Differences in Shedding of the Interleukin-11 Receptor by the Proteases ADAM9, ADAM10, ADAM17, Meprin α, Meprin β and MT1-MMP
Abstract
:1. Introduction
2. Results
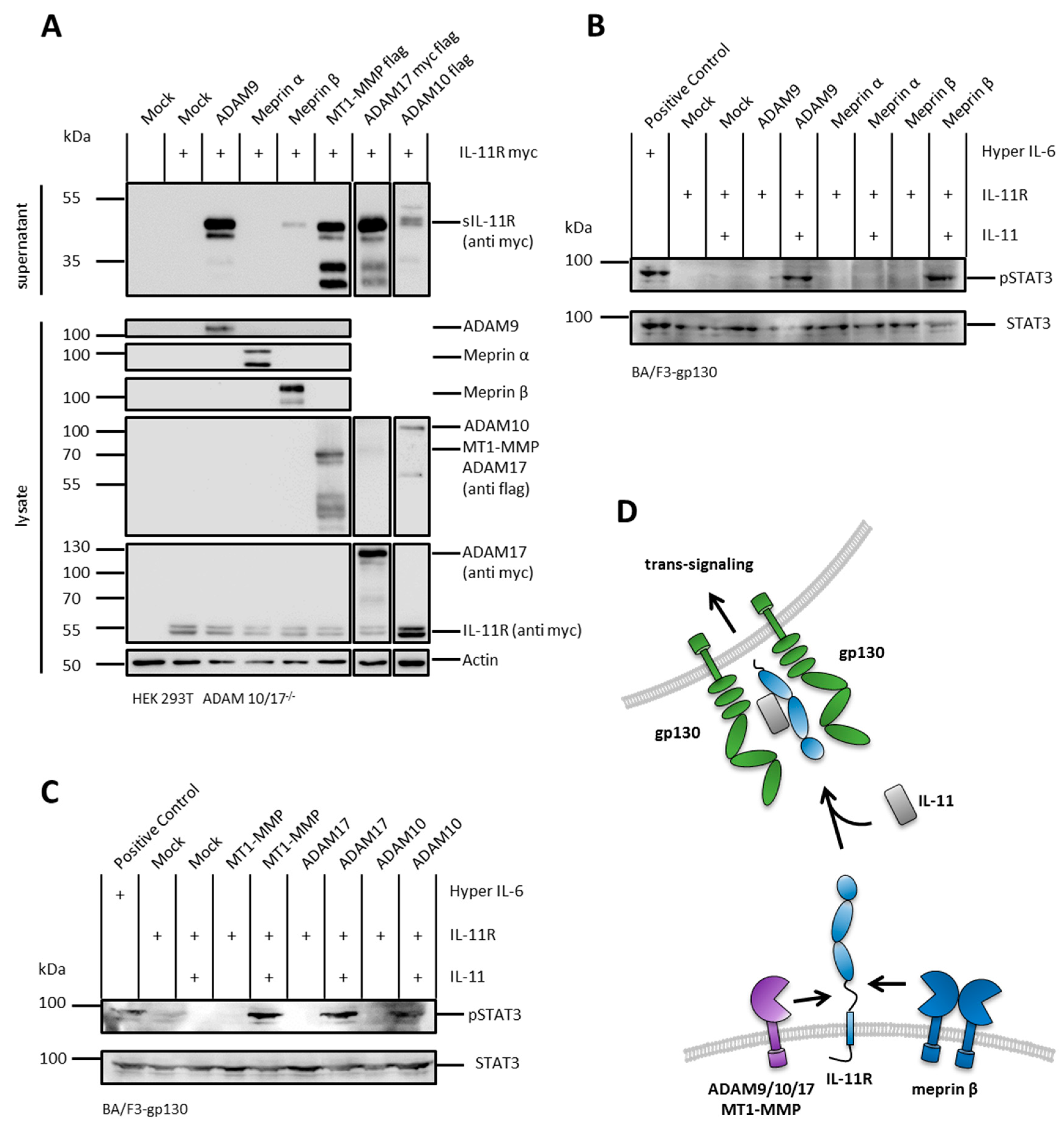
2.1. The IL-11R Is Cleaved by Different Metalloproteases
2.2. sIL-11R Shed by Different Metalloproteases Is Able to Induce Phosphorylation of STAT3
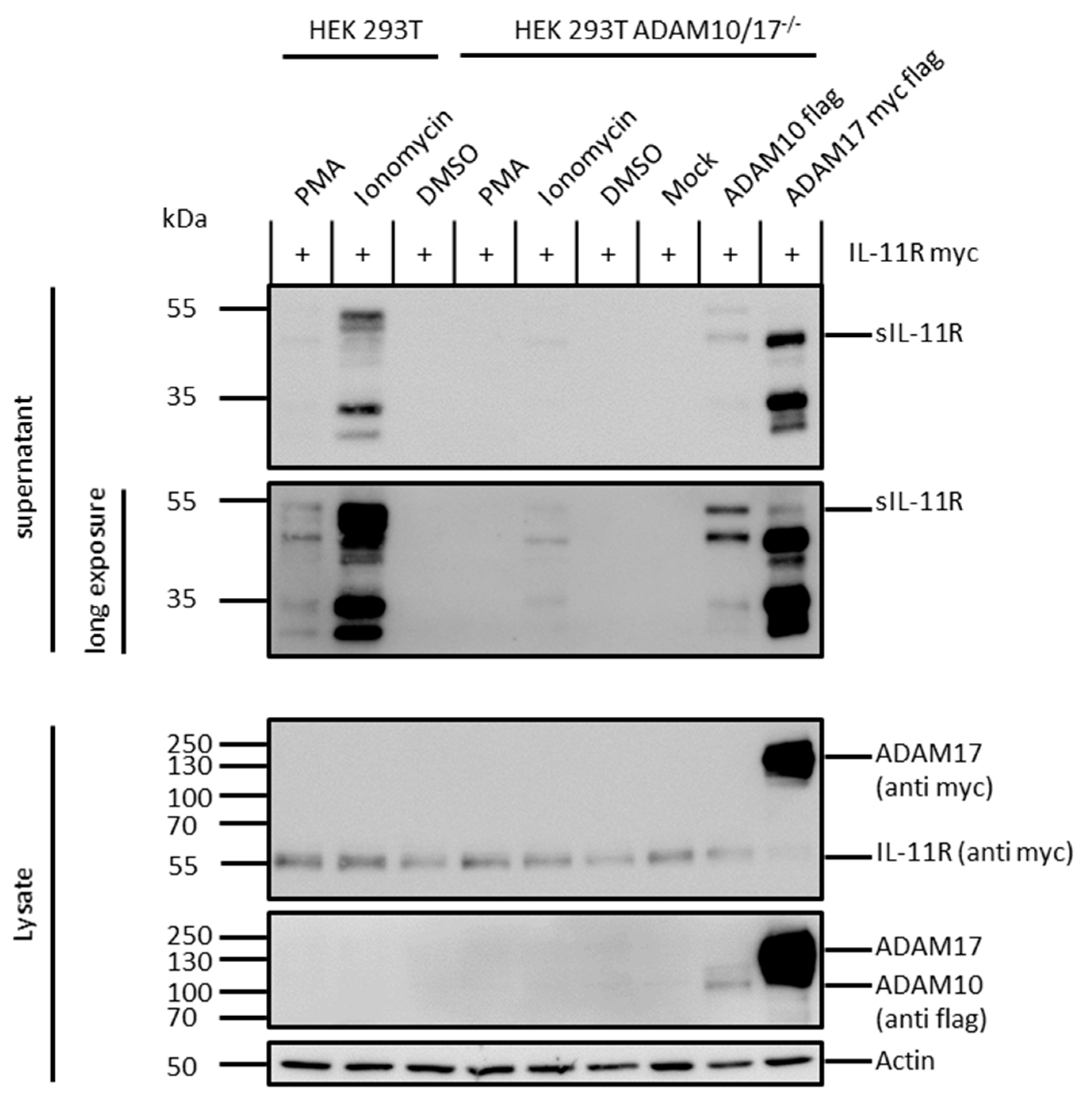
2.3. ADAM 10 and ADAM 17 Are Both Able to Shed the IL-11R
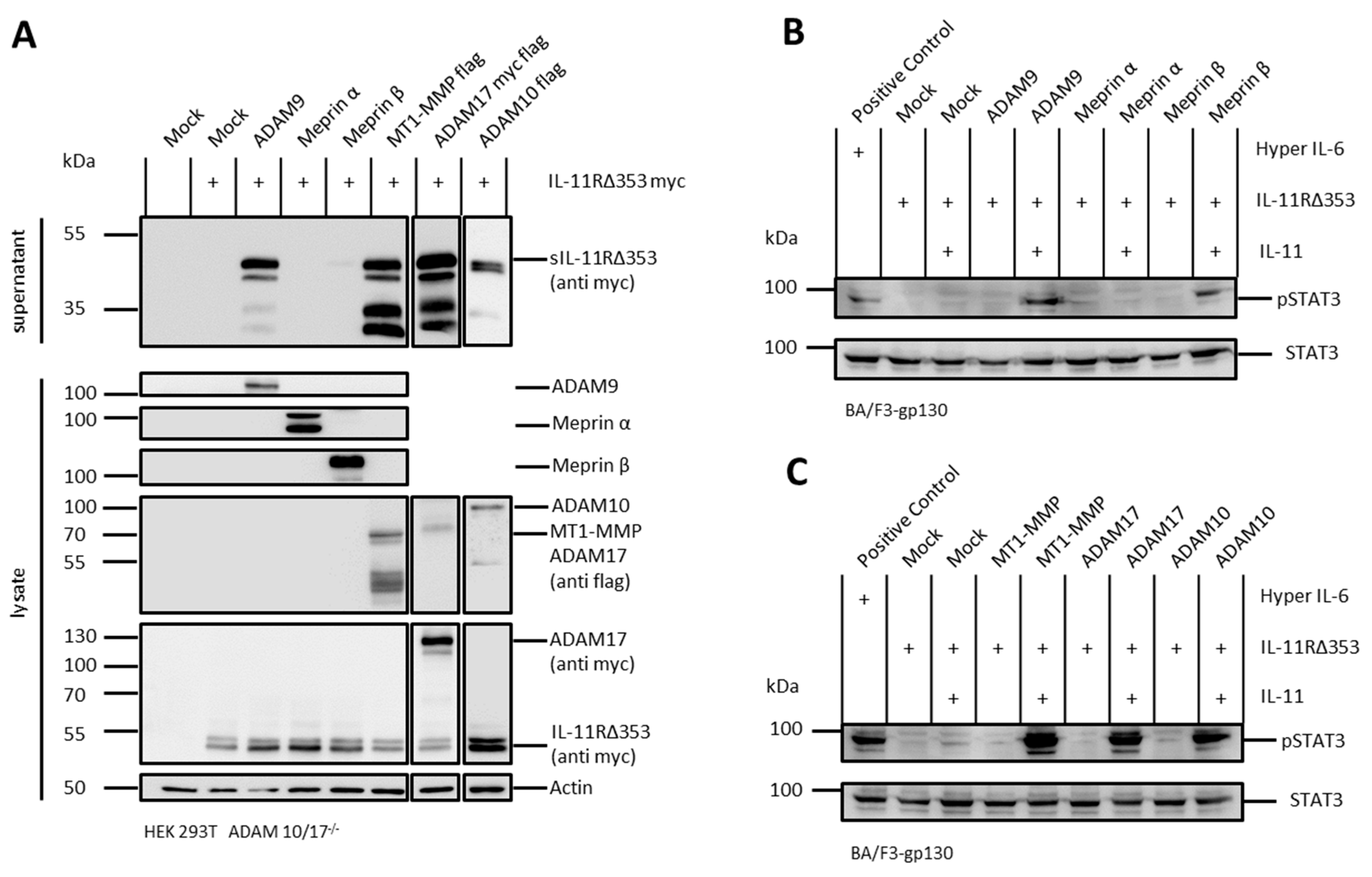
2.4. Arginine 355 Is not Obligatory for Ectodomain Shedding of the IL-11R
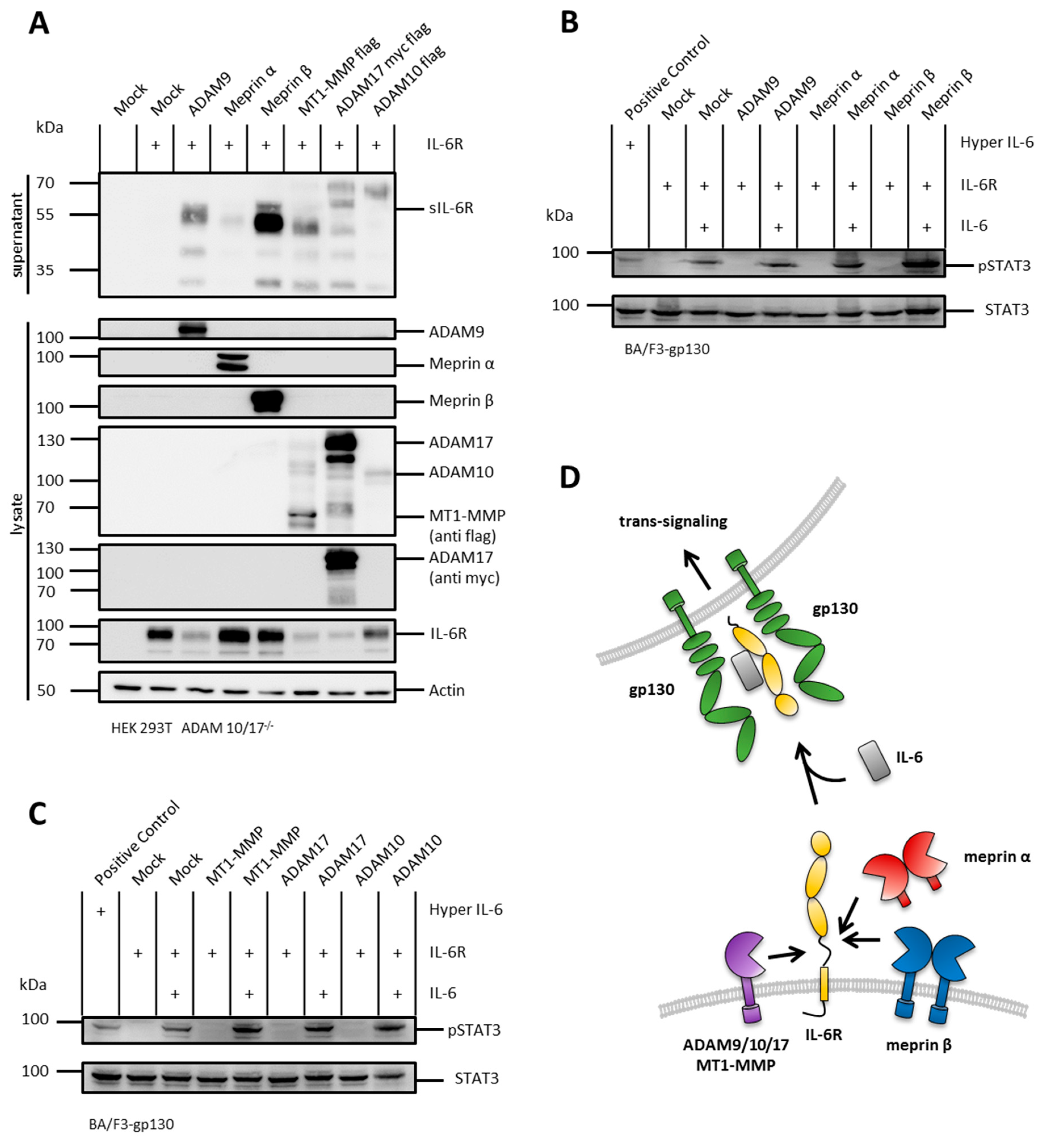
2.5. The IL-6R Is Cleaved by Several Metalloproteases
2.6. N-Terminal Cleavage of the IL-6R and the IL-11R Apart from the Stalk Regions
3. Discussion
4. Material and Methods
4.1. Chemicals
4.2. Cells
4.3. Plasmids
- pcDNA 3.1
- murine ADAM9 in pcDNA 3.1
- human ADAM10 flag-tagged in pcDNA4/TO
- human ADAM17 DDK in pCMV6: purchased on Origene
- human meprin α in pSG5
- human meprin β in pSG5
- human IL-11R N:myc, C:HA in pcDNA3.1
- human IL-11R in pcDNA 3.1 lacking H353-S362
- human MT1-MMP flag-tagged in pcDNA4/TO
- human IL-6R in pcDNA 3.1
- Sequences were confirmed by Sanger-sequencing.
4.4. Shedding Assay
4.5. ADAM10/17 Stimulation
4.6. Phosphorylation Assay
4.7. Cell Lysis, SDS-PAGE, and Immunoblot Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Garbers, C.; Scheller, J. Interleukin-6 and interleukin-11: Same same but different. Biol. Chem. 2013, 394, 1145–1161. [Google Scholar] [CrossRef]
- Agthe, M.; Garbers, Y.; Putoczki, T.; Garbers, C. Interleukin-11 classic but not trans-signaling is essential for fertility in mice. Placenta 2017, 57, 13–16. [Google Scholar] [CrossRef]
- Nieminen, P.; Morgan, N.V.; Fenwick, A.L.; Parmanen, S.; Veistinen, L.; Mikkola, M.L.; van der Spek, P.J.; Giraud, A.; Judd, L.; Arte, S.; et al. Inactivation of IL11 signaling causes craniosynostosis, delayed tooth eruption, and supernumerary teeth. Am. J. Hum. Genet. 2011, 89, 67–81. [Google Scholar] [CrossRef]
- Hilton, D.J.; Hilton, A.A.; Raicevic, A.; Rakar, S.; Harrison-Smith, M.; Gough, N.M.; Begley, C.G.; Metcalf, D.; Nicola, N.A.; Willson, T.A. Cloning of a murine IL-11 receptor alpha-chain; requirement for gp130 for high affinity binding and signal transduction. EMBO J. 1994, 13, 4765–4775. [Google Scholar] [CrossRef]
- Zhang, X.G.; Gu, J.J.; Lu, Z.Y.; Yasukawa, K.; Yancopoulos, G.D.; Turner, K.; Shoyab, M.; Taga, T.; Kishimoto, T.; Bataille, R.; et al. Ciliary neurotropic factor, interleukin 11, leukemia inhibitory factor, and oncostatin M are growth factors for human myeloma cell lines using the interleukin 6 signal transducer gp130. J. Exp. Med. 1994, 179, 1337–1342. [Google Scholar] [CrossRef]
- Garbers, C.; Hermanns, H.M.; Schaper, F.; Muller-Newen, G.; Grotzinger, J.; Rose-John, S.; Scheller, J. Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012, 23, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Nitz, R.; Agthe, M.; Monhasery, N.; Aparicio-Siegmund, S.; Schumacher, N.; Wolf, J.; Moller-Hackbarth, K.; Waetzig, G.H.; Grotzinger, J.; et al. Proteolytic Cleavage Governs Interleukin-11 Trans-signaling. Cell Rep. 2016, 14, 1761–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müllberg, J.; Schooltink, H.; Stoyan, T.; Günther, M.; Graeve, L.; Buse, G.; Mackiewicz, A.; Heinrich, P.; Rose-John, S. The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 1993, 23, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Garbers, C.; Heink, S.; Korn, T.; Rose-John, S. Interleukin-6: Designing specific therapeutics for a complex cytokine. Nat. Rev. Drug Discov. 2018, 17, 395–412. [Google Scholar] [CrossRef]
- Yawata, H.; Yasukawa, K.; Natsuka, S.; Murakami, M.; Yamasaki, K.; Hibi, M.; Taga, T.; Kishimoto, T. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J. 1993, 12, 1705–1712. [Google Scholar] [CrossRef]
- Nitz, R.; Lokau, J.; Aparicio-Siegmund, S.; Scheller, J.; Garbers, C. Modular organization of Interleukin-6 and Interleukin-11 alpha-receptors. Biochimie 2015, 119, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sprecher, C.A.; Grant, F.J.; Baumgartner, J.W.; Presnell, S.R.; Schrader, S.K.; Yamagiwa, T.; Whitmore, T.E.; O’Hara, P.J.; Foster, D.F. Cloning and characterization of a novel class I cytokine receptor. Biochem. Biophys. Res. Commun. 1998, 246, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.; Boll, I.; Rothaug, M.; Schumacher, N.; Schmidt, F.; Wichert, R.; Schneppenheim, J.; Lokau, J.; Pickhinke, U.; Koudelka, T.; et al. Meprin Metalloproteases Generate Biologically Active Soluble Interleukin-6 Receptor to Induce Trans-Signaling. Sci. Rep. 2017, 7, 44053. [Google Scholar] [CrossRef] [PubMed]
- Goth, C.K.; Halim, A.; Khetarpal, S.A.; Rader, D.J.; Clausen, H.; Schjoldager, K.T. A systematic study of modulation of ADAM-mediated ectodomain shedding by site-specific O-glycosylation. Proc. Natl. Acad. Sci. USA 2015, 112, 14623–14628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riethmueller, S.; Somasundaram, P.; Ehlers, J.C.; Hung, C.W.; Flynn, C.M.; Lokau, J.; Agthe, M.; Dusterhoft, S.; Zhu, Y.; Grotzinger, J.; et al. Proteolytic Origin of the Soluble Human IL-6R In Vivo and a Decisive Role of N-Glycosylation. PLoS Biol. 2017, 15, e2000080. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Rand, M.D.; Wu, X.; Sestan, N.; Wang, W.; Rakic, P.; Xu, T. Artavanis-Tsakonas, S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science 1999, 283, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Matthews, A.L.; Noy, P.J.; Reyat, J.S.; Tomlinson, M.G. Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: The emerging role of tetraspanins and rhomboids. Platelets 2017, 28, 333–341. [Google Scholar] [CrossRef]
- Janes, P.W.; Saha, N.; Barton, W.A.; Kolev, M.V.; Wimmer-Kleikamp, S.H.; Nievergall, E.; Blobel, C.P.; Himanen, J.P.; Lackmann, M.; Nikolov, D.B. Adam meets Eph: An ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell 2005, 123, 291–304. [Google Scholar] [CrossRef]
- Herzog, C.; Haun, R.S.; Kaushal, G.P. Role of meprin metalloproteinases in cytokine processing and inflammation. Cytokine 2019, 114, 18–25. [Google Scholar] [CrossRef]
- Prox, J.; Arnold, P.; Becker-Pauly, C. Meprin alpha and meprin beta: Procollagen proteinases in health and disease. Matrix Biol. 2015, 44–46, 7–13. [Google Scholar] [CrossRef]
- Broder, C.; Arnold, P.; Vadon-Le Goff, S.; Konerding, M.A.; Bahr, K.; Muller, S.; Overall, C.M.; Bond, J.S.; Koudelka, T.; Tholey, A.; et al. Metalloproteases meprin alpha and meprin beta are C- and N-procollagen proteinases important for collagen assembly and tensile strength. Proc. Natl. Acad. Sci. USA 2013, 110, 14219–14224. [Google Scholar] [CrossRef] [PubMed]
- Broder, C.; Becker-Pauly, C. The metalloproteases meprin alpha and meprin beta: unique enzymes in inflammation, neurodegeneration, cancer and fibrosis. Biochem. J. 2013, 450, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Jackle, F.; Schmidt, F.; Wichert, R.; Arnold, P.; Prox, J.; Mangold, M.; Ohler, A.; Pietrzik, C.U.; Koudelka, T.; Tholey, A.; et al. Metalloprotease meprin beta is activated by transmembrane serine protease matriptase-2 at the cell surface thereby enhancing APP shedding. Biochem. J. 2015, 470, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Wichert, R.; Ermund, A.; Schmidt, S.; Schweinlin, M.; Ksiazek, M.; Arnold, P.; Knittler, K.; Wilkens, F.; Potempa, B.; Rabe, B.; et al. Mucus Detachment by Host Metalloprotease Meprin beta Requires Shedding of Its Inactive Pro-form, which Is Abrogated by the Pathogenic Protease RgpB. Cell Rep. 2017, 21, 2090–2103. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.D.; Bond, J.S. Activation Mechanism of Meprins, Members of the Astacin Metalloendopeptidase Family. J. Biol. Chem. 1997, 272, 28126–28132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzog, C.; Haun, R.S.; Ludwig, A.; Shah, S.V.; Kaushal, G.P. ADAM10 is the major sheddase responsible for the release of membrane-associated meprin A. J. Biol. Chem. 2014, 289, 13308–13322. [Google Scholar] [CrossRef] [PubMed]
- Hahn, D.; Pischitzis, A.; Roesmann, S.; Hansen, M.K.; Leuenberger, B.; Luginbuehl, U.; Sterchi, E.E. Phorbol 12-myristate 13-acetate-induced ectodomain shedding and phosphorylation of the human meprinbeta metalloprotease. J. Biol. Chem. 2003, 278, 42829–42839. [Google Scholar] [CrossRef] [PubMed]
- Bedau, T.; Peters, F.; Prox, J.; Arnold, P.; Schmidt, F.; Finkernagel, M.; Kollmann, S.; Wichert, R.; Otte, A.; Ohler, A.; et al. Ectodomain shedding of CD99 within highly conserved regions is mediated by the metalloprotease meprin beta and promotes transendothelial cell migration. FASEB J. 2017, 31, 1226–1237. [Google Scholar] [CrossRef] [PubMed]
- Bedau, T.; Schumacher, N.; Peters, F.; Prox, J.; Arnold, P.; Koudelka, T.; Helm, O.; Schmidt, F.; Rabe, B.; Jentzsch, M.; et al. Cancer-associated mutations in the canonical cleavage site do not influence CD99 shedding by the metalloprotease meprin beta but alter cell migration in vitro. Oncotarget 2017, 8, 54873–54888. [Google Scholar] [CrossRef]
- Schutte, A.; Ermund, A.; Becker-Pauly, C.; Johansson, M.E.; Rodriguez-Pineiro, A.M.; Backhed, F.; Muller, S.; Lottaz, D.; Bond, J.S.; Hansson, G.C. Microbial-induced meprin beta cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc. Natl. Acad. Sci. USA 2014, 111, 12396–12401. [Google Scholar] [CrossRef]
- Keiffer, T.R.; Bond, J.S. Meprin metalloproteases inactivate interleukin 6. J. Biol. Chem. 2014, 289, 7580–7588. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.M.; Wong, H.L.; Jin, G.; Liu, B.; Cao, R.; Cao, Y.; Lehti, K.; Tryggvason, K.; Zhou, Z. MT1-MMP inactivates ADAM9 to regulate FGFR2 signaling and calvarial osteogenesis. Dev. Cell 2012, 22, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Zunke, F.; Rose-John, S. The shedding protease ADAM17: Physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Edwards, D.R.; Handsley, M.M.; Pennington, C.J. The ADAM metalloproteinases. Mol. Asp. Med. 2008, 29, 258–289. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Wandel, M.; Garbers, C. Enhancing Interleukin-6 and Interleukin-11 receptor cleavage. Int. J. Biochem. Cell Biol. 2017, 85, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Baumann, H.; Schendel, P. Interleukin-11 regulates the hepatic expression of the same plasma protein genes as interleukin-6. J. Biol. Chem. 1991, 266, 20424–20427. [Google Scholar] [PubMed]
- Balic, J.J.; Garbers, C.; Rose-John, S.; Yu, L.; Jenkins, B.J. Interleukin-11-driven gastric tumourigenesis is independent of trans-signalling. Cytokine 2017, 92, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Putoczki, T.L.; Thiem, S.; Loving, A.; Busuttil, R.A.; Wilson, N.J.; Ziegler, P.K.; Nguyen, P.M.; Preaudet, A.; Farid, R.; Edwards, K.M.; et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 2013, 24, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Shaarawy, M.; Zaki, S.; Sheiba, M.; El-Minawi, A.M. Circulating levels of osteoclast activating cytokines, interleukin-11 and transforming growth factor-beta2, as valuable biomarkers for the assessment of bone turnover in postmenopausal osteoporosis. Clin. Lab. 2003, 49, 625–636. [Google Scholar] [PubMed]
- Riethmueller, S.; Ehlers, J.C.; Lokau, J.; Dusterhoft, S.; Knittler, K.; Dombrowsky, G.; Grotzinger, J.; Rabe, B.; Rose-John, S.; Garbers, C. Cleavage Site Localization Differentially Controls Interleukin-6 Receptor Proteolysis by ADAM10 and ADAM17. Sci. Rep. 2016, 6, 25550. [Google Scholar] [CrossRef] [PubMed]
- Gearing, D.P.; Ziegler, S.F.; Comeau, M.R.; Friend, D.; Thoma, B.; Cosman, D.; Park, L.; Mosley, B. Proliferative responses and binding properties of hematopoietic cells transfected with low-affinity receptors for leukemia inhibitory factor, oncostatin M, and ciliary neurotrophic factor. Proc. Natl. Acad. Sci. USA 1994, 91, 1119–1123. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Agthe, M.; Garbers, C. Generation of Soluble Interleukin-11 and Interleukin-6 Receptors: A Crucial Function for Proteases during Inflammation. Mediat. Inflamm. 2016, 2016, 1785021. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sammel, M.; Peters, F.; Lokau, J.; Scharfenberg, F.; Werny, L.; Linder, S.; Garbers, C.; Rose-John, S.; Becker-Pauly, C. Differences in Shedding of the Interleukin-11 Receptor by the Proteases ADAM9, ADAM10, ADAM17, Meprin α, Meprin β and MT1-MMP. Int. J. Mol. Sci. 2019, 20, 3677. https://doi.org/10.3390/ijms20153677
Sammel M, Peters F, Lokau J, Scharfenberg F, Werny L, Linder S, Garbers C, Rose-John S, Becker-Pauly C. Differences in Shedding of the Interleukin-11 Receptor by the Proteases ADAM9, ADAM10, ADAM17, Meprin α, Meprin β and MT1-MMP. International Journal of Molecular Sciences. 2019; 20(15):3677. https://doi.org/10.3390/ijms20153677
Chicago/Turabian StyleSammel, Martin, Florian Peters, Juliane Lokau, Franka Scharfenberg, Ludwig Werny, Stefan Linder, Christoph Garbers, Stefan Rose-John, and Christoph Becker-Pauly. 2019. "Differences in Shedding of the Interleukin-11 Receptor by the Proteases ADAM9, ADAM10, ADAM17, Meprin α, Meprin β and MT1-MMP" International Journal of Molecular Sciences 20, no. 15: 3677. https://doi.org/10.3390/ijms20153677
APA StyleSammel, M., Peters, F., Lokau, J., Scharfenberg, F., Werny, L., Linder, S., Garbers, C., Rose-John, S., & Becker-Pauly, C. (2019). Differences in Shedding of the Interleukin-11 Receptor by the Proteases ADAM9, ADAM10, ADAM17, Meprin α, Meprin β and MT1-MMP. International Journal of Molecular Sciences, 20(15), 3677. https://doi.org/10.3390/ijms20153677





