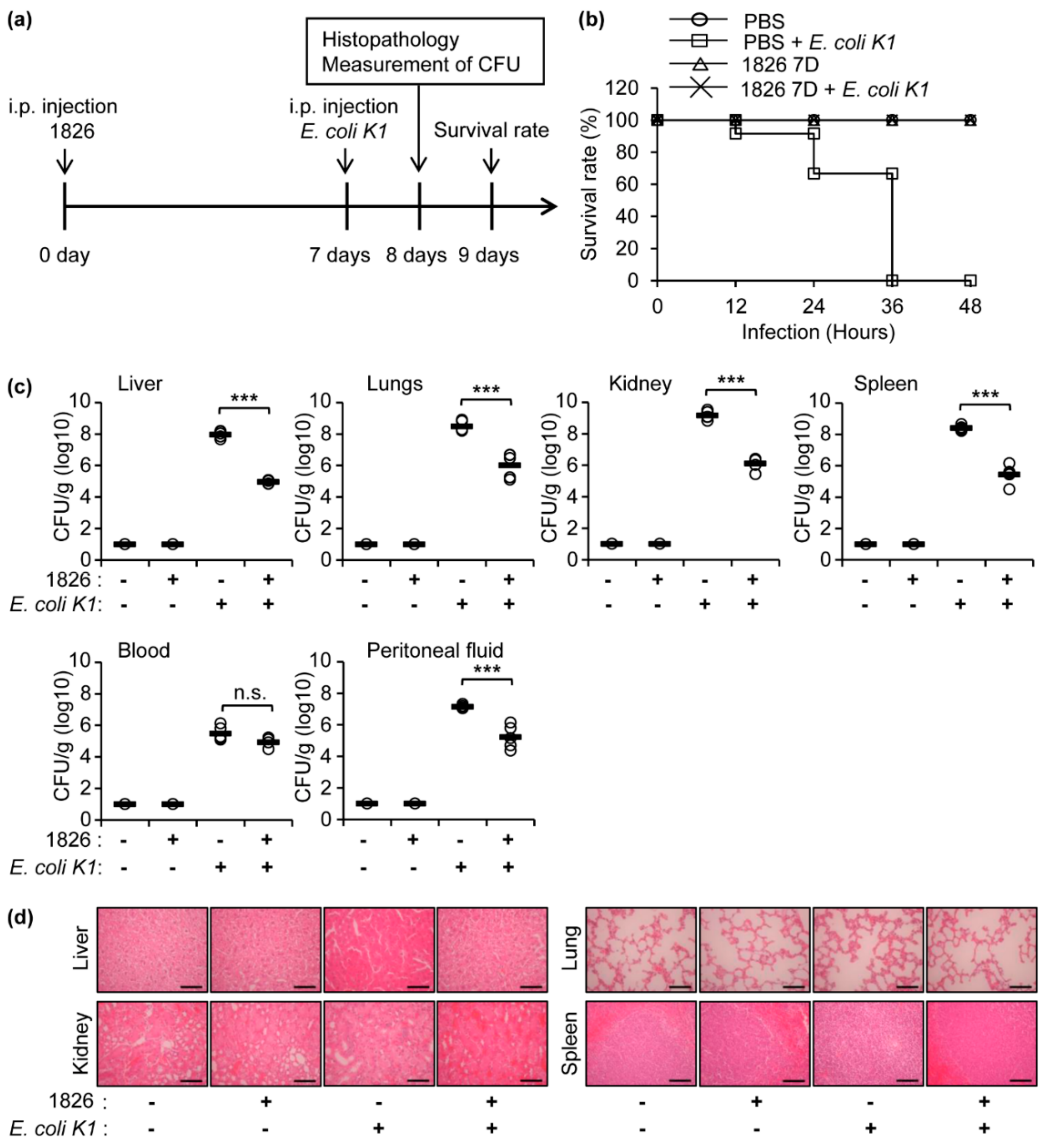
2.1. Intraperitoneal Administration of CpG-DNA Protects Mice against E. coli K1 Infection
2.2. Pre-Treatment of CpG-DNA Induces a Significant Recovery in the Mice after E. coli K1 Infection
2.3. Complement Activation by CpG-DNA Administration and/or E. coli K1 Infection
2.4. Depletion of C3 Causes a High Mortality Rate in the Mice against E. coli K1 Infection
2.5. Phagocytosis Is Not Increased by CpG-DNA Administration in the Mice Peritoneal Cavity against E. coli K1 Infection
3. Discussion
4. Materials and Methods
4.1. CpG-DNA
4.2. Mouse
4.3. Bacteria and In Vivo Infection Experiments
4.4. Hematoxylin and Eosin (H&E) Stain
4.5. Colony Forming Units in the Organs
4.6. Measurement of the Complement Components
4.7. C3 Depletion and In Vivo Experiments
4.8. ELISA
4.9. Analysis of E. coli K1 Uptake in the Mouse Peritoneal Cavity
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| C | Complement |
| CpG-DNA | Synthetic CpG-oligodeoxynucleotides |
| CFU | Colony forming units |
| CVF | Cobra venom factor |
| E. coli | Escherichia coli |
| ELISA | Enzyme-linked immunosorbent assay |
| TLR | Toll-like receptor |
References
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Krieg, A.M. Therapeutic potential of Toll-like receptor 9 activation. Nat. Rev. Drug Discov. 2006, 5, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Gilkeson, G.S.; Pisetsky, D.S. Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J. Immunol. 1991, 147, 1759–1764. [Google Scholar] [PubMed]
- Ballas, Z.K.; Rasmussen, W.L.; Krieg, A.M. Induction of NK activity in murine and human cells by CpG motifs in oligodeoxynucleotides and bacterial DNA. J. Immunol. 1996, 157, 1840–1845. [Google Scholar] [PubMed]
- Klinman, D.M.; Yi, A.K.; Beaucage, S.L.; Conover, J.; Krieg, A.M. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc. Natl. Acad. Sci. USA 1996, 93, 2879–2883. [Google Scholar] [CrossRef] [PubMed]
- Carson, D.A.; Raz, E. Oligonucleotide adjuvants for T helper 1 (Th1)-specific vaccination. J. Exp. Med. 1997, 186, 1621–1622. [Google Scholar] [CrossRef] [PubMed]
- Mangsbo, S.M.; Sanchez, J.; Anger, K.; Lambris, J.D.; Ekdahl, K.N.; Loskog, A.S.; Nilsson, B.; Totterman, T.H. Complement activation by CpG in a human whole blood loop system: Mechanisms and immunomodulatory effects. J. Immunol. 2009, 183, 6724–6732. [Google Scholar] [CrossRef]
- Bafica, A.; Scanga, C.A.; Feng, C.G.; Leifer, C.; Cheever, A.; Sher, A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 2005, 202, 1715–1724. [Google Scholar] [CrossRef]
- Bhan, U.; Lukacs, N.W.; Osterholzer, J.J.; Newstead, M.W.; Zeng, X.; Moore, T.A.; McMillan, T.R.; Krieg, A.M.; Akira, S.; Standiford, T.J. TLR9 is required for protective innate immunity in Gram-negative bacterial pneumonia: Role of dendritic cells. J. Immunol. 2007, 179, 3937–3946. [Google Scholar] [CrossRef]
- Noto, M.J.; Boyd, K.L.; Burns, W.J.; Varga, M.G.; Peek, R.M., Jr.; Skaar, E.P. Toll-Like Receptor 9 Contributes to Defense against Acinetobacter baumannii Infection. Infect. Immun. 2015, 83, 4134–4141. [Google Scholar] [CrossRef] [Green Version]
- Van der Meer, A.J.; Achouiti, A.; van der Ende, A.; Soussan, A.A.; Florquin, S.; de Vos, A.; Zeerleder, S.S.; van der Poll, T. Toll-like receptor 9 enhances bacterial clearance and limits lung consolidation in murine pneumonia caused by methicillin resistant Staphylococcus aureus. Mol. Med. 2016, 22, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.J.; Ito, S.; Tamura, T.; Hemmi, H.; Conover, J.; Ozato, K.; Akira, S.; Klinman, D.M. CpG-activated Thy1.2+ dendritic cells protect against lethal Listeria monocytogenes infection. Eur. J. Immunol. 2005, 35, 2397–2405. [Google Scholar]
- Mohamed, W.; Domann, E.; Chakraborty, T.; Mannala, G.; Lips, K.S.; Heiss, C.; Schnettler, R.; Alt, V. TLR9 mediates S. aureus killing inside osteoblasts via induction of oxidative stress. BMC Microbiol. 2016, 16, 230. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, A.; Lahiri, A.; Das, P.; Vani, J.; Shaila, M.S.; Chakravortty, D. TLR 9 activation in dendritic cells enhances salmonella killing and antigen presentation via involvement of the reactive oxygen species. PLoS ONE 2010, 5, e13772. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.C.; Moore, T.A.; Newstead, M.W.; Zeng, X.; Krieg, A.M.; Standiford, T.J. CpG oligodeoxynucleotides stimulate protective innate immunity against pulmonary Klebsiella infection. J. Immunol. 2004, 173, 5148–5155. [Google Scholar] [CrossRef] [PubMed]
- Ribes, S.; Meister, T.; Ott, M.; Redlich, S.; Janova, H.; Hanisch, U.K.; Nessler, S.; Nau, R. Intraperitoneal prophylaxis with CpG oligodeoxynucleotides protects neutropenic mice against intracerebral Escherichia coli K1 infection. J. Neuroinflamm. 2014, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Kim, D.; Gautam, A.; Lee, H.; Kwak, M.H.; Park, M.C.; Park, S.; Wu, G.; Lee, B.L.; Lee, Y.; et al. CpG-DNA exerts antibacterial effects by protecting immune cells and producing bacteria-reactive antibodies. Sci. Rep. 2018, 8, 16236. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Kim, D.; Lee, H.; Kwak, M.H.; Park, S.; Lee, Y.; Kwon, H.J. CpG-DNA induces bacteria-reactive IgM enhancing phagocytic activity against Staphylococcus aureus infection. Bmb Rep. 2019. [Epub ahead of print]. [Google Scholar]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Gomez-Aldapa, C.A.; Segovia-Cruz, J.A.; Cerna-Cortes, J.F.; Rangel-Vargas, E.; Salas-Rangel, L.P.; Gutierrez-Alcantara, E.J.; Castro-Rosas, J. Prevalence and behavior of multidrug-resistant shiga toxin-producing Escherichia coli, enteropathogenic E. coli and enterotoxigenic E. coli on coriander. Food Microbiol. 2016, 59, 97–103. [Google Scholar] [CrossRef]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Lariviere, S.; Harel, J. Antimicrobial resistance genes in enterotoxigenic Escherichia coli O149:K91 isolates obtained over a 23-year period from pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, T.J.; Ruiz, J.; Molina, M.; Del Valle, L.J.; Vargas, M.; Gil, A.I.; Ecker, L.; Barletta, F.; Hall, E.; Cleary, T.G.; et al. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am. J. Trop. Med. Hyg. 2009, 81, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Darsley, M.J.; Chakraborty, S.; DeNearing, B.; Sack, D.A.; Feller, A.; Buchwaldt, C.; Bourgeois, A.L.; Walker, R.; Harro, C.D. The oral, live attenuated enterotoxigenic Escherichia coli vaccine ACE527 reduces the incidence and severity of diarrhea in a human challenge model of diarrheal disease. Clin. Vaccine Immunol. 2012, 19, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, A.; Bourgeois, L.; Carlin, N.; Clements, J.; Gustafsson, B.; Hartford, M.; Holmgren, J.; Petzold, M.; Walker, R.; Svennerholm, A.M. Safety and immunogenicity of an improved oral inactivated multivalent enterotoxigenic Escherichia coli (ETEC) vaccine administered alone and together with dmLT adjuvant in a double-blind, randomized, placebo-controlled Phase I study. Vaccine 2014, 32, 7077–7084. [Google Scholar] [CrossRef] [Green Version]
- Ruan, X.; Sack, D.A.; Zhang, W. Genetic fusions of a CFA/I/II/IV MEFA (multiepitope fusion antigen) and a toxoid fusion of heat-stable toxin (STa) and heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) retain broad anti-CFA and antitoxin antigenicity. PLoS ONE 2015, 10, e0121623. [Google Scholar] [CrossRef] [PubMed]
- Elkins, K.L.; Rhinehart-Jones, T.R.; Stibitz, S.; Conover, J.S.; Klinman, D.M. Bacterial DNA containing CpG motifs stimulates lymphocyte-dependent protection of mice against lethal infection with intracellular bacteria. J. Immunol. 1999, 162, 2291–2298. [Google Scholar] [PubMed]
- Judy, B.M.; Taylor, K.; Deeraksa, A.; Johnston, R.K.; Endsley, J.J.; Vijayakumar, S.; Aronson, J.F.; Estes, D.M.; Torres, A.G. Prophylactic application of CpG oligonucleotides augments the early host response and confers protection in acute melioidosis. PLoS ONE 2012, 7, e34176. [Google Scholar] [CrossRef]
- Lubbers, R.; van Essen, M.F.; van Kooten, C.; Trouw, L.A. Production of complement components by cells of the immune system. Clin. Exp. Immunol. 2017, 188, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Galbraith, W.M.; Hobson, W.C.; Giclas, P.C.; Schechter, P.J.; Agrawal, S. Complement activation and hemodynamic changes following intravenous administration of phosphorothioate oligonucleotides in the monkey. Antisense Res. Dev. 1994, 4, 201–206. [Google Scholar] [CrossRef]
- Henry, S.P.; Giclas, P.C.; Leeds, J.; Pangburn, M.; Auletta, C.; Levin, A.A.; Kornbrust, D.J. Activation of the alternative pathway of complement by a phosphorothioate oligonucleotide: Potential mechanism of action. J. Pharm. Exp. 1997, 281, 810–816. [Google Scholar]
- Lupu, F.; Keshari, R.S.; Lambris, J.D.; Coggeshall, K.M. Crosstalk between the coagulation and complement systems in sepsis. Thromb. Res. 2014, 133 (Suppl. S1), S28–S31. [Google Scholar] [CrossRef] [Green Version]
- Hack, C.E.; Nuijens, J.H.; Felt-Bersma, R.J.; Schreuder, W.O.; Eerenberg-Belmer, A.J.; Paardekooper, J.; Bronsveld, W.; Thijs, L.G. Elevated plasma levels of the anaphylatoxins C3a and C4a are associated with a fatal outcome in sepsis. Am. J. Med. 1989, 86, 20–26. [Google Scholar] [CrossRef]
- Silasi-Mansat, R.; Zhu, H.; Popescu, N.I.; Peer, G.; Sfyroera, G.; Magotti, P.; Ivanciu, L.; Lupu, C.; Mollnes, T.E.; Taylor, F.B.; et al. Complement inhibition decreases the procoagulant response and confers organ protection in a baboon model of Escherichia coli sepsis. Blood 2010, 116, 1002–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uwatoko, S.; Mannik, M. The location of binding sites on C1q for DNA. J. Immunol. 1990, 144, 3484–3488. [Google Scholar] [PubMed]
- Xing, J.; Zhang, A.; Zhang, H.; Wang, J.; Li, X.C.; Zeng, M.S.; Zhang, Z. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun. 2017, 8, 945. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Weng, L.; Yuan, B.; Wang, Z.; Jia, L.; Jin, R.; Lu, H.; Li, X.C.; Liu, Y.J.; Zhang, Z. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat. Immunol. 2016, 17, 1373–1380. [Google Scholar] [CrossRef]
- Xing, J.; Zhang, A.; Minze, L.J.; Li, X.C.; Zhang, Z. TRIM29 Negatively Regulates the Type I IFN Production in Response to RNA Virus. J. Immunol. 2018, 201, 183–192. [Google Scholar] [CrossRef]
- Szalai, A.J.; Briles, D.E.; Volanakis, J.E. Role of complement in C-reactive-protein-mediated protection of mice from Streptococcus pneumoniae. Infect. Immun. 1996, 64, 4850–4853. [Google Scholar] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.H.; Park, J.; Kim, D.; Gautam, A.; Akauliya, M.; Kim, J.; Lee, H.; Park, S.; Lee, Y.; Kwon, H.-J. Anti-Bacterial Effect of CpG-DNA Involves Enhancement of the Complement Systems. Int. J. Mol. Sci. 2019, 20, 3397. https://doi.org/10.3390/ijms20143397
Kim TH, Park J, Kim D, Gautam A, Akauliya M, Kim J, Lee H, Park S, Lee Y, Kwon H-J. Anti-Bacterial Effect of CpG-DNA Involves Enhancement of the Complement Systems. International Journal of Molecular Sciences. 2019; 20(14):3397. https://doi.org/10.3390/ijms20143397
Chicago/Turabian StyleKim, Te Ha, Joongwon Park, Dongbum Kim, Avishekh Gautam, Madhav Akauliya, Jinsoo Kim, Hanseul Lee, Sangkyu Park, Younghee Lee, and Hyung-Joo Kwon. 2019. "Anti-Bacterial Effect of CpG-DNA Involves Enhancement of the Complement Systems" International Journal of Molecular Sciences 20, no. 14: 3397. https://doi.org/10.3390/ijms20143397




