Molecular Characterization and Functional Analysis of Three Autophagy Genes, BxATG5, BxATG9, and BxATG16, in Bursaphelenchus xylophilus
Abstract
:1. Introduction
2. Results
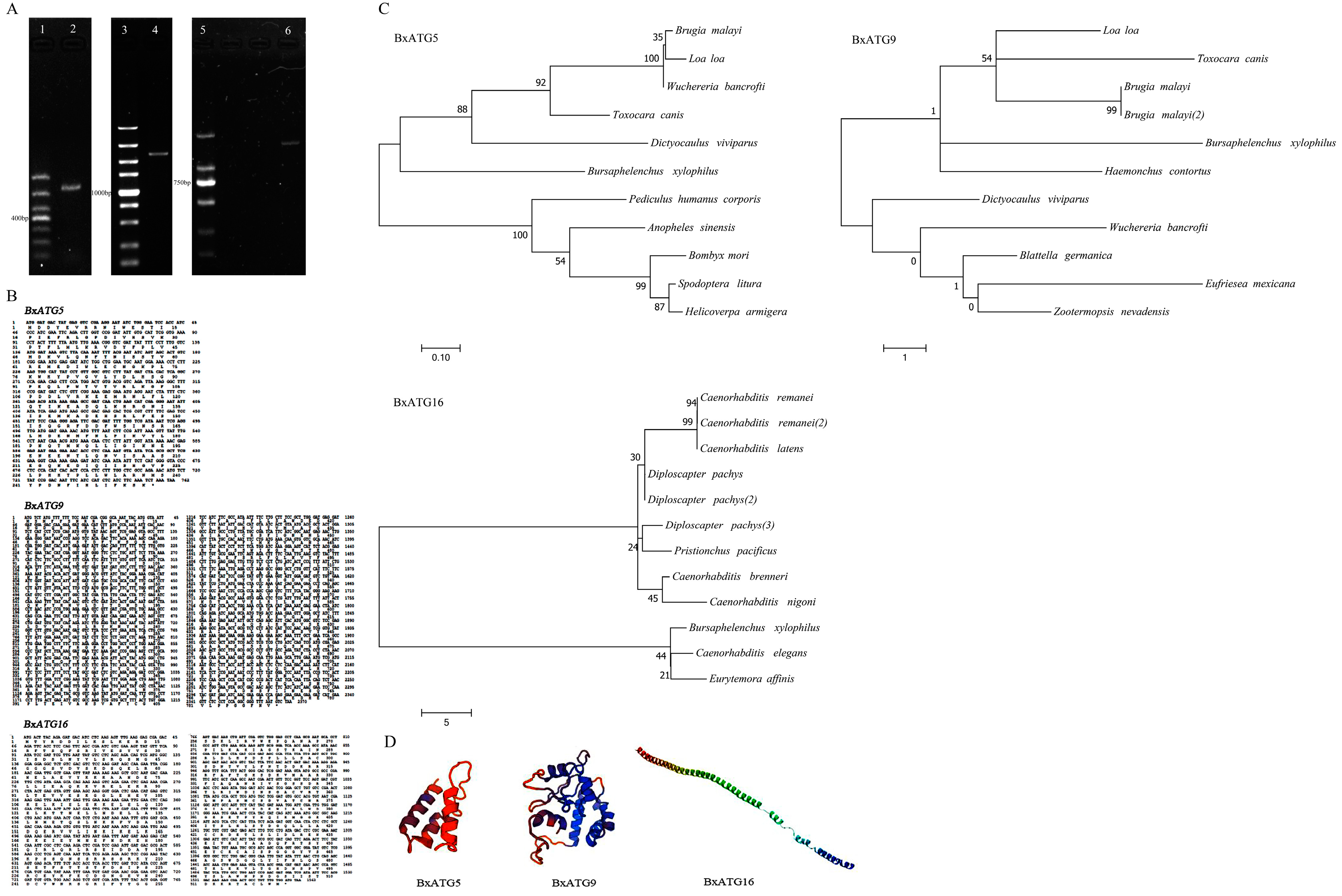
2.1. Cloning and Sequence Analysis of Three Autophagy Genes of PWN
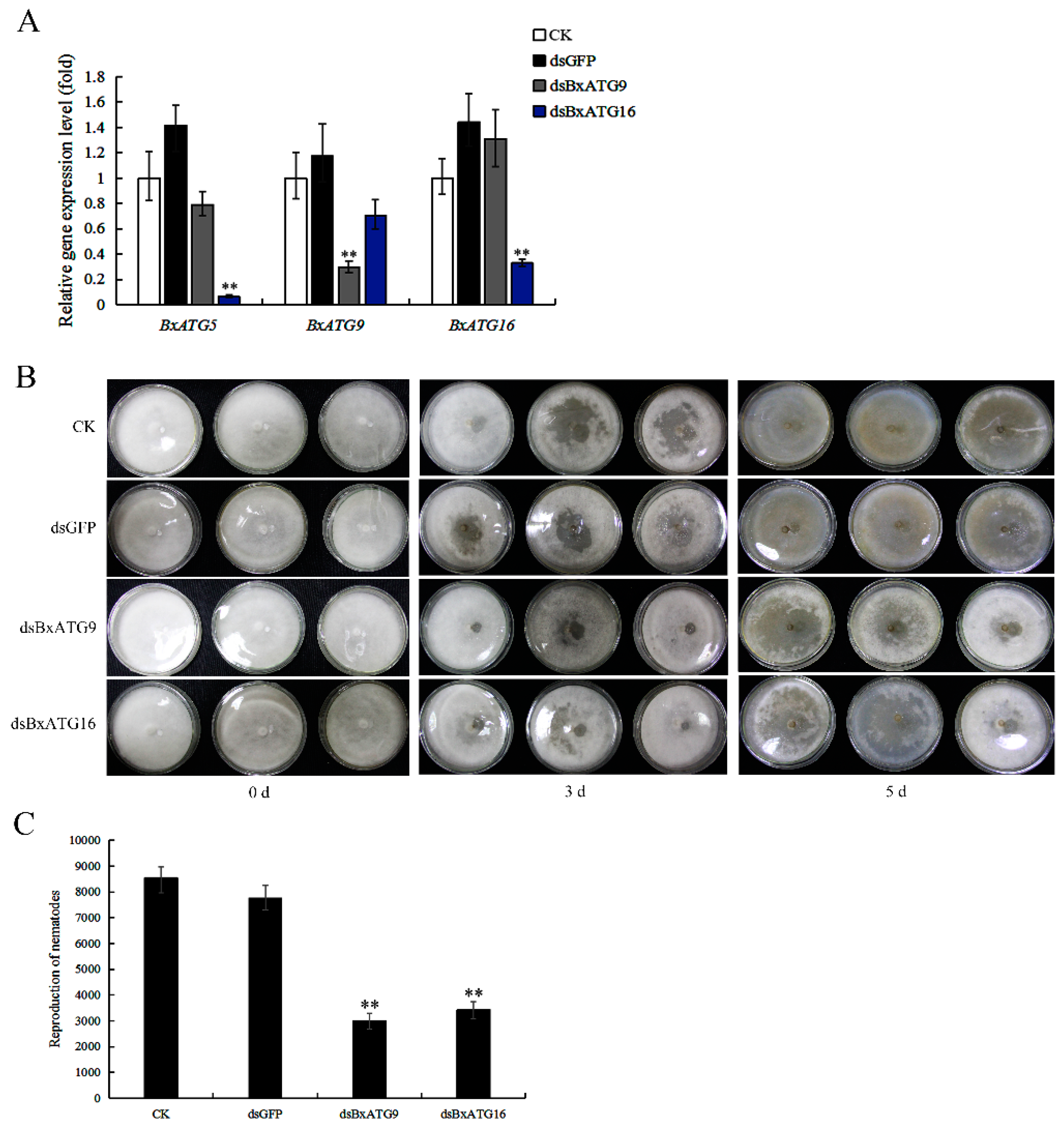
2.2. Silencing BxATG9 and BxATG16 Reduced PWN Feeding and Reproduction
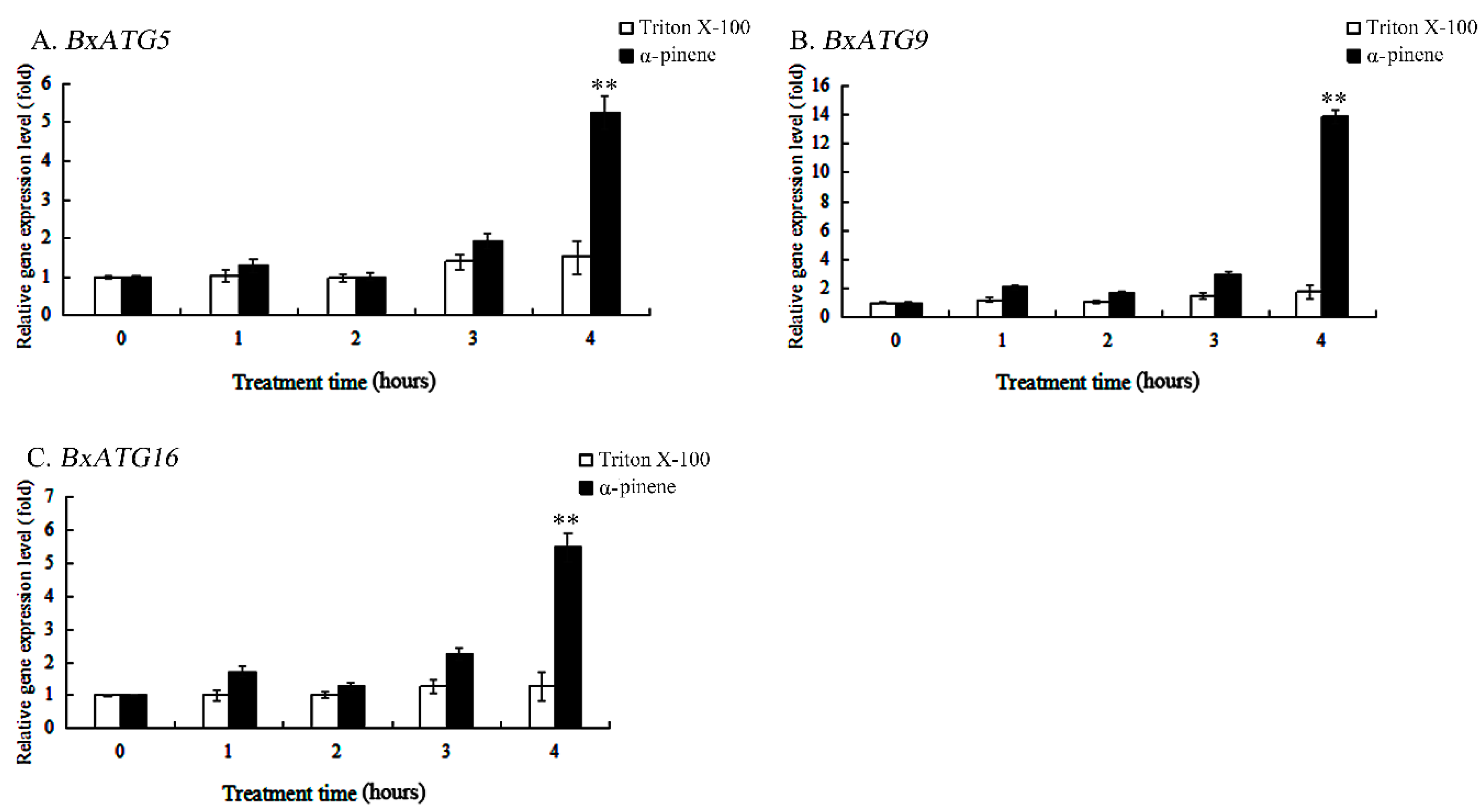
2.3. α-pinene Stress Increased the Expression Levels of Autophagy Genes in PWN
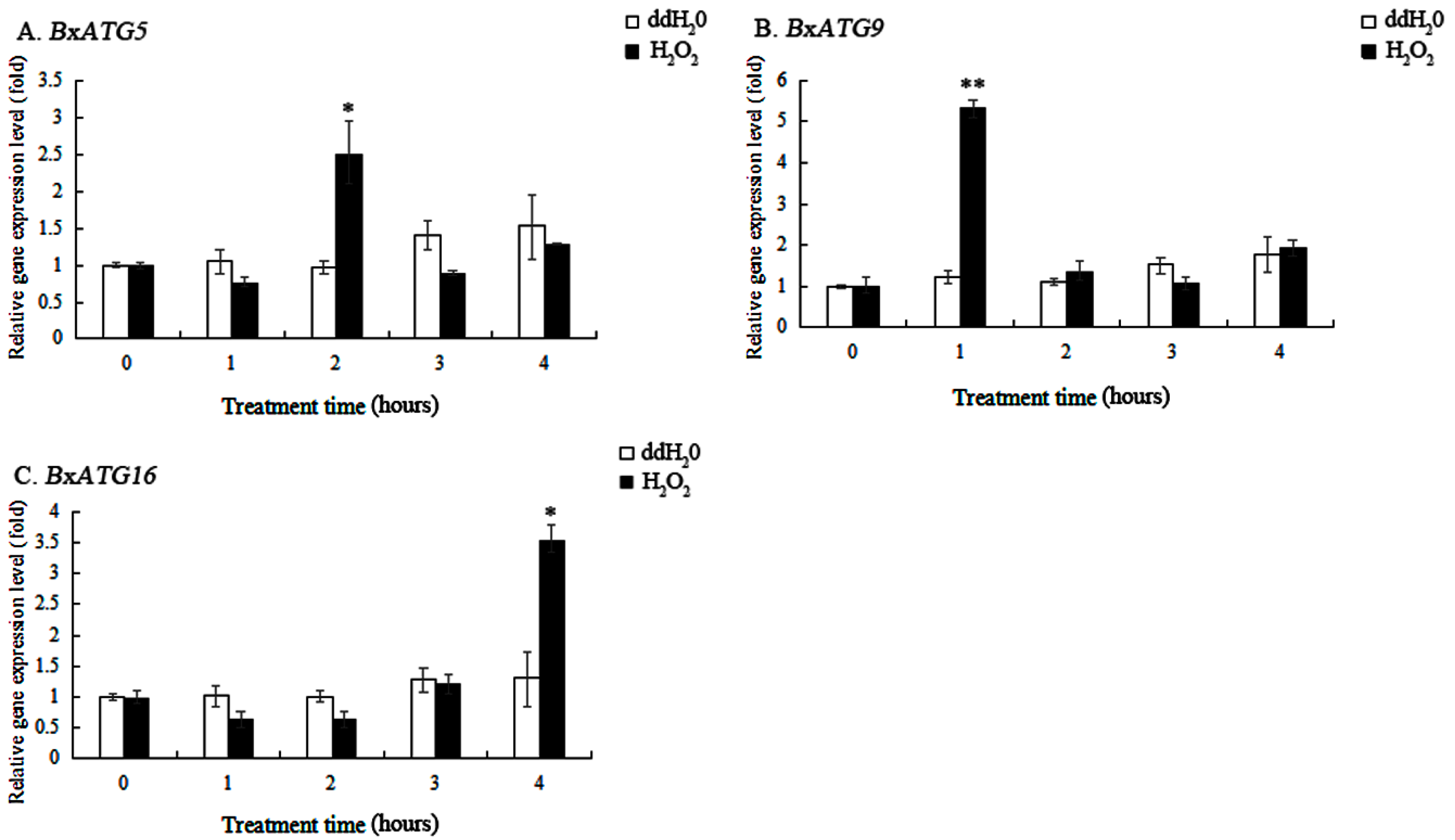
2.4. Response of Autophagy Genes in PWN to Oxidative Stress
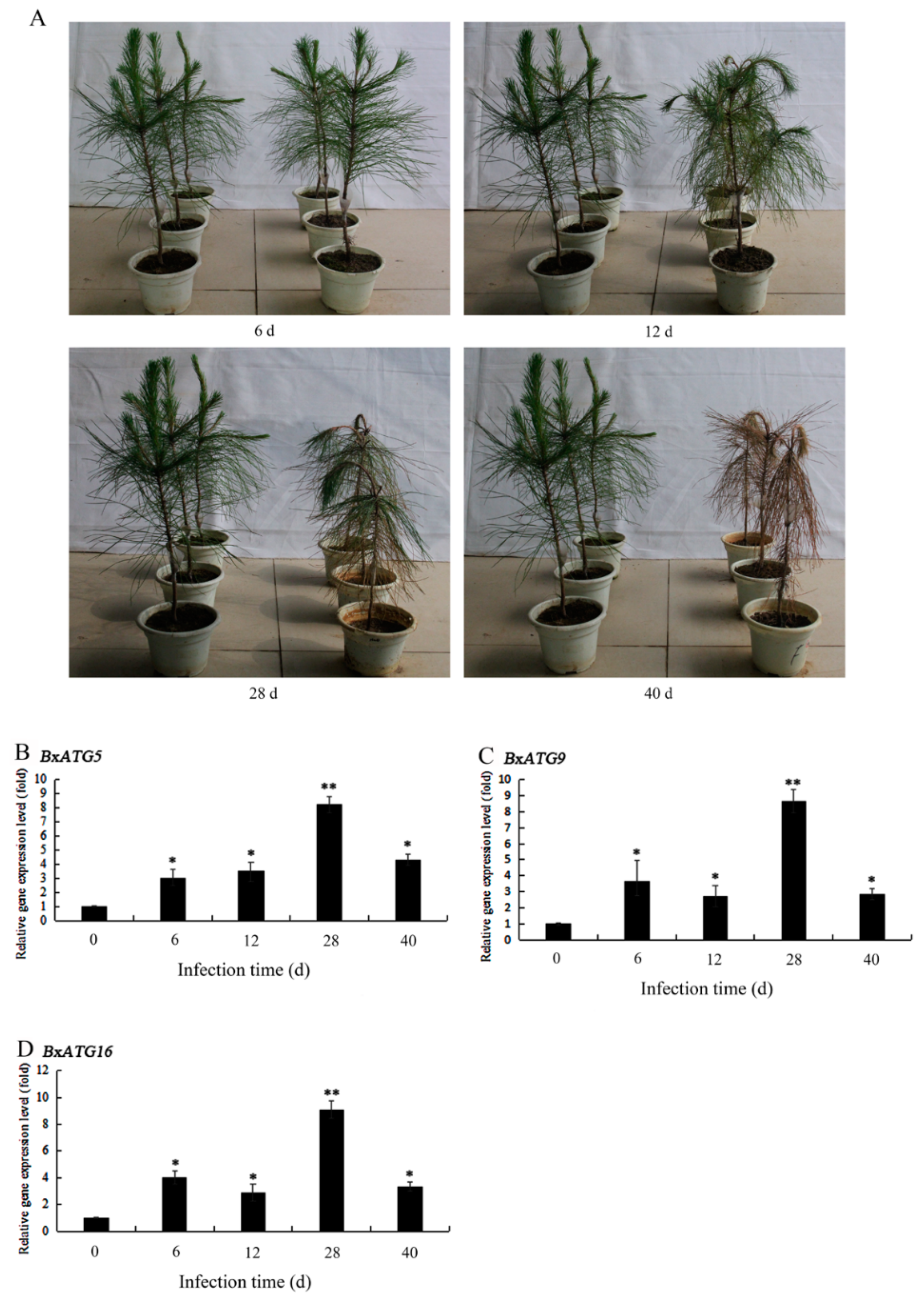
2.5. Expression Patterns of Autophagy Genes in PWN during the Development of PWD
3. Discussion
4. Materials and Methods
4.1. Biological Materials and Growth Conditions
4.2. PWN RNA Extraction and cDNA Synthesis
4.3. Cloning and Sequencing of Coding Sequences for the Autophagy Genes BxATG5, BxATG9, and BxATG16 from PWN
4.4. Bioinformatic Analysis of the Autophagy Genes BxATG5, BxATG9, and BxATG16 from PWN
4.5. Interference with the Autophagy Genes BxATG9 and BxATG16 Using Double-Stranded RNA
4.6. qRT-PCR
4.7. Analysis of Feeding and Reproduction of PWN after RNAi
4.8. Analysis of Expression Levels of BxATG5, BxATG9, and BxATG16 in PWN under Abiotic Stress and at Different Stages of PWD
4.9. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Mamiya, Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Ann. Rev. Phytopathol. 1983, 21, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.J.; Wang, Q.L. Distribution of the pinewood nematode in China and susceptibility of some Chinese and exotic pines to the nematode. Can. J. For. Res. 1989, 19, 1527–1530. [Google Scholar]
- Mota, M.M.; Braasch, H.; Bravo, M.A.; Penas, A.C.; Burgermeister, W.; Metge, K.; Sousa, E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar]
- Braasch, H.; Tomiczek, C.; Metge, K.; Hoyer, U.; Burgermeister, W.; Wulfert, I.; Schonfeld, U. Records of Bursaphelenchus spp. (Nematoda, Parasitaphelenchidae) in coniferous timber imported from the Asian part of Russia. For. Pathol. 2001, 31, 129–140. [Google Scholar] [CrossRef]
- Yang, B.J.; Pan, H.Y.; Tang, J.; Wang, Y.Y.; Wang, L.F. Pine Wood Nematode Disease; Forestry Publishing House: Beijing, China, 2003; pp. 45–48. [Google Scholar]
- Huang, L.; Wang, P.; Tian, M.Q.; Zhu, L.H.; Ye, J.R. Major sperm protein BxMSP10 is required for reproduction and egg hatching in Bursaphelenchus xylophilus. Exp. Parasitol. 2019, 197, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.Y.; Xu, M.; Xu, F.Y.; Ye, J.R. A comparative proteomics analysis of Pinus massoniana inoculated with Bursaphelenchus xylophilus. Pak. J. Bot. 2015, 47, 1271–1280. [Google Scholar]
- Li, L.; Tan, J.; Chen, F. Bacillus pumilus strain LYMC-3 shows nematicidal activity against Bursaphelenchus xylophilus via the production of a guanidine compound. Biocontrol Sci. Technol. 2018, 28, 1128–1139. [Google Scholar] [CrossRef]
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Palmisano, N.J.; Melendez, A. Autophagy in C. elegans development. Dev. Biol. 2019, 447, 103–125. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Meléndez, A.; TaUóczy, Z.; Seaman, M.; Eskelinen, E.L.; Hall, D.H.; Levine, B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003, 301, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005, 12, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.N.; Jeong, T.J. Overview of autophagy in plant cells. J. Life Sci. 2014, 24, 209–217. [Google Scholar] [CrossRef]
- Klionsky, D.J. Citing recent declines in the discovery of new ATG genes, some scientists now suggest that the end of autophagy research may be within sight. Autophagy 2014, 10, 715–716. [Google Scholar] [CrossRef] [Green Version]
- Lionaki, E.; Markaki, M.; Tavernarakis, N. Autophagy and ageing: Insights from invertebrate model organisms. Ageing Res. Rev. 2013, 12, 413–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Valent, B.; Farrall, L.; Chumley, F.G. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 1991, 127, 87–101. [Google Scholar]
- Fernández, J.M.F.; Ortega, A.G.; Cruz, R.R.; Camberos, E.P.; Alvarez, A.H.; Velazquez, M.M. Molecular cloning and characterization of two novel autophagy-related genes belonging to the ATG8 family from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Exp. Appl. Acarol. 2014, 64, 533–542. [Google Scholar] [CrossRef]
- Tindwa, H.; Jo, Y.H.; Patnaik, B.B.; Lee, Y.S.; Kang, S.S.; Han, Y.S. Molecular cloning and characterization of autophagy-related gene TmATG8 in Listeria-invaded hemocytes of Tenebrio molitor. Dev. Comp. Immunol. 2015, 51, 88–98. [Google Scholar] [CrossRef]
- Deng, L.N.; Wu, X.Q.; Ye, J.R.; Xue, Q. Identification of autophagy in the pine wood nematode Bursaphelenchus xylophilus and the molecular characterization and functional analysis of two novel autophagy-related genes, BxATG1 and BxATG8. Int. J. Mol. Sci. 2016, 17, 279. [Google Scholar] [CrossRef]
- Wu, F.; Deng, L.N.; Wu, X.Q.; Liu, H.B.; Ye, J.R. Expression profiling of autophagy genes BxATG1 and BxATG8 under biotic and abiotic stresses in pine wood nematode Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2017, 18, 2639. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Wu, F.; Wu, X.Q.; Ye, J.R. Differential effects of rapamycin on Bursaphelenchus xylophilus with different virulence and differential expression of autophagy genes under stresses in nematodes. Acta Biochim. Biophys. Sin. 2019, 51, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Wu, X.Q.; Feng, Y.Q.; Rui, L. Autophagy contributes to the feeding, reproduction, and mobility of Bursaphelenchus xylophilus at low temperatures. Acta Biochim. Biophys. Sin. 2019. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Suzuki, N.N.; Obara, K.; Fujioka, Y.; Ohsumi, Y.; Inagaki, F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007, 282, 6763–6772. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zulak, K.G.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant. Biol. 2010, 52, 8697. [Google Scholar] [CrossRef]
- Li, Y.X.; Meng, F.L.; Deng, X.; Wang, X.; Feng, Y.Q.; Zhang, W.; Pan, L.; Zhang, X.Y. Comparative transcriptome analysis of the pinewood nematode Bursaphelenchus xylophilus reveals the molecular mechanism underlying its defense response to host-derived α-pinene. Int. J. Mol. Sci. 2019, 20, 911. [Google Scholar] [CrossRef]
- Vellosillo, T.; Vicente, J.; Kulasekaran, S.; Hamberg, M.; Castresana, C. Emerging complexity in reactive oxygen species production and signaling during the response of plants to pathogens. Plant. Physiol. 2010, 154, 444–448. [Google Scholar] [CrossRef]
- Yu, L.Z.; Wu, X.Q.; Ye, J.R.; Zhang, S.N. Relationships between nitric oxide response signal and external factors during the early interaction between Pinus thunbergii and Bursaphelenchus xylophilus. Chin. J. Appl. Ecol. 2013, 24, 646–652. [Google Scholar]
- Boya, R.; Reggiori, R.; Codogno, P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013, 15, 713–720. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Martinet, W.; Agostinis, P.; Vanhoecke, B.; Dewaele, M.; De Meyer, G.R.Y. Autophagy in disease: A double-edged sword with therapeutic potential. Clin. Sci. 2009, 116, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Kuma, A.; Kobayashi, Y.; Yamamoto, A.; Matsubae, M.; Takao, T.; Natsume, T.; Ohsumi, Y.; Yoshimori, T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003, 116, 1679–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wesley, S.V.; Helliwell, C.A.; Smith, N.A.; Wang, M.B.; Rouse, D.T.; Liu, Q.; Gooding, P.S.; Singh, S.P.; Abbott, D.; Stoutjesdijk, P.A.; et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001, 27, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Wang, H.I.; Lu, H.C.; Chen, C.E.; Chen, H.H.; Yeh, H.H.; Tang, C.Y. An efficient RNA interference screening strategy for gene functional analysis. BMC Genom. 2012, 13, 491. [Google Scholar] [CrossRef]
- Ma, H.B.; Lu, Q.; Liang, J.; Zhang, X.Y. Functional analysis of the cellulose gene of the pine wood nematode, Bursaphelenchus xylophilus, using RNA interference. Genet. Mol. Res. 2011, 10, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Koh, Y.H.; Moon, Y.S.; Lee, S.H. Molecular properties of a venom allergen-like protein suggest a parasitic function in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 2012, 42, 63–70. [Google Scholar] [CrossRef]
- Cheng, X.Y.; Dai, S.M.; Xiao, L.; Xie, B.Y. Influence of cellulase gene knockdown by dsRNA interference on the development and reproduction of the pine wood nematode Bursaphelenchus xylophilus. Nematology 2010, 12, 225–233. [Google Scholar] [CrossRef]
- Xu, X.L.; Wu, X.Q.; Ye, J.R.; Huang, L. Molecular characterization and functional analysis of three pathogenesis-related Cytochrome P450 genes from Bursaphelenchus xylophilus (Tylenchida: Aphelenchoidoidea). Int. J. Mol. Sci. 2015, 16, 5216–5234. [Google Scholar] [CrossRef]
- Seybold, S.J.; Huber, D.P.W.; Lee, J.C.; Graves, A.D.; Bohlmann, J. Pine monoterpenes and pine bark beetles: A marriage of convenience for defense and chemical communication. Phytochem. Rev. 2006, 5, 143–178. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Kanzaki, N.; Futai, K. Volatile compounds in pine stands suffering from pine wilt disease: Qualitative and quantitative evaluation. Nematology 2006, 8, 869–880. [Google Scholar]
- Rafatian, G.; Khodagholi, F.; Farimani, M.M.; Abraki, S.B.; Gardaneh, M. Increase of autophagy and attenuation of apoptosis by Salvigenin promote survival of SH-SY5Y cells following treatment with H2O2. Mol. Cell. Biochem. 2012, 371, 9–22. [Google Scholar] [CrossRef] [PubMed]
- He, W.Q.; Xie, B.S.; He, D.Q.; Mao, Z.B. Autophagy has protective effects on hydrogen peroxide-induced 2BS premature aging cells. Chin. J. Biochem. Mol. Biol. 2009, 25, 943–948. [Google Scholar]
- Liu, J.; Wu, X.Q.; Ying, C.X.; He, L.X.; Ye, J.R. The difference of progenitive power and superoxide anion production in Bursaphelenchus xylophilus and B. mucronatus. J. Nanjing For. Univ. 2009, 33, 5–8. [Google Scholar]
- Scherz-Shouval, R.; Shvets, E.; Fass, E.; Shorer, H.; Gil, L.; Elazar, Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007, 26, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Asakura, M.; Ninomiya, S.; Sugimoto, M.; Oku, M.; Yamashita, S.I.; Okuno, T.; Sakai, Y.; Takano, Y. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbicular. Plant Cell 2009, 21, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Maehara, N.; Hata, K.; Futai, K. Effect of blue-stain fungi on the number of Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae) carried by Monochamus alternatus (Coleoptera: Cerambycidae). Nematology 2005, 7, 161–167. [Google Scholar] [CrossRef]
- Wu, X.Q.; Yuan, W.M.; Tian, X.J.; Fan, B.; Fang, X.; Ye, J.R.; Ding, X.L. Specific and functional diversity of endophytic bacteria from pine wood nematode Bursaphelenchus xylophilus with different virulence. Int. J. Biol. Sci. 2013, 9, 34–44. [Google Scholar] [CrossRef]
- Viglierchio, D.R.; Schmitt, R.V. On the methodology of nematode extraction from field samples: Baermann funnel modifications. J. Nematol. 1983, 15, 438–444. [Google Scholar]
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant-Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Primers | Sequence (5′ to 3′) |
|---|---|
| BxATG5-F | ATGGATGACTATGAGGTCCG |
| BxATG5-R | TTATTTAGATTTGAAGATGAGATGG |
| BxATG9-F | ATGTCTATGTTTTTTTCCAATCG |
| BxATG9-R | TTAGACATTAAACCCGCCTG |
| BxATG16-F | ATGACTTACAGAGATGACATCCT |
| BxATG16-R | TTACATCCACAAACAGGCA |
| M13F (−47) | CGCCAGGGTTTTCCCAGTCACGAC |
| M13R (−48) | AGCGGATAACAATTTCACACAGGA |
| iBxATG9-F | TAATACGACTCACTATAGGGAGAATTACAATGAGTTGGATCACG |
| iBxATG9-R | TAATACGACTCACTATAGGGAGATGATCGGCATAACAGGG |
| iBxATG16-F | TAATACGACTCACTATAGGGAGAAATGATGAACTTTTGGCTCT |
| iBxATG16-R | TAATACGACTCACTATAGGGAGACTTCCGGGAGCTTCTTC |
| iBxATG9-qF | AAGACTGAAGTTGAGACA |
| iBxATG9-qR | ATTATGGCGAAGATGGAT |
| iBxATG16-qF | AACTACTAACGAATTGCTAA |
| iBxATG16-qR | ATCAACACCACTCTTTCT |
| Actin-F | GCAACACGGAGTTCGTTGTAGA |
| Actin-R | GTATCGTCACCAACTGGGATGA |
| qBxATG5-F | CAAACGATGAAACAACTCCTTAT |
| qBxATG5-R | CGAAGCCGCTGATATTACA |
| qBxATG9-F | GCGGTCAAGTATATGGAT |
| qBxATG9-R | ATTATGGCGAAGATGGAT |
| qBxATG16-F | CCAGTTCTCATTACGATAA |
| qBxATG16-R | AGTAGTTGACCATCTGTA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.-B.; Rui, L.; Feng, Y.-Q.; Wu, X.-Q. Molecular Characterization and Functional Analysis of Three Autophagy Genes, BxATG5, BxATG9, and BxATG16, in Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2019, 20, 3769. https://doi.org/10.3390/ijms20153769
Liu H-B, Rui L, Feng Y-Q, Wu X-Q. Molecular Characterization and Functional Analysis of Three Autophagy Genes, BxATG5, BxATG9, and BxATG16, in Bursaphelenchus xylophilus. International Journal of Molecular Sciences. 2019; 20(15):3769. https://doi.org/10.3390/ijms20153769
Chicago/Turabian StyleLiu, Hong-Bin, Lin Rui, Ya-Qi Feng, and Xiao-Qin Wu. 2019. "Molecular Characterization and Functional Analysis of Three Autophagy Genes, BxATG5, BxATG9, and BxATG16, in Bursaphelenchus xylophilus" International Journal of Molecular Sciences 20, no. 15: 3769. https://doi.org/10.3390/ijms20153769





