The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress
Abstract
1. Introduction
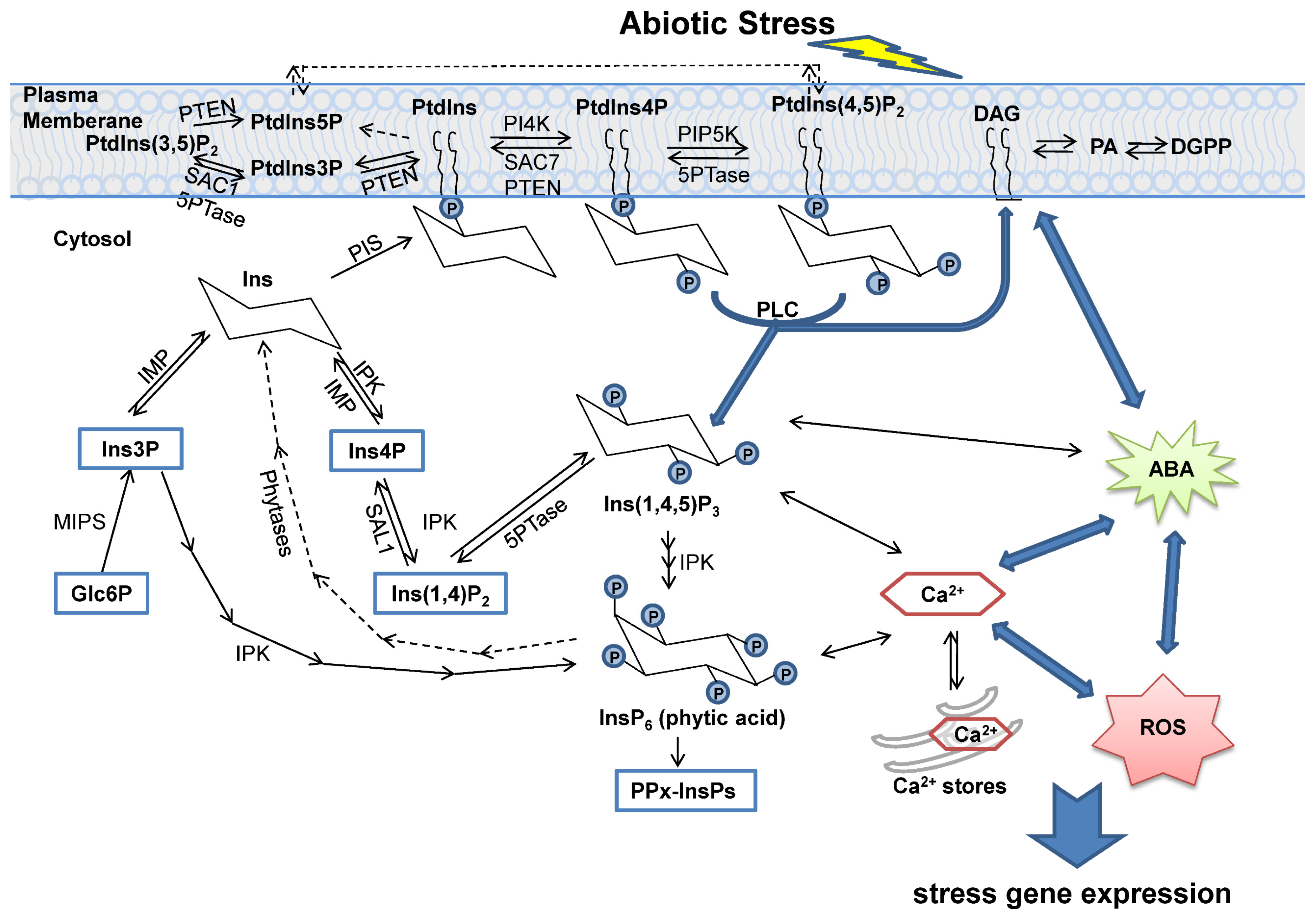
2. The Biosynthesis and Degradation of Inositol and Its Derivatives
3. Phosphatases in Inositol Signaling Pathways
4. Function of Inositol Phosphatases under Abiotic Stress
4.1. 5PTases and Plant Responses to Abiotic Stress
4.2. SAL1 and Plant Responses to Abiotic Stress
4.3. IMPs and Plant Responses to Abiotic Stress
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | abscisic acid |
| ABRE | abscisic acid responsiveness |
| BST1 | BRISTLED1 |
| CBF | CRT-binding factor |
| CVL1 | CVP2-like1 |
| CVP2 | cotyledon vascular pattern2 |
| DAG | diacylglycerol |
| DGPP | diacylglycerol pyrophosphate |
| DREB2A | dehydration-responsive element-binding protein 2A |
| ER | endoplasmic reticulum |
| FRA3 | fragile fiber 3 |
| FRA7 | fragile fiber 7 |
| FRY1 | FIERY1 |
| Glc6P | glucose-6-phosphate |
| GARE | gibberellins responsive |
| HOS2 | high expression of osmotic stress-regulated gene expression 2 |
| IMP | myo-inositol monophosphatase |
| IMPL | inositol monophosphatase-like |
| Ins | inositol |
| InsP 5-ptase | inositol polyphosphate 5-phosphatase |
| IP | inositol phosphate |
| IP3 | Inositol(1,4,5)trisphosphate |
| IPK | inositol polyphosphate multi kinase |
| IPPc | inositol polyphosphate phosphatase catalytic |
| LTR | low-temperature responsiveness |
| MBS | MYB binding site |
| MDA | malondialdehyde |
| MRH3 | root hair morphogenesis 3 |
| MIPS | myo-inositol-3-phosphate synthase |
| P | phosphate |
| PA | phosphatidic acid |
| PAP | 3′-phosphoadenosine 5’-phosphate |
| PAPS | 2’-PAP and 3’-phosphoadenosine 5’-phosphosulfate |
| PI | phosphoinositide |
| PIP5K | PtIns4P 5-kinase |
| PIP4K | PtIns4P 4-kinase |
| PIS | phosphatidylinositol synthase |
| PKC | protein kinase C |
| PLC | phospholipase C |
| PPx-InsPs | pyrophosphates |
| PTEN | phosphatase and tensin homologue deleted on chromosome 10 |
| PtdIns | phosphatidylinositol |
| RHD4 | root hair defective 4 |
| RON1 | rotunda 1 |
| ROS | reactive oxygen species |
| SAC | suppressor of actin |
| SnRK1.1 | sucrose nonfermenting-1-related kinase |
| TCA | salicylic acid responsiveness |
| VTC4 | vitamin C 4 |
| XRN | 5′ to 3′ exoribonuclease |
| 5PTases | inositol polyphosphate 5-phosphatases |
References
- Valluru, R.; Van den Ende, W. myo-inositol and beyond—Emerging networks under stress. Plant Sci. 2011, 181, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Michell, R.H. Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, M.; Heilmann, I. Plant phosphoinositides—Complex networks controlling growth and adaptation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2015, 1851, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, G.E. The cellular language of myo-inositol signaling. New Phytol. 2011, 192, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T.; Vermeer, J.E.M. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 2010, 33, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ufer, G.; Bartels, D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016, 39, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.-W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef]
- Michell, R.H. Inositol and its derivatives: Their evolution and functions. Adv. Enzyme Regul. 2011, 51, 84–90. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Gillaspy, G.E.; Keddie, J.S.; Oda, K.; Gruissem, W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell 1995, 7, 2175–2185. [Google Scholar]
- Sato, Y.; Yazawa, K.; Yoshida, S.; Tamaoki, M.; Nakajima, N.; Iwai, H.; Ishii, T.; Satoh, S. Expression and functions of myo-inositol monophosphatase family genes in seed development of Arabidopsis. J. Plant Res. 2011, 124, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Heilmann, I. Phosphoinositide signaling in plant development. Development 2016, 143, 2044–2055. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.-C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, W.; Ökrész, L.; Bögre, L.; Munnik, T. Learning the lipid language of plant signalling. Trends Plant Sci. 2004, 9, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.J.; Yang, G. Inositol 1,4,5-trisphosphate 3-kinases: Functions and regulations. Cell Res. 2005, 15, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.P.; Gillaspy, G.E.; Perera, I.Y. Biosynthesis and possible functions of inositol pyrophosphates in plants. Front. Plant Sci. 2015, 6, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Luo, Z.; Kong, S.; Zhao, Y.; Zhang, C.; Zhang, W.; Yuan, H.; Cheng, L. Expansion and Functional Divergence of Inositol Polyphosphate 5-Phosphatases in Angiosperms. Genes 2019, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Majerus, P.W.; Kisseleva, M.V.; Norris, F.A. The Role of Phosphatases in Inositol Signaling Reactions. J. Biol. Chem. 1999, 274, 10669–10672. [Google Scholar] [CrossRef] [PubMed]
- Berdy, S.E.; Kudla, J.; Gruissem, W.; Gillaspy, G.E. Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol. 2001, 126, 801–810. [Google Scholar] [CrossRef]
- Sanchez, J.-P.; Chua, N.-H. Arabidopsis PLC1 Is Required for Secondary Responses to Abscisic Acid Signals. Plant Cell 2001, 13, 1143–1154. [Google Scholar] [CrossRef]
- Burnette, R.N. An Arabidopsis Inositol 5-Phosphatase Gain-of-Function Alters Abscisic Acid Signaling. Plant Physiol. 2003, 132, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Despres, B.; Bouissonnié, F.; Wu, H.-J.; Gomord, V.; Guilleminot, J.; Grellet, F.; Berger, F.; Delseny, M.; Devic, M. Three SAC1-like genes show overlapping patterns of expression in Arabidopsis but are remarkably silent during embryo development. Plant J. 2003, 34, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.-H. The SAC Domain-Containing Protein Gene Family in Arabidopsis. Plant Physiol. 2003, 132, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Williams, M.E.; Torabinejad, J.; Cohick, E.; Parker, K.; Drake, E.J.; Thompson, J.E.; Hortter, M.; Dewald, D.B. Mutations in the Arabidopsis phosphoinositide phosphatase gene SAC9 lead to overaccumulation of PtdIns(4,5)P2 and constitutive expression of the stress-response pathway. Plant Physiol. 2005, 138, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Thole, J.M.; Vermeer, J.E.M.; Zhang, Y.; Gadella, T.W.J.; Nielsen, E. Root hair defective4 encodes a phosphatidylinositol-4-phosphate phosphatase required for proper root hair development in Arabidopsis thaliana. Plant Cell 2008, 20, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Nováková, P.; Hirsch, S.; Feraru, E.; Tejos, R.; van Wijk, R.; Viaene, T.; Heilmann, M.; Lerche, J.; De Rycke, R.; Feraru, M.I.; et al. SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 2818–2823. [Google Scholar] [CrossRef] [PubMed]
- Ercetin, M.E.; Gillaspy, G.E. Molecular characterization of an Arabidopsis gene encoding a phospholipid-specific inositol polyphosphate 5-phosphatase. Plant Physiol. 2004, 135, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Gunesekera, B.; Torabinejad, J.; Robinson, J.; Gillaspy, G.E. Inositol Polyphosphate 5-Phosphatases 1 and 2 Are Required for Regulating Seedling Growth. Plant Physiol. 2007, 143, 1408–1417. [Google Scholar] [CrossRef]
- Ercetin, M.E. Molecular Characterization and Loss-of-Function Analysis of an Arabidopsis thaliana Gene Encoding a Phospholipid-Specific Inositol Polyphosphate 5-Phosphatase. Ph.D. Thesis, Virginia Polytechnic Institute and State University, Blacksburg, VA, USA, 19 May 2005. [Google Scholar]
- Jones, M.A.; Raymond, M.J.; Smirnoff, N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 2006, 45, 83–100. [Google Scholar] [CrossRef]
- Parker, J.S.; Cavell, A.C.; Dolan, L.; Roberts, K.; Grierson, C.S. Genetic Interactions during Root Hair Morphogenesis in Arabidopsis. Plant Cell 2000, 12, 1961–1974. [Google Scholar] [CrossRef]
- Carland, F.M.; Nelson, T. COTYLEDON VASCULAR PATTERN2–Mediated Inositol (1,4,5) Triphosphate Signal Transduction Is Essential for Closed Venation Patterns of Arabidopsis Foliar Organs. Plant Cell 2004, 16, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Carland, F.; Nelson, T. CVP2- and CVL1-mediated phosphoinositide signaling as a regulator of the ARF GAP SFC/VAN3 in establishment of foliar vein patterns. Plant J. 2009, 59, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Villalon, A.; Gujas, B.; van Wijk, R.; Munnik, T.; Hardtke, C.S. Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 2015, 142, 1437–1446. [Google Scholar] [CrossRef] [PubMed]
- Kaye, Y.; Golani, Y.; Singer, Y.; Leshem, Y.; Cohen, G.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol Polyphosphate 5-Phosphatase7 Regulates the Production of Reactive Oxygen Species and Salt Tolerance in Arabidopsis. Plant Physiol. 2011, 157, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Golani, Y.; Kaye, Y.; Gilhar, O.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol Polyphosphate Phosphatidylinositol 5-Phosphatase9 (At5PTase9) Controls Plant Salt Tolerance by Regulating Endocytosis. Mol. Plant 2013, 6, 1781–1794. [Google Scholar] [CrossRef] [PubMed]
- Ercetin, M.E.; Ananieva, E.A.; Safaee, N.M.; Torabinejad, J.; Robinson, J.Y.; Gillaspy, G.E. A phosphatidylinositol phosphate-specific myo-inositol polyphosphate 5-phosphatase required for seedling growth. Plant Mol. Biol. 2008, 67, 375–388. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.-H. Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1720–1728. [Google Scholar] [CrossRef]
- Wang, Y.; Chu, Y.-J.; Xue, H.-W. Inositol polyphosphate 5-phosphatase-controlled Ins(1,4,5)P3/Ca2+ is crucial for maintaining pollen dormancy and regulating early germination of pollen. Development 2012, 139, 2221–2233. [Google Scholar] [CrossRef]
- Chen, X.; Lin, W.-H.; Wang, Y.; Luan, S.; Xue, H.-W. An Inositol Polyphosphate 5-Phosphatase Functions in PHOTOTROPIN1 Signaling in Arabidopis by Altering Cytosolic Ca2+. Plant Cell 2008, 20, 353–366. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Gillaspy, G.E.; Ely, A.; Burnette, R.N.; Erickson, F.L. Interaction of the WD40 Domain of a myoinositol Polyphosphate 5-Phosphatase with SnRK1 Links Inositol, Sugar, and Stress Signaling. Plant Physiol. 2008, 148, 1868–1882. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, W.-H.; Chen, X.; Xue, H.-W. The role of Arabidopsis 5PTase13 in root gravitropism through modulation of vesicle trafficking. Cell Res. 2009, 19, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Wang, Y.; Mueller-Roeber, B.; Brearley, C.A.; Xu, Z.-H.; Xue, H.-W. At5PTase13 Modulates Cotyledon Vein Development through Regulating Auxin Homeostasis. Plant Physiol. 2005, 139, 1677–1691. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Burk, D.H.; Morrison, W.H.; Ye, Z.-H. FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 2004, 16, 3242–3259. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Burk, D.H.; Nairn, C.J.; Wood-Jones, A.; Morrison, W.H.; Ye, Z.-H. Mutation of SAC1, an Arabidopsis SAC domain phosphoinositide phosphatase, causes alterations in cell morphogenesis, cell wall synthesis, and actin organization. Plant Cell 2005, 17, 1449–1466. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, A.H.; Youssef, N.N.; DeWald, D.B. Unique cell wall abnormalities in the putative phosphoinositide phosphatase mutant AtSAC9. Planta 2011, 234, 993–1005. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Garciadeblás, B.; Rodríguez-Navarro, A. The SAL1 gene of Arabidopsis, encoding an enzyme with 3’(2’),5’-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 1996, 8, 529–537. [Google Scholar] [PubMed]
- Xiong, L.; Lee, B.; Ishitani, M.; Lee, H.; Zhang, C.; Zhu, J.K. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001, 15, 1971–1984. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, B.; Hicks, L.M.; Xiong, L. A Nucleotide Metabolite Controls Stress-Responsive Gene Expression and Plant Development. PLoS ONE 2011, 6, e26661. [Google Scholar] [CrossRef]
- Kim, B.-H.; von Arnim, A.G. FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J. 2009, 58, 208–219. [Google Scholar] [CrossRef]
- Wilson, P.B.; Estavillo, G.M.; Field, K.J.; Pornsiriwong, W.; Carroll, A.J.; Howell, K.A.; Woo, N.S.; Lake, J.A.; Smith, S.M.; Harvey Millar, A.; et al. The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 2009, 58, 299–317. [Google Scholar] [CrossRef]
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for a SAL1-PAP Chloroplast Retrograde Pathway That Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.X.; Mabbitt, P.D.; Phua, S.Y.; Mueller, J.W.; Nisar, N.; Gigolashvili, T.; Stroeher, E.; Grassl, J.; Arlt, W.; Estavillo, G.M.; et al. Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase. Proc. Natl. Acad. Sci. USA 2016, 113, E4567–E4576. [Google Scholar] [CrossRef] [PubMed]
- Robles, P.; Fleury, D.; Candela, H.; Cnops, G.; Alonso-Peral, M.M.; Anami, S.; Falcone, A.; Caldana, C.; Willmitzer, L.; Ponce, M.R.; et al. The RON1/FRY1/SAL1 gene is required for leaf morphogenesis and venation patterning in Arabidopsis. Plant Physiol. 2010, 152, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Vanneste, S.; Brewer, P.B.; Michniewicz, M.; Grones, P.; Kleine-Vehn, J.; Löfke, C.; Teichmann, T.; Bielach, A.; Cannoot, B.; et al. Inositol trisphosphate-induced Ca2+ signaling modulates auxin transport and PIN polarity. Dev. Cell 2011, 20, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Gil-Mascarell, R.; López-Coronado, J.M.; Bellés, J.M.; Serrano, R.; Rodríguez, P.L. The Arabidopsis HAL2-like gene family includes a novel sodium-sensitive phosphatase. Plant J. 1999, 17, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Torabinejad, J.; Donahue, J.L.; Gunesekera, B.N.; Allen-Daniels, M.J.; Gillaspy, G.E. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, A.; Collakova, E.; Gillaspy, G.E. Characterization of the inositol monophosphatase gene family in Arabidopsis. Front. Plant Sci. 2015, 5, 725. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Zhou, L.-Z.; Fox, E.; Pao, J.; Sun, W.; Zhou, C.; McCormick, S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011, 68, 1081–1092. [Google Scholar] [CrossRef]
- Gupta, R.; Ting, J.T.L.; Sokolov, L.N.; Johnson, S.A.; Luan, S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 2002, 14, 2495–2507. [Google Scholar] [CrossRef]
- Pribat, A.; Sormani, R.; Rousseau-Gueutin, M.; Julkowska, M.M.; Testerink, C.; Joubès, J.; Castroviejo, M.; Laguerre, M.; Meyer, C.; Germain, V.; et al. A novel class of PTEN protein in Arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 2012, 441, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Xu, H.; Xu, W.; He, Z.; Xu, W.; Ma, M. A SAL1 Loss-of-Function Arabidopsis Mutant Exhibits Enhanced Cadmium Tolerance in Association with Alleviation of Endoplasmic Reticulum Stress. Plant Cell Physiol. 2016, 57, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Litthauer, S.; Chan, K.X.; Jones, M.A. 3’-Phosphoadenosine 5’-Phosphate Accumulation Delays the Circadian System. Plant Physiol. 2018, 176, 3120–3135. [Google Scholar] [CrossRef] [PubMed]
- Phua, S.Y.; Yan, D.; Chan, K.X.; Estavillo, G.M.; Nambara, E.; Pogson, B.J. The Arabidopsis SAL1-PAP Pathway: A Case Study for Integrating Chloroplast Retrograde, Light and Hormonal Signaling in Modulating Plant Growth and Development? Front. Plant Sci. 2018, 9, 1171. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.N.; Marineo, S.; Mandala, S.; Davids, F.; Sewell, B.T.; Ingle, R.A. The Missing Link in Plant Histidine Biosynthesis: Arabidopsis myoinositol monophosphatase-like2 Encodes a Functional Histidinol-Phosphate Phosphatase. Plant Physiol. 2010, 152, 1186–1196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.-X.; Qin, L.-J.; Zhao, D.-G. Overexpression of the OsIMP Gene Increases the Accumulation of Inositol and Confers Enhanced Cold Tolerance in Tobacco through Modulation of the Antioxidant Enzymes’ Activities. Genes 2017, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Joshi-Saha, A.; Reddy, K.S. Repeat length variation in the 5’UTR of myo-inositol monophosphatase gene is related to phytic acid content and contributes to drought tolerance in chickpea (Cicer arietinum L.). J. Exp. Bot. 2015, 66, 5683–5690. [Google Scholar] [CrossRef]
- Saxena, S.C.; Salvi, P.; Kaur, H.; Verma, P.; Petla, B.P.; Rao, V.; Kamble, N.; Majee, M. Differentially expressed myo-inositol monophosphatase gene (CaIMP) in chickpea (Cicer arietinum L.) encodes a lithium-sensitive phosphatase enzyme with broad substrate specificity and improves seed germination and seedling growth under abiotic stresses. J. Exp. Bot. 2013, 64, 5623–5639. [Google Scholar] [CrossRef]
- Perera, I.Y.; Hung, C.-Y.; Moore, C.D.; Stevenson-Paulik, J.; Boss, W.F. Transgenic Arabidopsis Plants Expressing the Type 1 Inositol 5-Phosphatase Exhibit Increased Drought Tolerance and Altered Abscisic Acid Signaling. Plant Cell 2008, 20, 2876–2893. [Google Scholar] [CrossRef]
- Lin, W.H.; Ye, R.; Ma, H.; Xu, Z.H.; Xue, H.W. DNA chip-based expression profile analysis indicates involvement of the phosphatidylinositol signaling pathway in multiple plant responses to hormone and abiotic treatments. Cell Res. 2004, 14, 34–45. [Google Scholar] [CrossRef]
- Xiong, L.; Lee, H.; Huang, R.; Zhu, J.-K. A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J. 2004, 40, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Gašparič, M.B.; Lenassi, M.; Gostinčar, C.; Rotter, A.; Plemenitaš, A.; Gunde-Cimerman, N.; Gruden, K.; Žel, J. Insertion of a Specific Fungal 3′-phosphoadenosine-5′-phosphatase Motif into a Plant Homologue Improves Halotolerance and Drought Tolerance of Plants. PLoS ONE 2013, 8, e81872. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.-S.; Koo, N.S.-C.; Li, F.W.-Y.; Li, M.-W.; Wang, H.; Tsai, S.-N.; Sun, F.; Lim, B.L.; Ko, W.-H.; Lam, H.-M. GmSAL1 hydrolyzes inositol-1,4,5-trisphosphate and regulates stomatal closure in detached leaves and ion compartmentalization in plant cells. PLoS ONE 2013, 8, e78181. [Google Scholar] [CrossRef] [PubMed]
- Ruszkowski, M.; Dauter, Z. Structural Studies of Medicago truncatula Histidinol Phosphate Phosphatase from Inositol Monophosphatase Superfamily Reveal Details of Penultimate Step of Histidine Biosynthesis in Plants. J. Biol. Chem. 2016, 291, 9960–9973. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, V.; Parida, S.K.; Chattopadhyay, D. A repeat length variation in myo-inositol monophosphatase gene contributes to seed size trait in chickpea. Sci. Rep. 2017, 7, 4764. [Google Scholar] [CrossRef] [PubMed]
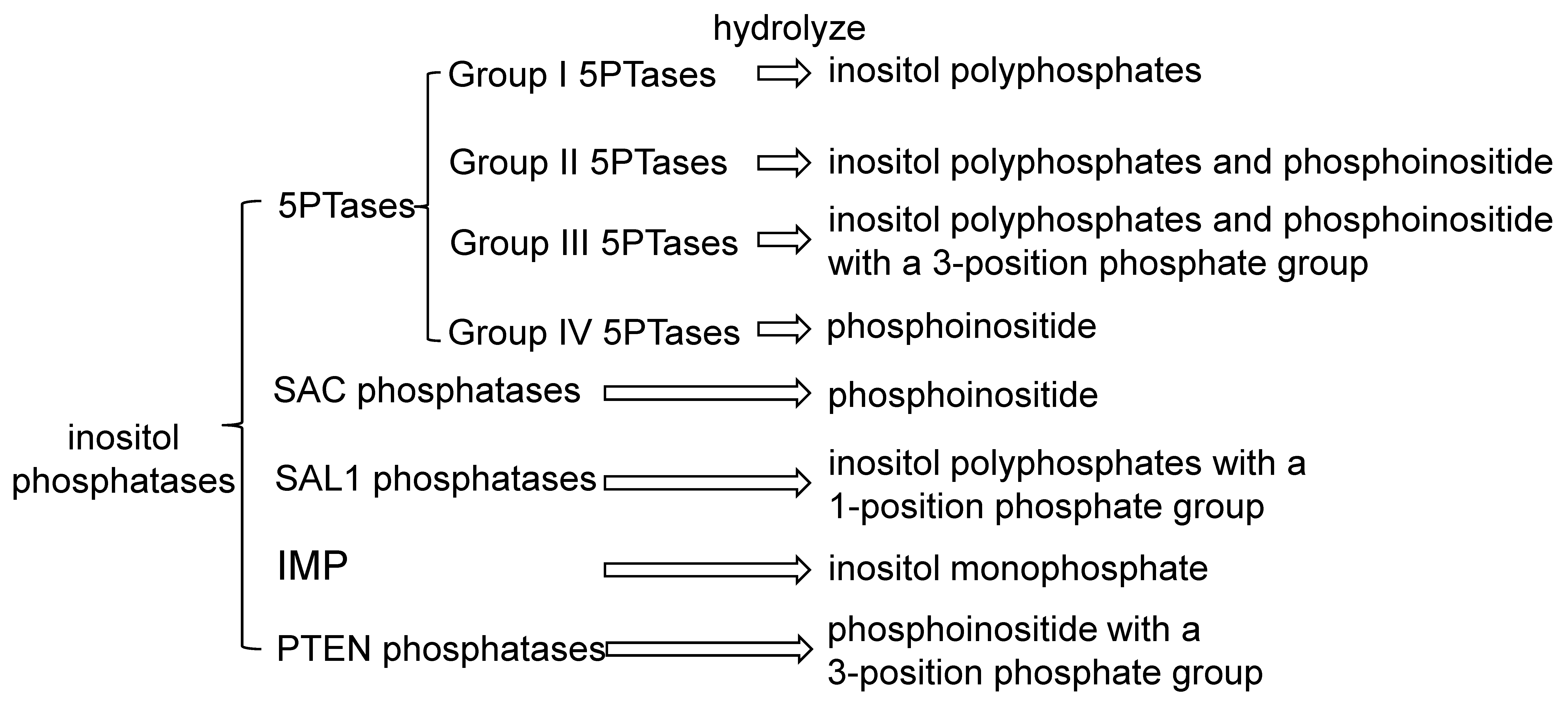
| Name | Gene ID | Substrates | Cellular Localization | Expression Patterns | Function | References |
|---|---|---|---|---|---|---|
| 5PTase—hydrolyze inositol-5-phosphate | ||||||
| At5TPase1 | At1G34120 | Group II Ins(1,4,5)P3, Ins(1,3,4,5)P4, PtdIns(4,5)P2 | - | leaf, flower, bolt, seedling | alter ABA and light signaling, stomatal opening, seedling development | [19,21,27,28] |
| At5TPase2 | At4G18010 | Group II Ins(1,4,5)P3, Ins(1,3,4,5)P4, PtdIns(4,5)P2 | - | leaf, flower, bolt, seedling | alter ABA signaling, seedling development | [20,27,28] |
| At5TPase3 | At1G71710 | Group II PtdIns(4,5)P2, PtdIns(3,4,5)P3, Ins(1,4,5)P3, Ins(1,3,4,5)P4, | - | - | - | [29] |
| At5TPase4 | At3G63240 | Group IV PtdIns(4,5)P2 | - | - | - | [29] |
| At5TPase5/MRH3/BST1 | At5G65090 | - | - | - | root hair development | [30,31] |
| At5TPase6/CVP2 | At1G05470 | Group IV PtdIns(4,5)P2, PtdIns(3,4,5)P3 | - | vascular system | foliar vein patterning, root branching | [32,33,34] |
| At5TPase7/ CVL1 | At2G32010 | Group IV PtdIns(4,5)P2, PtdIns(3,4,5)P3 | plasma membrane, nuclear speckles | vascular system | foliar vein patterning, root branching, salt tolerance, and ROS production | [33,34,35] |
| At5TPase8 | At2G37440 | - | - | - | - | - |
| At5TPase9 | At2G01900 | Group IV PtdIns(4,5)P2, PtdIns(3,4,5)P3 | - | root | salt tolerance and ROS production endocytosis | [36] |
| At5TPase10 | At5G04980 | - | - | - | - | - |
| At5TPase11 | At1G47510 | Group IV PtdIns(4,5)P2, PtdIns(3,5)P2, PtdIns(3,4,5)P3 | cell surface or plasma membrane | flower, leaf, root, silique, bolt, seedling | seedling development | [27,37] |
| At5TPase12 | At2G43900 | Group I Ins(1,4,5)P3 | - | pollen grain, leaf and flower (mostly); root, stem and young seedling (weakly) | pollen dormancy/germination | [38,39] |
| At5TPase13 | At1G05630 | Group I Ins(1,4,5)P3, | nucleus | young seedlings, flowers | cotyledon vein development, alter auxin, ABA, sugar and PHOTOTROPIN1 signaling, root gravitropism, vesicle trafficking | [38,39,40,41,42,43] |
| At5TPase14 | At2G31830 | Group II PtdIns(4,5)P2, PtdIns(3,4,5)P3, Ins(1,4,5)P3 | - | pollen grain | - | [38,39] |
| At5TPase15/ FRA3 | At1G65580 | Group II PtdIns(4,5)P2, PtdIns(3,4,5)P3, Ins(1,4,5)P3 | - | seedling, stem, root, flower, mature leaf (weak) | secondary wall synthesis and actin organization | [44] |
| SAC—hydrolyze phosphatidylinositol phosphates | ||||||
| SAC1/FRA7 | At1G22620 | PtdIns(3,5)P2 | Golgi | ubiquitous, predominant in vascular tissues and fibers of stems | cell morphogenesis, cell wall synthesis, actin organization | [23,45] |
| SAC2 | At3G14205 | - | tonoplast | ubiquitous | vacuolar function | [23,26] |
| SAC3 | At3G43220 | - | tonoplast | ubiquitous | vacuolar function | [23,26] |
| SAC4 | At5G20840 | - | tonoplast | ubiquitous | vacuolar function | [23,26] |
| SAC5 | At1G17340 | - | tonoplast | ubiquitous | vacuolar function | [23,26] |
| SAC6/SAC1b | At5G66020 | - | endoplasmic reticulum | pollen grain | embryo development | [22,23] |
| SAC7/SAC1c/RHD4 | At3G51460 | PtdIns4P | endoplasmic reticulum | most tissues (strong) | embryo development, root hair development | [22,23,25] |
| SAC8/ AtSAC1a | At3G51830 | - | endoplasmic reticulum | hypocotyls of seedlings, pollen grain, most tissues (week), | embryo development | [22,23] |
| SAC9 | At3G59770 | - | - | root (strong), leaf and shoot (weak) | cell wall formation, stress response | [23,24,46] |
| SAL—hydrolyze inositol-1-phosphate | ||||||
| AtSAL1/ AtFIERY1 (AtFRY1)/ HOS2/RON1 | At5G63980 | Ins(1,4)P2, Ins(1,3,4)P3, PAP, PAPS | chloroplast, mitochondria | vascular tissue | alter ABA, auxin and stress signaling (cold, drought, salt, lithium, high light, cadmium), venation patterning | [47,48,49,50,51,52,53,54,55] |
| AtSAL2 | At5G64000 | Ins(1,4)P2, PAP, | - | - | - | [56] |
| IMP—hydrolyze inositol-3-phosphate, inositol-4-phosphate | ||||||
| IMP/VTC4 | At3G02870 | Ins3P, Ins1P, L-Galactose-1-P | cytosol | photosynthetictissues | seed development, ascorbate biosynthesis, alter cold, salt and ABA responses | [11,57,58,59] |
| IMPL1 | At1G31190 | Ins3P, Ins1P, Ins2P, L-Galactose-1-P | chloroplast | ubiquitous | seed development | [11,57,59] |
| IMPL2 | At4G39120 | Histidinol 1-P | chloroplast | root (strong), hypocotyl (weak) | seed development, histidinebiosynthesis | [11,57,59] |
| PTEN—hydrolyze inositol-3-phosphate | ||||||
| PTEN1 | At5G39400 | PtdIns(3,4,5)P3, phosphotyrosin | vesicles, autophagic body | pollen grain | pollen development | [60,61] |
| PTEN2a | At3G19420 | PtdIns3P, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns4P, PtdIns(3,4,5)P3, phosphotyrosin | - | seedling, leaf, flower, silique | - | [62] |
| PTEN2b | At3G50110 | PtdIns3P, phosphotyrosin | - | seedling, leaf, flower, silique | - | [62] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Q.; Kong, D.; Li, Q.; Sun, S.; Song, J.; Zhu, Y.; Liang, K.; Ke, Q.; Lin, W.; Huang, J. The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress. Int. J. Mol. Sci. 2019, 20, 3999. https://doi.org/10.3390/ijms20163999
Jia Q, Kong D, Li Q, Sun S, Song J, Zhu Y, Liang K, Ke Q, Lin W, Huang J. The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress. International Journal of Molecular Sciences. 2019; 20(16):3999. https://doi.org/10.3390/ijms20163999
Chicago/Turabian StyleJia, Qi, Defeng Kong, Qinghua Li, Song Sun, Junliang Song, Yebao Zhu, Kangjing Liang, Qingming Ke, Wenxiong Lin, and Jinwen Huang. 2019. "The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress" International Journal of Molecular Sciences 20, no. 16: 3999. https://doi.org/10.3390/ijms20163999
APA StyleJia, Q., Kong, D., Li, Q., Sun, S., Song, J., Zhu, Y., Liang, K., Ke, Q., Lin, W., & Huang, J. (2019). The Function of Inositol Phosphatases in Plant Tolerance to Abiotic Stress. International Journal of Molecular Sciences, 20(16), 3999. https://doi.org/10.3390/ijms20163999





