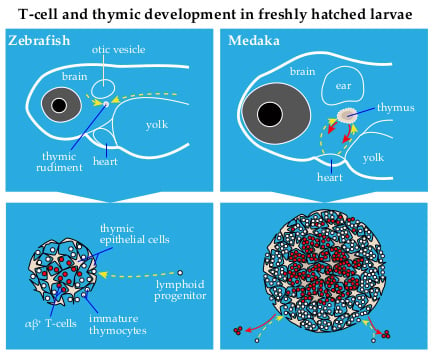
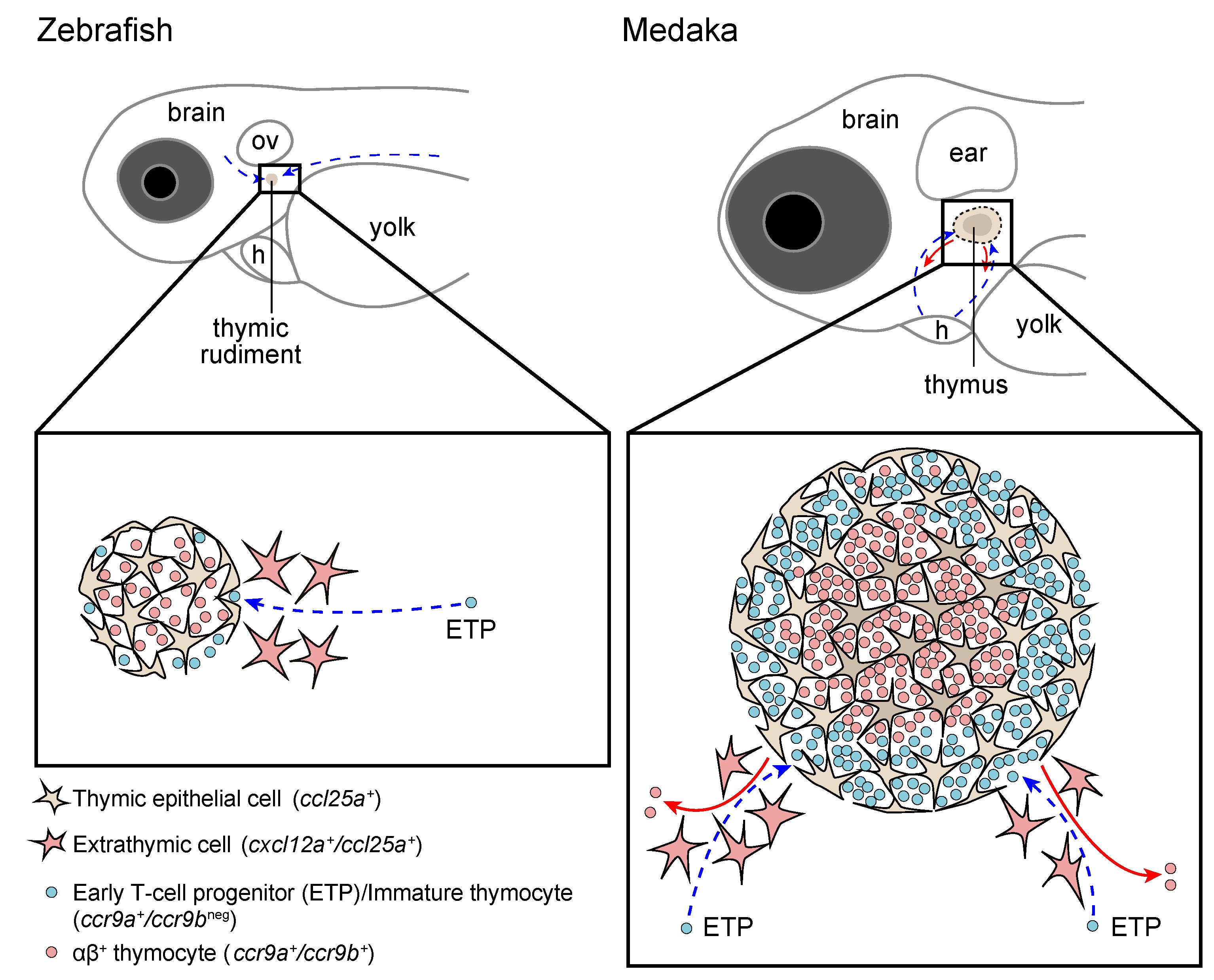
Zebrafish and Medaka: Two Teleost Models of T-Cell and Thymic Development
Abstract
:1. Introduction
2. Thymus Development in Zebrafish and Medaka: Similarities and Differences
3. T-Cell Development in Zebrafish and Medaka
3.1. The Entry of Lymphoid Progenitors into the Thymus
3.2. Commitment of Thymocytes
3.3. Selection of Functional and Non-self-reactive Thymocytes
3.4. Egress of Thymocytes
4. Genetic Tools to Study T-cell Development in Zebrafish and Medaka
5. Zebrafish as Model System for T-Cell Diseases
6. Conclusions
- To what extent are the underlying mechanisms of T-cell commitment and exit into the periphery conserved between teleosts and mammals?
- How is the choice between αβ and γδ T-cell sublineages regulated in teleosts?
- How does the morphological transformation of the thymus in adolescent fish change the cellular composition and spatial gene expression patterns within the thymus?
- Why does age-related thymus involution not occur in some teleost species?
- In mammals, naïve T-cells can differentiate into various subtypes, including cytotoxic T-cells, helper T-cells (e.g., Th1, Th2, Th17), and regulatory T-cells (Treg). What is the full diversity of T-cell subtypes in teleosts?
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AIRE | Autoimmune regulator |
| dpf | Days post-fertilization |
| ETP | Early T-cell progenitor |
| RTE | Recent thymic emigrants |
| T-ALL | T-cell acute lymphoblastic leukemia |
| TCR | T-cell receptor |
| TEC | Thymic epithelial cell |
| T-LBL | T-lymphoblastic lymphoma |
| VLR | Variable lymphocyte receptor |
| wpf | Weeks post-fertilization |
References
- Bell, J.J.; Bhandoola, A. The earliest thymic progenitors for t cells possess myeloid lineage potential. Nature 2008, 452, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Boehm, T. Thymus development and function. Curr. Opin. Immunol. 2008, 20, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhao, Y. Evolution of thymus organogenesis. Dev. Comp. Immunol. 2013, 39, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Rodewald, H.-R. Thymus organogenesis. Annu. Rev. Immunol. 2008, 26, 355–388. [Google Scholar] [CrossRef] [PubMed]
- Takahama, Y. Journey through the thymus: Stromal guides for t-cell development and selection. Nat. Rev. Immunol. 2006, 6, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Bunting, M.D.; Comerford, I.; McColl, S.R. Finding their niche: Chemokines directing cell migration in the thymus. Immunol. Cell Biol. 2011, 89, 185–196. [Google Scholar] [CrossRef]
- Dzhagalov, I.L.; Chen, K.G.; Herzmark, P.; Robey, E.A. Elimination of self-reactive t cells in the thymus: A timeline for negative selection. Plos. Biol. 2013, 11, e1001566. [Google Scholar] [CrossRef]
- Klein, L.; Hinterberger, M.; Wirnsberger, G.; Kyewski, B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009, 9, 833–844. [Google Scholar] [CrossRef]
- Langenau, D.M.; Zon, L.I. The zebrafish: A new model of t-cell and thymic development. Nat. Rev. Immunol. 2005, 5, 307–317. [Google Scholar] [CrossRef]
- Boehm, T.; Swann, J.B. Origin and evolution of adaptive immunity. Annu. Rev. Anim. Biosci. 2014, 2, 259–283. [Google Scholar] [CrossRef]
- Boehm, T.; Iwanami, N.; Hess, I. Evolution of the immune system in the lower vertebrates. Annu. Rev. Genom. Hum. Genet. 2012, 13, 127–149. [Google Scholar] [CrossRef]
- Boehm, T.; Hess, I.; Swann, J.B. Evolution of lymphoid tissues. Trends Immunol. 2012, 33, 315–321. [Google Scholar] [CrossRef]
- Hirano, M.; Das, S.; Guo, P.; Cooper, M.D. The evolution of adaptive immunity in vertebrates. Adv. Immunol. 2011, 109, 125–157. [Google Scholar]
- Pancer, Z.; Cooper, M.D. The evolution of adaptive immunity. Annu Rev Immunol 2006, 24, 497–518. [Google Scholar] [CrossRef]
- Cooper, M.D.; Alder, M.N. The evolution of adaptive immune systems. Cell 2006, 124, 815–822. [Google Scholar] [CrossRef]
- Boehm, T.; McCurley, N.; Sutoh, Y.; Schorpp, M.; Kasahara, M.; Cooper, M.D. Vlr-based adaptive immunity. Annu. Rev. Immunol. 2012, 30, 203–220. [Google Scholar] [CrossRef]
- Das, S.; Li, J.; Holland, S.J.; Iyer, L.M.; Hirano, M.; Schorpp, M.; Aravind, L.; Cooper, M.D.; Boehm, T. Genomic donor cassette sharing during vlra and vlrc assembly in jawless vertebrates. Proc. Natl. Acad. Sci. USA 2014, 111, 14828–14833. [Google Scholar] [CrossRef]
- Holland, S.J.; Gao, M.; Hirano, M.; Iyer, L.M.; Luo, M.; Schorpp, M.; Cooper, M.D.; Aravind, L.; Mariuzza, R.A.; Boehm, T. Selection of the lamprey vlrc antigen receptor repertoire. Proc. Natl. Acad. Sci. USA 2014, 111, 14834–14839. [Google Scholar] [CrossRef]
- Kasahara, M.; Sutoh, Y. Two forms of adaptive immunity in vertebrates: Similarities and differences. Adv. Immunol. 2014, 122, 59–90. [Google Scholar]
- Das, S.; Hirano, M.; Aghaallaei, N.; Bajoghli, B.; Boehm, T.; Cooper, M.D. Organization of lamprey variable lymphocyte receptor c locus and repertoire development. Proc. Natl. Acad. Sci. USA 2013, 110, 6043–6048. [Google Scholar] [CrossRef]
- Bajoghli, B.; Guo, P.; Aghaallaei, N.; Hirano, M.; Strohmeier, C.; McCurley, N.; Bockman, D.E.; Schorpp, M.; Cooper, M.D.; Boehm, T. A thymus candidate in lampreys. Nature 2011, 470, 90–94. [Google Scholar] [CrossRef]
- Bowden, T.J.; Cook, P.; Rombout, J.H. Development and function of the thymus in teleosts. Fish Shellfish Immunol. 2005, 19, 413–427. [Google Scholar] [CrossRef]
- Zhang, Y.; Wiest, D.L. Using the zebrafish model to study t cell development. Methods Mol. Biol. 2016, 1323, 273–292. [Google Scholar]
- Li, J.; Iwanami, N.; Hoa, V.Q.; Furutani-Seiki, M.; Takahama, Y. Noninvasive intravital imaging of thymocyte dynamics in medaka. J. Immunol. 2007, 179, 1605–1615. [Google Scholar] [CrossRef]
- Iwanami, N.; Takahama, Y.; Kunimatsu, S.; Li, J.; Takei, R.; Ishikura, Y.; Suwa, H.; Niwa, K.; Sasado, T.; Morinaga, C.; et al. Mutations affecting thymus organogenesis in medaka, oryzias latipes. Mech. Dev. 2004, 121, 779–789. [Google Scholar] [CrossRef]
- Langenau, D.M.; Ferrando, A.A.; Traver, D.; Kutok, J.L.; Hezel, J.P.; Kanki, J.P.; Zon, L.I.; Look, A.T.; Trede, N.S. In vivo tracking of t cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. USA 2004, 101, 7369–7374. [Google Scholar] [CrossRef]
- Trede, N.S.; Ota, T.; Kawasaki, H.; Paw, B.H.; Katz, T.; Demarest, B.; Hutchinson, S.; Zhou, Y.; Hersey, C.; Zapata, A.; et al. Zebrafish mutants with disrupted early t-cell and thymus development identified in early pressure screen. Dev. Dyn. 2008, 237, 2575–2584. [Google Scholar] [CrossRef]
- Bajoghli, B. Evolution and function of chemokine receptors in the immune system of lower vertebrates. Eur. J. Immunol. 2013, 43, 1686–1692. [Google Scholar] [CrossRef]
- Trede, N.S.; Zapata, A.; Zon, L.I. Fishing for lymphoid genes. Trends Immunol. 2001, 22, 302–307. [Google Scholar] [CrossRef]
- Schorpp, M.; Leicht, M.; Nold, E.; Hammerschmidt, M.; Haas-Assenbaum, A.; Wiest, W.; Boehm, T. A zebrafish orthologue (whnb) of the mouse nude gene is expressed in the epithelial compartment of the embryonic thymic rudiment. Mech. Dev. 2002, 118, 179–185. [Google Scholar] [CrossRef]
- Hess, I.; Boehm, T. Intravital imaging of thymopoiesis reveals dynamic lympho-epithelial interactions. Immunity 2012, 36, 298–309. [Google Scholar] [CrossRef]
- Bajoghli, B.; Aghaallaei, N.; Hess, I.; Rode, I.; Netuschil, N.; Tay, B.H.; Venkatesh, B.; Yu, J.K.; Kaltenbach, S.L.; Holland, N.D.; et al. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell 2009, 138, 186–197. [Google Scholar] [CrossRef]
- Bleul, C.C.; Corbeaux, T.; Reuter, A.; Fisch, P.; Monting, J.S.; Boehm, T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature 2006, 441, 992–996. [Google Scholar] [CrossRef]
- Swann, J.B.; Weyn, A.; Nagakubo, D.; Bleul, C.C.; Toyoda, A.; Happe, C.; Netuschil, N.; Hess, I.; Haas-Assenbaum, A.; Taniguchi, Y.; et al. Conversion of the thymus into a bipotent lymphoid organ by replacement of foxn1 with its paralog, foxn4. Cell Rep. 2014, 8, 1184–1197. [Google Scholar] [CrossRef]
- Koch, U.; Fiorini, E.; Benedito, R.; Besseyrias, V.; Schuster-Gossler, K.; Pierres, M.; Manley, N.R.; Duarte, A.; Macdonald, H.R.; Radtke, F. Delta-like 4 is the essential, nonredundant ligand for notch1 during thymic t cell lineage commitment. J. Exp. Med. 2008, 205, 2515–2523. [Google Scholar] [CrossRef]
- Hozumi, K.; Mailhos, C.; Negishi, N.; Hirano, K.; Yahata, T.; Ando, K.; Zuklys, S.; Hollander, G.A.; Shima, D.T.; Habu, S. Delta-like 4 is indispensable in thymic environment specific for t cell development. J. Exp. Med. 2008, 205, 2507–2513. [Google Scholar] [CrossRef]
- Ferrero, I.; Koch, U.; Claudinot, S.; Favre, S.; Radtke, F.; Luther, S.A.; MacDonald, H.R. Dl4-mediated notch signaling is required for the development of fetal alphabeta and gammadelta t cells. Eur. J. Immunol. 2013, 43, 2845–2853. [Google Scholar] [CrossRef]
- Schlake, T.; Schorpp, M.; Nehls, M.; Boehm, T. The nude gene encodes a sequence-specific DNA binding protein with homologs in organisms that lack an anticipatory immune system. Proc. Natl. Acad. Sci. USA 1997, 94, 3842–3847. [Google Scholar] [CrossRef]
- Lam, S.H.; Chua, H.L.; Gong, Z.; Wen, Z.; Lam, T.J.; Sin, Y.M. Morphologic transformation of the thymus in developing zebrafish. Dev. Dyn. 2002, 225, 87–94. [Google Scholar] [CrossRef]
- Romano, N.; Fanelli, M.; Maria Del Papa, G.; Scapigliati, G.; Mastrolia, L. Histological and cytological studies on the developing thymus of sharpsnout seabream, diplodus puntazzo. J. Anat. 1999, 194, 39–50. [Google Scholar] [CrossRef]
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453. [Google Scholar] [CrossRef]
- O’Neill, J.G. Ontogeny of the lymphoid organs in an antarctic teleost, harpagifer antarcticus (notothenioidei: Perciformes). Dev. Comp. Immunol. 1989, 13, 25–33. [Google Scholar] [CrossRef]
- Bajoghli, B.; Kuri, P.; Inoue, D.; Aghaallaei, N.; Hanelt, M.; Thumberger, T.; Rauzi, M.; Wittbrodt, J.; Leptin, M. Noninvasive in toto imaging of the thymus reveals heterogeneous migratory behavior of developing t cells. J. Immunol. 2015, 195, 2177–2186. [Google Scholar] [CrossRef]
- Boehm, T.; Swann, J.B. Thymus involution and regeneration: Two sides of the same coin? Nat. Rev. Immunol. 2013, 13, 831–838. [Google Scholar] [CrossRef]
- Lynch, H.E.; Goldberg, G.L.; Chidgey, A.; Van den Brink, M.R.; Boyd, R.; Sempowski, G.D. Thymic involution and immune reconstitution. Trends Immunol. 2009, 30, 366–373. [Google Scholar] [CrossRef] [Green Version]
- Deanesly, R. Memoirs: The structure and development of the thymus in fish, with special reference to salmo fario. J. Cell. Sci. 1927, 2, 113–145. [Google Scholar]
- O’Neill, J.G. Thymic development in two species of marine teleost; an antarctic silverfish, pleuragramma antarcticum boulenger, and a warmer-water sea bass, dicentrarchus labrax (linnaeus). Proc. NIPR Symp. Polar Biol. 1989, 2, 83–93. [Google Scholar]
- Ghoneum, M.M.; Egami, N. Age related changes in morphology of the thymus of the fish, oryzias latipes. Exp. Gerontol. 1982, 17, 33–40. [Google Scholar] [CrossRef]
- Kissa, K.; Murayama, E.; Zapata, A.; Cortes, A.; Perret, E.; Machu, C.; Herbomel, P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 2007, 111, 1147–1156. [Google Scholar] [CrossRef]
- Calderon, L.; Boehm, T. Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc. Natl. Acad. Sci. USA 2011, 108, 7517–7522. [Google Scholar] [CrossRef] [Green Version]
- Radtke, F.; Wilson, A.; Mancini, S.J.C.; MacDonald, H.R. Notch regulation of lymphocyte development and function. Nat. Immunol. 2004, 5, 247–253. [Google Scholar] [CrossRef]
- Radtke, F.; Fasnacht, N.; Macdonald, H.R. Notch signaling in the immune system. Immunity 2010, 32, 14–27. [Google Scholar] [CrossRef]
- Koch, U.; Radtke, F. Mechanisms of t cell development and transformation. Annu. Rev. Cell Dev. Biol. 2011, 27, 539–562. [Google Scholar] [CrossRef]
- Garbe, A.I.; von Boehmer, H. Tcr and notch synergize in alphabeta versus gammadelta lineage choice. Trends Immunol. 2007, 28, 124–131. [Google Scholar] [CrossRef]
- Kreslavsky, T.; Garbe, A.I.; Krueger, A.; von Boehmer, H. T cell receptor–instructed αβ versus γδ lineage commitment revealed by single-cell analysis. J. Exp. Med. 2008, 205, 1173–1186. [Google Scholar] [CrossRef]
- Kreslavsky, T.; Gleimer, M.; Garbe, A.I.; von Boehmer, H. Alphabeta versus gammadelta fate choice: Counting the t-cell lineages at the branch point. Immunol. Rev. 2010, 238, 169–181. [Google Scholar] [CrossRef]
- Kim, A.D.; Melick, C.H.; Clements, W.K.; Stachura, D.L.; Distel, M.; Panakova, D.; MacRae, C.; Mork, L.A.; Crump, J.G.; Traver, D. Discrete notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 2014, 33, 2363–2373. [Google Scholar] [CrossRef]
- Ciofani, M.; Zuniga-Pflucker, J.C. Determining gammadelta versus alphass t cell development. Nat. Rev. Immunol. 2010, 10, 657–663. [Google Scholar] [CrossRef]
- Smelty, P.; Marchal, C.; Renard, R.; Sinzelle, L.; Pollet, N.; Dunon, D.; Jaffredo, T.; Sire, J.Y.; Fellah, J.S. Identification of the pre-t-cell receptor alpha chain in nonmammalian vertebrates challenges the structure-function of the molecule. Proc. Natl. Acad. Sci. USA 2010, 107, 19991–19996. [Google Scholar] [CrossRef]
- Proudhon, C.; Hao, B.; Raviram, R.; Chaumeil, J.; Skok, J.A. Long-range regulation of v(d)j recombination. Adv. Immunol. 2015, 128, 123–182. [Google Scholar]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the t cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef]
- Le Borgne, M.; Ladi, E.; Dzhagalov, I.; Herzmark, P.; Liao, Y.F.; Chakraborty, A.K.; Robey, E.A. The impact of negative selection on thymocyte migration in the medulla. Nat. Immunol. 2009, 10, 823–830. [Google Scholar] [CrossRef] [Green Version]
- Mathis, D.; Benoist, C. Aire. Annu. Rev. Immunol. 2009, 27, 287–312. [Google Scholar] [CrossRef]
- Matloubian, M.; Lo, C.G.; Cinamon, G.; Lesneski, M.J.; Xu, Y.; Brinkmann, V.; Allende, M.L.; Proia, R.L.; Cyster, J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on s1p receptor 1. Nature 2004, 427, 355–360. [Google Scholar] [CrossRef]
- Tobia, C.; Chiodelli, P.; Nicoli, S.; Dell’era, P.; Buraschi, S.; Mitola, S.; Foglia, E.; van Loenen, P.B.; Alewijnse, A.E.; Presta, M. Sphingosine-1-phosphate receptor-1 controls venous endothelial barrier integrity in zebrafish. Arter. Thromb Vasc. Biol. 2012, 32, e104–e116. [Google Scholar] [CrossRef]
- Mendelson, K.; Zygmunt, T.; Torres-Vazquez, J.; Evans, T.; Hla, T. Sphingosine 1-phosphate receptor signaling regulates proper embryonic vascular patterning. J. Biol. Chem. 2013, 288, 2143–2156. [Google Scholar] [CrossRef]
- Hisano, Y.; Ota, S.; Takada, S.; Kawahara, A. Functional cooperation of spns2 and fibronectin in cardiac and lower jaw development. Biol. Open 2013, 2, 789–794. [Google Scholar] [CrossRef] [Green Version]
- Gaengel, K.; Niaudet, C.; Hagikura, K.; Lavina, B.; Muhl, L.; Hofmann, J.J.; Ebarasi, L.; Nystrom, S.; Rymo, S.; Chen, L.L.; et al. The sphingosine-1-phosphate receptor s1pr1 restricts sprouting angiogenesis by regulating the interplay between ve-cadherin and vegfr2. Dev. Cell 2012, 23, 587–599. [Google Scholar] [CrossRef]
- Bajoghli, B.; Aghaallaei, N.; Heimbucher, T.; Czerny, T. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev. Biol. 2004, 271, 416–430. [Google Scholar] [CrossRef] [Green Version]
- Punwani, D.; Zhang, Y.; Yu, J.; Cowan, M.J.; Rana, S.; Kwan, A.; Adhikari, A.N.; Lizama, C.O.; Mendelsohn, B.A.; Fahl, S.P.; et al. Multisystem anomalies in severe combined immunodeficiency with mutant bcl11b. N. Engl. J. Med. 2016, 375, 2165–2176. [Google Scholar] [CrossRef]
- Driever, W.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Malicki, J.; Stemple, D.L.; Stainier, D.Y.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46. [Google Scholar]
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Petrie-Hanson, L.; Hohn, C.; Hanson, L. Characterization of rag1 mutant zebrafish leukocytes. BMC Immunol. 2009, 10, 8. [Google Scholar] [CrossRef]
- Iwanami, N.; Mateos, F.; Hess, I.; Riffel, N.; Soza-Ried, C.; Schorpp, M.; Boehm, T. Genetic evidence for an evolutionarily conserved role of il-7 signaling in t cell development of zebrafish. J. Immunol. 2011, 186, 7060–7066. [Google Scholar] [CrossRef]
- Soza-Ried, C.; Hess, I.; Netuschil, N.; Schorpp, M.; Boehm, T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc. Natl. Acad. Sci. USA 2010, 107, 17304–17308. [Google Scholar] [CrossRef]
- Schorpp, M.; Bialecki, M.; Diekhoff, D.; Walderich, B.; Odenthal, J.; Maischein, H.M.; Zapata, A.G.; Boehm, T. Conserved functions of ikaros in vertebrate lymphocyte development: Genetic evidence for distinct larval and adult phases of t cell development and two lineages of b cells in zebrafish. J. Immunol. 2006, 177, 2463–2476. [Google Scholar] [CrossRef]
- Monnich, M.; Hess, I.; Wiest, W.; Bachrati, C.; Hickson, I.D.; Schorpp, M.; Boehm, T. Developing t lymphocytes are uniquely sensitive to a lack of topoisomerase iii alpha. Eur. J. Immunol. 2010, 40, 2379–2384. [Google Scholar] [CrossRef]
- Iwanami, N.; Sikora, K.; Richter, A.S.; Monnich, M.; Guerri, L.; Soza-Ried, C.; Lawir, D.F.; Mateos, F.; Hess, I.; O’Meara, C.P.; et al. Forward genetic screens in zebrafish identify pre-mrna-processing pathways regulating early t cell development. Cell Rep. 2016, 17, 2259–2270. [Google Scholar] [CrossRef]
- Lawir, D.F.; Iwanami, N.; Schorpp, M.; Boehm, T. A missense mutation in zbtb17 blocks the earliest steps of t cell differentiation in zebrafish. Sci. Rep. 2017, 7, 44145. [Google Scholar] [CrossRef]
- Iwanami, N.; Higuchi, T.; Sasano, Y.; Fujiwara, T.; Hoa, V.Q.; Okada, M.; Talukder, S.R.; Kunimatsu, S.; Li, J.; Saito, F.; et al. Wdr55 is a nucleolar modulator of ribosomal rna synthesis, cell cycle progression, and teleost organ development. PLoS Genet. 2008, 4, e1000171. [Google Scholar] [CrossRef]
- Reischauer, S.; Stone, O.A.; Villasenor, A.; Chi, N.; Jin, S.W.; Martin, M.; Lee, M.T.; Fukuda, N.; Marass, M.; Witty, A.; et al. Cloche is a bhlh-pas transcription factor that drives haemato-vascular specification. Nature 2016, 535, 294–298. [Google Scholar] [CrossRef]
- Wu, R.S.; Lam, II.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A rapid method for directed gene knockout for screening in g0 zebrafish. Dev. Cell 2018, 46, 112–125. [Google Scholar] [CrossRef]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a crispr nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef]
- Gutierrez-Triana, J.A.; Tavhelidse, T.; Thumberger, T.; Thomas, I.; Wittbrodt, B.; Kellner, T.; Anlas, K.; Tsingos, E.; Wittbrodt, J. Efficient single-copy hdr by 5′ modified long dsdna donors. eLife 2018, 7, e39468. [Google Scholar] [CrossRef]
- Auer, T.O.; Duroure, K.; De Cian, A.; Concordet, J.P.; Del Bene, F. Highly efficient crispr/cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014, 24, 142–153. [Google Scholar] [CrossRef]
- Aghaallaei, N.; Bajoghli, B. Making thymus visible: Understanding t-cell development from a new perspective. Front. Immunol. 2018, 9, 375. [Google Scholar] [CrossRef]
- Kasheta, M.; Painter, C.A.; Moore, F.E.; Lobbardi, R.; Bryll, A.; Freiman, E.; Stachura, D.; Rogers, A.B.; Houvras, Y.; Langenau, D.M.; et al. Identification and characterization of t reg-like cells in zebrafish. J. Exp. Med. 2017, 214, 3519–3530. [Google Scholar] [CrossRef]
- Yamada, A.; Takaki, S.; Hayashi, F.; Georgopoulos, K.; Perlmutter, R.M.; Takatsu, K. Identification and characterization of a transcriptional regulator for the lck proximal promoter. J. Biol. Chem. 2001, 276, 18082–18089. [Google Scholar] [CrossRef]
- Durinck, K.; Goossens, S.; Peirs, S.; Wallaert, A.; Van Loocke, W.; Matthijssens, F.; Pieters, T.; Milani, G.; Lammens, T.; Rondou, P.; et al. Novel biological insights in t-cell acute lymphoblastic leukemia. Exp. Hematol. 2015, 43, 625–639. [Google Scholar] [CrossRef]
- Neumann, M.; Vosberg, S.; Schlee, C.; Heesch, S.; Schwartz, S.; Gokbuget, N.; Hoelzer, D.; Graf, A.; Krebs, S.; Bartram, I.; et al. Mutational spectrum of adult t-all. Oncotarget 2015, 6, 2754–2766. [Google Scholar] [CrossRef]
- Blackburn, J.S.; Liu, S.; Raiser, D.M.; Martinez, S.A.; Feng, H.; Meeker, N.D.; Gentry, J.; Neuberg, D.; Look, A.T.; Ramaswamy, S.; et al. Notch signaling expands a pre-malignant pool of t-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia 2012, 26, 2069–2078. [Google Scholar] [CrossRef]
- Gutierrez, A.; Grebliunaite, R.; Feng, H.; Kozakewich, E.; Zhu, S.; Guo, F.; Payne, E.; Mansour, M.; Dahlberg, S.E.; Neuberg, D.S.; et al. Pten mediates myc oncogene dependence in a conditional zebrafish model of t cell acute lymphoblastic leukemia. J. Exp. Med. 2011, 208, 1595–1603. [Google Scholar] [CrossRef]
- Langenau, D.M.; Feng, H.; Berghmans, S.; Kanki, J.P.; Kutok, J.L.; Look, A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced t cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2005, 102, 6068–6073. [Google Scholar] [CrossRef]
- Reynolds, C.; Roderick, J.E.; LaBelle, J.L.; Bird, G.; Mathieu, R.; Bodaar, K.; Colon, D.; Pyati, U.; Stevenson, K.E.; Qi, J.; et al. Repression of bim mediates survival signaling by myc and akt in high-risk t-cell acute lymphoblastic leukemia. Leukemia 2014, 28, 1819–1827. [Google Scholar] [CrossRef]
- Borga, C.; Park, G.; Foster, C.; Burroughs-Garcia, J.; Marchesin, M.; Shah, R.; Hasan, A.; Ahmed, S.T.; Bresolin, S.; Batchelor, L.; et al. Simultaneous b and t cell acute lymphoblastic leukemias in zebrafish driven by transgenic myc: Implications for oncogenesis and lymphopoiesis. Leukemia 2019, 33, 333–347. [Google Scholar] [CrossRef]
- Mansour, M.R.; He, S.; Li, Z.; Lobbardi, R.; Abraham, B.J.; Hug, C.; Rahman, S.; Leon, T.E.; Kuang, Y.Y.; Zimmerman, M.W.; et al. Jdp2: An oncogenic bzip transcription factor in t cell acute lymphoblastic leukemia. J. Exp. Med. 2018, 215, 1929–1945. [Google Scholar] [CrossRef]
- Leong, W.Z.; Tan, S.H.; Ngoc, P.C.T.; Amanda, S.; Yam, A.W.Y.; Liau, W.S.; Gong, Z.; Lawton, L.N.; Tenen, D.G.; Sanda, T. Arid5b as a critical downstream target of the tal1 complex that activates the oncogenic transcriptional program and promotes t-cell leukemogenesis. Genes Dev. 2017, 31, 2343–2360. [Google Scholar] [CrossRef]
- Frazer, J.K.; Meeker, N.D.; Rudner, L.; Bradley, D.F.; Smith, A.C.; Demarest, B.; Joshi, D.; Locke, E.E.; Hutchinson, S.A.; Tripp, S.; et al. Heritable t-cell malignancy models established in a zebrafish phenotypic screen. Leukemia 2009, 23, 1825–1835. [Google Scholar] [CrossRef]
- Moore, J.C.; Mulligan, T.S.; Yordan, N.T.; Castranova, D.; Pham, V.N.; Tang, Q.; Lobbardi, R.; Anselmo, A.; Liwski, R.S.; Berman, J.N.; et al. T cell immune deficiency in zap70 mutant zebrafish. Mol. Cell Biol. 2016, 36, 2868–2876. [Google Scholar] [CrossRef]
- Gennery, A. Recent advances in understanding rag deficiencies. F1000Res 2019, 8. [Google Scholar] [CrossRef]
- Tang, Q.; Abdelfattah, N.S.; Blackburn, J.S.; Moore, J.C.; Martinez, S.A.; Moore, F.E.; Lobbardi, R.; Tenente, I.M.; Ignatius, M.S.; Berman, J.N.; et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat. Methods 2014, 11, 821–824. [Google Scholar] [CrossRef] [Green Version]
- Kasahara, M.; Naruse, K.; Sasaki, S.; Nakatani, Y.; Qu, W.; Ahsan, B.; Yamada, T.; Nagayasu, Y.; Doi, K.; Kasai, Y.; et al. The medaka draft genome and insights into vertebrate genome evolution. Nature 2007, 447, 714–719. [Google Scholar] [CrossRef] [Green Version]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bajoghli, B.; Dick, A.M.; Claasen, A.; Doll, L.; Aghaallaei, N. Zebrafish and Medaka: Two Teleost Models of T-Cell and Thymic Development. Int. J. Mol. Sci. 2019, 20, 4179. https://doi.org/10.3390/ijms20174179
Bajoghli B, Dick AM, Claasen A, Doll L, Aghaallaei N. Zebrafish and Medaka: Two Teleost Models of T-Cell and Thymic Development. International Journal of Molecular Sciences. 2019; 20(17):4179. https://doi.org/10.3390/ijms20174179
Chicago/Turabian StyleBajoghli, Baubak, Advaita M. Dick, Annisa Claasen, Larissa Doll, and Narges Aghaallaei. 2019. "Zebrafish and Medaka: Two Teleost Models of T-Cell and Thymic Development" International Journal of Molecular Sciences 20, no. 17: 4179. https://doi.org/10.3390/ijms20174179
APA StyleBajoghli, B., Dick, A. M., Claasen, A., Doll, L., & Aghaallaei, N. (2019). Zebrafish and Medaka: Two Teleost Models of T-Cell and Thymic Development. International Journal of Molecular Sciences, 20(17), 4179. https://doi.org/10.3390/ijms20174179