Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism
Abstract
1. Introduction
2. Results
2.1. Improvement of Spirometry of Asthmatic Models and Quality Assessment on RWC
2.2. Alleviatory Effect of RWC on AHR and Asthmatic Inflammation
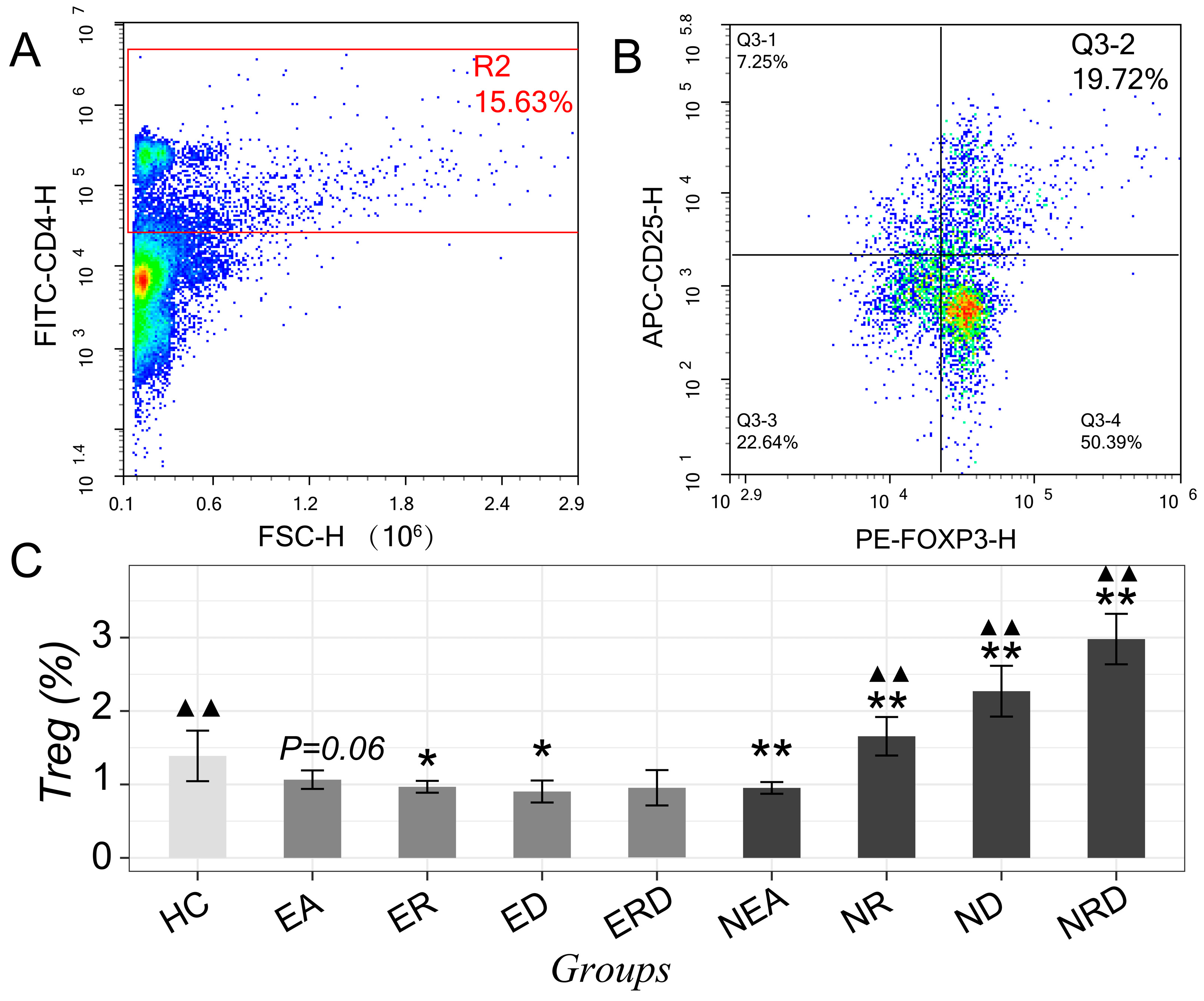
2.3. Improvement of Th2/Th1 Imbalance and Regulatory Effect on Th17/Tregs
2.4. Influence on Th1/Th2/Th17-Related Cytokines
2.5. α-Diversity and β-Diversity of the Microbiota in Lower Airway
2.6. Taxonomic Differences in Microbiota after Treatment
2.7. Difference of Systematic and Pulmonary Metabolic Pattern
2.8. Perturbed Metabolic Pathways
2.9. Multiomics Joint Exploration and Pharmacological Network of RWC
3. Discussion
4. Materials and Methods
4.1. Quantification on Salidroside in RWC Injection with HPLC
4.2. Experimental Animal and Procedure of Model Establishment
4.3. Spirometry and AHR Examination
4.4. H&E Staining
4.5. Flow Cytometry of Th1/Th2/Th17 and Tregs
4.6. ELISA Assay
4.7. DNA Extraction and 16S rDNA Sequencing
4.8. Sample Preparation for Metabolomics and UPLC-Q/TOF-MS/MS Procedure
4.9. Omics Data Processing
4.10. Bioinformatic and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AHR | airway hyperresponsiveness |
| AR | analytical reagent |
| BALF | bronchoalveolar lavage fluid |
| Cdyn | dynamic compliance |
| CFA | complete Freund’s adjuvant |
| DEX | dexamethasone |
| EA | eosinophilic asthma |
| ED | eosinophilic asthma treated with dexamethasone |
| ER | eosinophilic asthma treated with Rhodiola wallichiana var. cholaensis |
| ERD | eosinophilic asthma treated with Rhodiola wallichiana var. cholaensis and dexamethasone |
| FDR | false discovery rate |
| FEV0.1 | forced expiratory volume in 0.1 s |
| FRC | functional residual capacity |
| HC | healthy control |
| HPLC | high-performance liquid chromatography |
| H&E | hematoxylin and eosin |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | KEGG orthology |
| LDA | linear discriminant analysis |
| LEFse | linear discriminant analysis effect size |
| NEA | non-eosinophilic asthma |
| ND | non-eosinophilic asthma treated with dexamethasone |
| NMDS | non-metric multidimensional scaling |
| NR | non-eosinophilic asthma treated with Rhodiola wallichiana var. cholaensis |
| NRD | non-eosinophilic asthma treated with Rhodiola wallichiana var. cholaensis and dexamethasone |
| OBS | observed species |
| OPLS-DA | orthogonal projections to latent structures discriminant analysis |
| OTU | operational taxonomic units |
| OVA | ovalbumin |
| PCA | principal component analysis |
| PCoA | principal coordinates analysis |
| PD | phylogenetic diversity |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States |
| PMA | phorbol-12-myristate-13-acetate |
| QC | quality control |
| Q/TOF-MS | quadrupole time-of-flight mass spectrometry |
| RI | resistance of inspiration |
| RSD | relative standard deviation |
| RT | retention time |
| RWC | Rhodiola wallichiana var. cholaensis |
| SPF | specific pathogen-free |
| Tregs | regulatory T cells |
| UPLC | ultra-performance liquid chromatography |
References
- Gibson, P.G. Inflammatory phenotypes in adult asthma: Clinical applications. Clin. Respir. J. 2009, 3, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Ortega, H.G.; Liu, M.C.; Pavord, I.D.; Brusselle, G.G.; FitzGerald, J.M.; Chetta, A.; Humbert, M.; Katz, L.E.; Keene, O.N.; Yancey, S.W.; et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N. Engl. J. Med. 2014, 371, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.R.; van Dalen, C.J.; Hermans, I.F.; Gibson, P.G.; Simpson, J.L.; Douwes, J. Sputum basophils are increased in eosinophilic asthma compared with non-eosinophilic asthma phenotypes. Allergy 2017, 72, 1583–1586. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, P.G. Subtypes of asthma defined by epithelial cell expression of messenger RNA and microRNA. Ann. Am. Thorac Soc. 2013, 10 (Suppl.), S186–S189. [Google Scholar] [CrossRef] [PubMed]
- Hastie, A.T.; Moore, W.C.; Meyers, D.A.; Vestal, P.L.; Li, H.; Peters, S.P.; Bleecker, E.R.; National Heart, L.; Blood Institute Severe Asthma Research, P. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J. Allergy Clin. Immunol. 2010, 125, 1028.e13–1036.e13. [Google Scholar] [CrossRef] [PubMed]
- Khatry, D.B.; Gossage, D.L.; Geba, G.P.; Parker, J.M.; Jarjour, N.N.; Busse, W.W.; Molfino, N.A. Discriminating sputum-eosinophilic asthma: Accuracy of cutoffs in blood eosinophil measurements versus a composite index, ELEN. J. Allergy Clin. Immunol. 2015, 136, 812.e2–814.e2. [Google Scholar] [CrossRef]
- Brooks, C.R.; van Dalen, C.J.; Zacharasiewicz, A.; Simpson, J.L.; Harper, J.L.; Le Gros, G.; Gibson, P.G.; Pearce, N.; Douwes, J. Absence of airway inflammation in a large proportion of adolescents with asthma. Respirology 2016, 21, 460–466. [Google Scholar] [CrossRef]
- Douwes, J.; Gibson, P.; Pekkanen, J.; Pearce, N. Non-eosinophilic asthma: Importance and possible mechanisms. Thorax 2002, 57, 643–648. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, G.; Gibson, P.; Guan, X.; Zhang, W.; Zheng, R.; Chen, F.; Wang, Z.; Wang, F. Airway Microbiome in Different Inflammatory Phenotypes of Asthma: A Cross-Sectional Study in Northeast China. Int. J. Med. Sci. 2019, 16, 477–485. [Google Scholar] [CrossRef]
- Wang, L.; Zha, B.; Shen, Q.; Zou, H.; Cheng, C.; Wu, H.; Liu, R. Sevoflurane Inhibits the Th2 Response and NLRP3 Expression in Murine Allergic Airway Inflammation. J. Immunol. Res. 2018, 2018, 9021037. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Hammad, H. The immunology of asthma. Nat. Immunol. 2015, 16, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.K.; Kim, S.W.; Park, C.S.; Kim, B.I.; Kang, H.; Koh, Y.Y. Bronchoalveolar lavage cytokine profiles in acute asthma and acute bronchiolitis. J. Allergy Clin. Immunol. 2003, 112, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Essilfie, A.T.; Simpson, J.L.; Horvat, J.C.; Preston, J.A.; Dunkley, M.L.; Foster, P.S.; Gibson, P.G.; Hansbro, P.M. Haemophilus influenzae infection drives IL-17-mediated neutrophilic allergic airways disease. PLoS Pathog. 2011, 7, e1002244. [Google Scholar] [CrossRef] [PubMed]
- Lewkowich, I.P.; Herman, N.S.; Schleifer, K.W.; Dance, M.P.; Chen, B.L.; Dienger, K.M.; Sproles, A.A.; Shah, J.S.; Kohl, J.; Belkaid, Y.; et al. CD4+CD25+ T cells protect against experimentally induced asthma and alter pulmonary dendritic cell phenotype and function. J. Exp. Med. 2005, 202, 1549–1561. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Corren, J.; Pavord, I.D.; Maspero, J.; Wenzel, S.; Rabe, K.F.; Busse, W.W.; Ford, L.; Sher, L.; FitzGerald, J.M.; et al. Dupilumab Efficacy and Safety in Moderate-to-Severe Uncontrolled Asthma. N. Engl. J. Med. 2018, 378, 2486–2496. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.; Wenzel, S.; Rabe, K.F.; Bourdin, A.; Lugogo, N.L.; Kuna, P.; Barker, P.; Sproule, S.; Ponnarambil, S.; Goldman, M.; et al. Oral Glucocorticoid-Sparing Effect of Benralizumab in Severe Asthma. N. Engl. J. Med. 2017, 376, 2448–2458. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Q.; Tsring, N.; Yu, T.Y.; Wu, J.C.; Wong, S.; Chen, G.Y.; Dekyi, P.; Pan, F.; Xian, S.T.; Rinchen, D.; et al. Protective effects of traditional Tibetan medicine Zuo-Mu-A Decoction () on the blood parameters and myocardium of high altitude polycythemia model rats. Chin. J. Integr. Med. 2017, 23, 908–915. [Google Scholar] [CrossRef]
- Yan, G.H.; Choi, Y.H. Salidroside attenuates allergic airway inflammation through negative regulation of nuclear factor-kappa B and p38 mitogen-activated protein kinase. J. Pharmacol. Sci. 2014, 126, 126–135. [Google Scholar] [CrossRef]
- Goleva, E.; Jackson, L.P.; Harris, J.K.; Robertson, C.E.; Sutherland, E.R.; Hall, C.F.; Good, J.T., Jr.; Gelfand, E.W.; Martin, R.J.; Leung, D.Y. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am. J. Respir. Crit. Care Med. 2013, 188, 1193–1201. [Google Scholar] [CrossRef]
- Bhavsar, P.; Hew, M.; Khorasani, N.; Torrego, A.; Barnes, P.J.; Adcock, I.; Chung, K.F. Relative corticosteroid insensitivity of alveolar macrophages in severe asthma compared with non-severe asthma. Thorax 2008, 63, 784–790. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, X.D.; Kolosov, V.P.; Perelman, J.M. Salidroside reduces cold-induced mucin production by inhibiting TRPM8 activation. Int. J. Mol. Med. 2013, 32, 637–646. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Je, I.G.; Kim, D.S.; Kim, S.W.; Lee, S.; Lee, H.S.; Park, E.K.; Khang, D.; Kim, S.H. Tyrosol Suppresses Allergic Inflammation by Inhibiting the Activation of Phosphoinositide 3-Kinase in Mast Cells. PLoS ONE 2015, 10, e0129829. [Google Scholar] [CrossRef] [PubMed]
- Scarfone, R.J.; Friedlaender, E. Corticosteroids in acute asthma: Past, present, and future. Pediat. Emerg. Care 2003, 19, 355–361. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microb. 2015, 17, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, C.C.; Oliveira, A.S.; Santos, M.; Rudnitskaya, A.; Todo-Bom, A.; Bousquet, J.; Rocha, S.M. Urinary metabolomic profiling of asthmatics can be related to clinical characteristics. Allergy 2016, 71, 1362–1365. [Google Scholar] [CrossRef] [PubMed]
- Comhair, S.A.; McDunn, J.; Bennett, C.; Fettig, J.; Erzurum, S.C.; Kalhan, S.C. Metabolomic Endotype of Asthma. J. Immunol. 2015, 195, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Wang, G.; Wang, C.; Zhang, W.; Liu, J.; Wang, F. Serum Metabolomics Analysis of Asthma in Different Inflammatory Phenotypes: A Cross-Sectional Study in Northeast China. Biomed. Res. Int. 2018, 2018, 2860521. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, M.; Armstrong AJ, S.; Phelan, V.V.; Reisdorph, N.; Lozupone, C.A. Microbiome and metabolome data integration provides insight into health and disease. Transl. Res. 2017, 189, 51–64. [Google Scholar] [CrossRef]
- Haspeslagh, E.; Debeuf, N.; Hammad, H.; Lambrecht, B.N. Murine Models of Allergic Asthma. Methods Mol. Biol. 2017, 1559, 121–136. [Google Scholar]
- Park, J.; Lee, H.J.; Song, D.; Gi, M.; Jo, S.; Jeon, D.K.; Seo, Y.; Kim, B.; Lee, H.; Namkung, W.; et al. Novel pendrin inhibitor attenuates airway hyperresponsiveness and mucin expression in experimental murine asthma. J. Allergy Clin. Immunol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, P.; Naessens, T.; De Koker, S.; Hennuy, B.; Hacha, J.; Smet, M.; Cataldo, D.; Di Valentin, E.; Piette, J.; Tournoy, K.G.; et al. Inflammatory signatures for eosinophilic vs. neutrophilic allergic pulmonary inflammation reveal critical regulatory checkpoints. Am. J. Physiol. Lung Cell Mol. Physiol. 2011, 300, L679–L690. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jin, R.G.; Xiao, L.; Wang, Q.J.; Yan, T.H. Anti-asthma effects of synthetic salidroside through regulation of Th1/Th2 balance. Chin. J. Nat. Med. 2014, 12, 500–504. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Lim, S.; Oates, T.; Chung, K.F. Increase in airway neutrophils after oral but not inhaled corticosteroid therapy in mild asthma. Respir. Med. 2005, 99, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Yang, B.; Warrington, R.J.; Mink, S.; Kalicinsky, C.; Becker, A.B.; Simons, E.; Peng, Z. Myeloid-derived suppressor cells: Roles and relations with Th2, Th17 and Treg cells in asthma. Allergy 2019. [Google Scholar] [CrossRef]
- Wang, W.; Liu, Q.B.; Jing, W. Astragalus membranaceus improves therapeutic efficacy of asthmatic children by regulating the balance of Treg/Th17 cells. Chin. J. Nat. Med. 2019, 17, 252–263. [Google Scholar] [CrossRef]
- Poulakos, M.N.; Cargill, S.M.; Waineo, M.F.; Wolford, A.L., Jr. Mepolizumab for the treatment of severe eosinophilic asthma. Am. J. Health Syst. Pharm. 2017, 74, 963–969. [Google Scholar] [CrossRef]
- Wills-Karp, M.; Luyimbazi, J.; Xu, X.; Schofield, B.; Neben, T.Y.; Karp, C.L.; Donaldson, D.D. Interleukin-13: Central mediator of allergic asthma. Science 1998, 282, 2258–2261. [Google Scholar] [CrossRef]
- Zhao, J.; Lloyd, C.M.; Noble, A. Th17 responses in chronic allergic airway inflammation abrogate regulatory T-cell-mediated tolerance and contribute to airway remodeling. Mucosal Immunol. 2013, 6, 335–346. [Google Scholar] [CrossRef]
- Kudo, M.; Melton, A.C.; Chen, C.; Engler, M.B.; Huang, K.E.; Ren, X.; Wang, Y.; Bernstein, X.; Li, J.T.; Atabai, K.; et al. IL-17A produced by alphabeta T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat. Med. 2012, 18, 547–554. [Google Scholar] [CrossRef]
- Agache, I.; Akdis, C.A. Endotypes of allergic diseases and asthma: An important step in building blocks for the future of precision medicine. Allergol. Int. 2016, 65, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.W.; Kang, O.H.; Lee, Y.S.; Han, S.H.; Lee, S.W.; Cha, S.W.; Seo, Y.S.; Mun, S.H.; Gong, R.; Shin, D.W.; et al. Anti-inflammatory effect of salidroside on phorbol-12-myristate-13-acetate plus A23187-mediated inflammation in HMC-1 cells. Int. J. Mol. Med. 2016, 38, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Amini, S.E.; Gouyer, V.; Portal, C.; Gottrand, F.; Desseyn, J.L. Muc5b is mainly expressed and sialylated in the nasal olfactory epithelium whereas Muc5ac is exclusively expressed and fucosylated in the nasal respiratory epithelium. Histochem. Cell Biol. 2019, 152, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.K.; Yang, R.; Dunitz, J.M.; Boyer, H.C.; Hunter, R.C. 16S rRNA gene sequencing reveals site-specific signatures of the upper and lower airways of cystic fibrosis patients. J. Cyst. Fibros. 2018, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Srinivas, G.; Kuenzel, S.; Linnenbrink, M.; Alnahas, S.; Bruce, K.D.; Steinhoff, U.; Baines, J.F.; Schaible, U.E. Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS ONE 2014, 9, e113466. [Google Scholar] [CrossRef]
- Ickovic, M.R.; Henocq, E.; Relyveld, E.H.; Bizzini, B. Effect of immunostimulation with the Corynebacterium granulosum derived immunomodulator P40 on patients with recurring respiratory infections. J. Asthma. 1984, 21, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Lopes Dos Santos Santiago, G.; Brusselle, G.; Dauwe, K.; Deschaght, P.; Verhofstede, C.; Vaneechoutte, D.; Deschepper, E.; Jordens, P.; Joos, G.; Vaneechoutte, M. Influence of chronic azithromycin treatment on the composition of the oropharyngeal microbial community in patients with severe asthma. BMC Microbiol. 2017, 17, 109. [Google Scholar] [CrossRef]
- Larsen, J.M.; Musavian, H.S.; Butt, T.M.; Ingvorsen, C.; Thysen, A.H.; Brix, S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology 2015, 144, 333–342. [Google Scholar] [CrossRef]
- Itthitaetrakool, U.; Pinlaor, P.; Pinlaor, S.; Chomvarin, C.; Dangtakot, R.; Chaidee, A.; Wilailuckana, C.; Sangka, A.; Lulitanond, A.; Yongvanit, P. Chronic Opisthorchis viverrini Infection Changes the Liver Microbiome and Promotes Helicobacter Growth. PLoS ONE 2016, 11, e0165798. [Google Scholar] [CrossRef]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.K.; Yun, B.S.; Matsuzaki, K.; Furukawa, M.; Min, H.K.; Bajaj, J.S.; et al. Bile Acid 7alpha-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2019, 26, 27.e4–34.e4. [Google Scholar] [CrossRef]
- Colldahl, H. The Intestinal Flora in Patients with Bronchial Asthma and Rheumatoid Arthritis. Acta Allergol. 1965, 20, 94–104. [Google Scholar] [PubMed]
- Reynolds, J.M.; Lee, Y.H.; Shi, Y.; Wang, X.; Angkasekwinai, P.; Nallaparaju, K.C.; Flaherty, S.; Chang, S.H.; Watarai, H.; Dong, C. Interleukin-17B Antagonizes Interleukin-25-Mediated Mucosal Inflammation. Immunity 2015, 42, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt Yilmaz, H.E.; Kupeli, E.; Sen, N.; Arer, I.; Caliskan, K.; Akcay, S.; Haberal, M. Acute Respiratory Failure in Renal Transplant Recipients: A Single Center Experience. Exp. Clin. Transplant. 2019, 17 (Suppl. 1), 172–174. [Google Scholar] [CrossRef]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Essack, S.Y. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob. Resist Infect. Control 2018, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Cephus, J.Y.; Stier, M.T.; Fuseini, H.; Yung, J.A.; Toki, S.; Bloodworth, M.H.; Zhou, W.; Goleniewska, K.; Zhang, J.; Garon, S.L.; et al. Testosterone Attenuates Group 2 Innate Lymphoid Cell-Mediated Airway Inflammation. Cell Rep. 2017, 21, 2487–2499. [Google Scholar] [CrossRef] [PubMed]
- Drbalova, K.; Matucha, P.; Matejkova-Behanova, M.; Bilek, R.; Kriz, L.; Kazihnitkova, H.; Hampl, R. Immunoprotective steroids and SHBG in non-treated hypothyroidism and their relationship to autoimmune thyroid disorders. Physiol. Res. 2008, 57 (Suppl. 1), S119–S125. [Google Scholar]
- Willart, M.A.; van Nimwegen, M.; Grefhorst, A.; Hammad, H.; Moons, L.; Hoogsteden, H.C.; Lambrecht, B.N.; Kleinjan, A. Ursodeoxycholic acid suppresses eosinophilic airway inflammation by inhibiting the function of dendritic cells through the nuclear farnesoid X receptor. Allergy 2012, 67, 1501–1510. [Google Scholar] [CrossRef]
- Shaik, F.B.; Panati, K.; Narasimha, V.R.; Narala, V.R. Chenodeoxycholic acid attenuates ovalbumin-induced airway inflammation in murine model of asthma by inhibiting the T(H)2 cytokines. Biochem. Biophys. Res. Commun. 2015, 463, 600–605. [Google Scholar] [CrossRef]
- Delport, R.; Ubbink, J.B.; Serfontein, W.J.; Becker, P.J.; Walters, L. Vitamin B6 nutritional status in asthma: The effect of theophylline therapy on plasma pyridoxal-5’-phosphate and pyridoxal levels. Int. J. Vitam. Nutr. Res. 1988, 58, 67–72. [Google Scholar]
- Lee-Sarwar, K.A.; Kelly, R.S.; Lasky-Su, J.; Zeiger, R.S.; O’Connor, G.T.; Sandel, M.T.; Bacharier, L.B.; Beigelman, A.; Laranjo, N.; Gold, D.R.; et al. Integrative analysis of the intestinal metabolome of childhood asthma. J. Allergy Clin. Immunol. 2019, 144, 442–454. [Google Scholar] [CrossRef]
- Pretorius, E.; Wallner, B.; Marx, J. Cortisol resistance in conditions such as asthma and the involvement of 11beta-HSD-2: A hypothesis. Horm. Metab. Res. Horm. Stoffwechselforschung Horm. Metab. 2006, 38, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Wang, J.H. The effect of icariin on immunity and its potential application. Am. J. Clin. Exp. Immunol. 2018, 7, 50–56. [Google Scholar] [PubMed]
- Frati, F.; Salvatori, C.; Incorvaia, C.; Bellucci, A.; Di Cara, G.; Marcucci, F.; Esposito, S. The Role of the Microbiome in Asthma: The Gut(-)Lung Axis. Int. J. Mol. Sci. 2018, 20, 123. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.S.; Cheng, S.W.; Zhu, J.; Atli, K.; Chan, B.C.; Liu, D.; Chan, H.Y.; Sun, X.; Chu, I.M.; Hon, K.L.; et al. Anti-Inflammatory Activities of Pentaherbs formula and Its Influence on Gut Microbiota in Allergic Asthma. Molecules 2018, 23, 2776. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.P.; Xia, L.X.; Bao, Z.Q.; Zhang, H.; Xu, Z.W.; Mao, Y.Y.; Cao, C.; Che, L.Q.; Liu, J.K.; Li, W.; et al. Bcl-2 inhibitors reduce steroid-insensitive airway inflammation. J. Allergy Clin. Immunol. 2017, 140, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Chang, H.S.; Bae, D.J.; Song, H.J.; Kim, M.S.; Park, J.S.; Jun, J.A.; Lee, S.Y.; Uh, S.T.; Kim, S.H.; et al. Role of S100A9 in the development of neutrophilic inflammation in asthmatics and in a murine model. Clin. Immunol. 2017, 183, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Sert, N.P.d.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2019: Updated guidelines for reporting animal research. bioRxiv 2019, 2019, 703181. [Google Scholar]
- Kim, R.Y.; Pinkerton, J.W.; Essilfie, A.T.; Robertson AA, B.; Baines, K.J.; Brown, A.C.; Mayall, J.R.; Ali, M.K.; Starkey, M.R.; Hansbro, N.G.; et al. Role for NLRP3 Inflammasome-mediated, IL-1beta-Dependent Responses in Severe, Steroid-Resistant Asthma. Am. J. Respir. Crit. Care Med. 2017, 196, 283–297. [Google Scholar] [CrossRef]
- Hansbro, P.M.; Kim, R.Y.; Starkey, M.R.; Donovan, C.; Dua, K.; Mayall, J.R.; Liu, G.; Hansbro, N.G.; Simpson, J.L.; Wood, L.G.; et al. Mechanisms and treatments for severe, steroid-resistant allergic airway disease and asthma. Immunol. Rev. 2017, 278, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, Y.; Favoreto, S., Jr.; Zhen, G.; Park, S.W.; Nguyenvu, L.T.; Kuperman, D.A.; Dolganov, G.M.; Huang, X.; Boushey, H.A.; Avila, P.C.; et al. The epithelial anion transporter pendrin is induced by allergy and rhinovirus infection, regulates airway surface liquid, and increases airway reactivity and inflammation in an asthma model. J. Immunol. 2008, 181, 2203–2210. [Google Scholar] [CrossRef]
- Van Hoecke, L.; Job, E.R.; Saelens, X.; Roose, K. Bronchoalveolar Lavage of Murine Lungs to Analyze Inflammatory Cell Infiltration. J. Vis. Exp. 2017, 123. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Wang, G.; Ran, N.; Lin, H.; Wang, Z.; Guan, X.; Yuan, Y.; Fang, K.; Liu, J.; Wang, F. Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS). Int. J. Mol. Sci. 2018, 19, 2894. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.; Dillon, M.; Bokulich, N.; Abnet, C.; Al-Ghalith, G.; Alexander, H.; Alm, E.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Peer J. Preprints 2018, 6, e27295v2. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Montenegro-Burke, J.R.; Ivanisevic, J.; Thomas, A.; Sidibe, J.; Teav, T.; Guijas, C.; Aisporna, A.E.; Rinehart, D.; Hoang, L.; et al. XCMS-MRM and METLIN-MRM: A cloud library and public resource for targeted analysis of small molecules. Nat. Methods 2018, 15, 681–684. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]











| Species | P Values | FDR | ND | NDR | NEA | NR | LDA Score |
|---|---|---|---|---|---|---|---|
| Proteus mirabilis | 6.44 × 10−4 | 0.036724 | 116,110 | 162,152 | 14,237 | 9937 | 4.88 |
| Lactobacillus hamsteri | 0.0014144 | 0.040311 | 108,397 | 73,790 | 3508 | 3417 | 4.72 |
| M | Mass | RT | Compound Name | KEGG_ID | Δm | Pathways | Sample |
|---|---|---|---|---|---|---|---|
| 1 | 327.2008 | 2.04 | Hydroxydehydroepiandrosterone/ Hydroxytestosterone | C18045/C05139/C05291/C05294 | 2 | Steroid hormone biosynthesis | Serum |
| 2 | 327.2008 | 2.04 | LysoPC(18:3(9Z,12Z,15Z)) | C04230 | 2 | Glycerophospholipid metabolism | Serum |
| 3 | 371.2275 | 3.02 | Dihydroxypregnenolone | C05487/C05478 | 0 | Steroid hormone biosynthesis | Serum |
| 4 | 248.0278 | 5.67 | Pyridoxal 5’-phosphate | C00018 | 0 | Vitamin B6 metabolism | Serum |
| 5 | 745.416 | 6.11 | Octaprenyl diphosphate | C04146 | 4 | Terpenoid backbone biosynthesis | Serum |
| 6 | 435.2578 | 6.87 | LysoPA (18:2(9Z,12Z)/0:0) | C00416 | 17 | Glycerophospholipid metabolism | Serum |
| 7 | 465.247 | 6.87 | Testosterone glucuronide | C11134 | 2 | Steroid hormone biosynthesis | Serum |
| 8 | 655.3898 | 8.1 | All-trans-heptaprenyl diphosphate | C04216 | 2 | Terpenoid backbone biosynthesis | Serum |
| 9 | 339.1873 | 8.12 | Hydroxyretinoic/Epoxyretinoic acid/Deoxy-PGJ2 | C16677/C16679/C16680/C14717 | 17 | Retinol metabolism & Arachidonic acid | Serum |
| 10 | 361.2005 | 8.29 | Cortisone/Aldosterone | C00762/C01780 | 1 | Steroid hormone biosynthesis | Serum |
| 11 | 318.3007 | 12.79 | Phytosphingosine | C12144 | 1 | Sphingolipid metabolism | Serum |
| 12 | 302.3065 | 14.99 | Sphinganine | C00836 | 4 | Sphingolipid metabolism | BALF |
| 13 | 303.2323 | 18.12 | Bovinic acid/Linoleic acid/Retinyl ester | C04056 | 1 | Linoleic acid metabolism | Serum |
| 14 | 298.1327 | 18.42 | Phenethylamine glucuronide | C03033 | 13 | Starch and sucrose metabolism | BALF |
| 15 | 349.2352 | 19.13 | Dihydroxypregnanedione | C05478 | 1 | Steroid hormone biosynthesis | Serum |
| 16 | 626.2956 | 19.19 | Leukotriene C4 | C02166 | 20 | Arachidonic acid metabolism | Serum |
| 17 | 609.3009 | 20.04 | All-trans-hexaprenyl diphosphate | C01230 | 6 | Terpenoid backbone biosynthesis | Serum |
| 18 | 499.2297 | 20.3 | Retinoyl b-glucuronide | C11061 | 1 | Retinol metabolism | Serum |
| 19 | 450.3213 | 22.01 | Chenodeoxycholic acid glycine conjugate | C05466 | 0 | Primary bile acid biosynthesis | BALF |
| 20 | 265.2516 | 22.01 | 1-Hexadecanol | C00823 | 4 | Fatty acid metabolism | BALF |
| 21 | 433.2227 | 23.08 | LysoPA (16:0/0:0) | C00416 | 1 | Glycerophospholipid metabolism | BALF |
| 22 | 555.2908 | 23.12 | 5b-Cyprinol sulfate | C05468 | 4 | Primary bile acid biosynthesis | Serum |
| 23 | 349.2357 | 23.13 | 11b,21-Dihydroxy-5b-pregnane-3,20-dione | C05475 | 2 | Steroid hormone biosynthesis | Serum |
| 24 | 469.1846 | 23.44 | Estrone glucuronide | C11133 | 2 | Steroid hormone biosynthesis | Serum |
| 25 | 335.2568 | 24.1 | Tetrahydrodeoxycorticosterone | C13713 | 3 | Steroid hormone biosynthesis | Serum |
| 26 | 742.5645 | 26.69 | PC (18:2(9Z,12Z)/P-16:0) | C00157 | 13 | Glycerophospholipid metabolism | Serum |
| 27 | 760.5777 | 27.04 | PC (18:1(9Z)/16:0)/PE (22:1(13Z)/15:0) | C00157/C00350 | 10 | Glycerophospholipid metabolism | BALF |
| 28 | 788.6045 | 27.06 | PC (14:0/22:1(13Z)) | C00157 | 15 | Linoleic acid metabolism | Serum |
| 29 | 798.585 | 27.1 | PE (22:0/16:0) | C00350 | 17 | Glycerophospholipid metabolism | Serum |
| 30 | 703.5725 | 27.3 | SM( d18:1/16:0) | C00550 | 3 | Sphingolipid metabolism | Serum |
| 31 | 782.5662 | 27.56 | PC (18:4(6Z,9Z,12Z,15Z)/18:0)/PE (22:1(13Z)/15:0) | C00157/C00350 | 1 | Glycerophospholipid metabolism | BALF |
| 32 | 732.5517 | 27.75 | PE (15:0/20:1(11Z)) | C00350 | 3 | Glycerophospholipid metabolism | BALF |
| 33 | 349.2334 | 28.62 | Dihydroxypregnanedione | C05475/C05478/C05487 | 4 | Steroid hormone biosynthesis | BALF |
| 34 | 425.341 | 28.7 | Hydroxycholesterol | C05500/C05502/C15519/C03594/ C13550/C05451 | 0 | Steroid hormone & Primary bile acid biosynthesis | Serum |
| Groups | Treatments | Routes of Administration | Number (n) |
|---|---|---|---|
| Control | N/A | N/A | 18 |
| EA | NS | i.g | 18 |
| RWC | i.g | 18 | |
| DEX | i.p | 18 | |
| RWC + DEX | i.g + i.p | 18 | |
| NEA | NS | i.g | 18 |
| RWC | i.g | 18 | |
| DEX | i.p | 18 | |
| RWC + DEX | i.g + i.p | 18 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Z.; Ran, N.; Yuan, Y.; Wang, C.; Wang, G.; Lin, H.; Hsu, A.C.-Y.; Liu, J.; Wang, F. Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism. Int. J. Mol. Sci. 2019, 20, 4216. https://doi.org/10.3390/ijms20174216
Pang Z, Ran N, Yuan Y, Wang C, Wang G, Lin H, Hsu AC-Y, Liu J, Wang F. Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism. International Journal of Molecular Sciences. 2019; 20(17):4216. https://doi.org/10.3390/ijms20174216
Chicago/Turabian StylePang, Zhiqiang, Nan Ran, Yuze Yuan, Cuizhu Wang, Guoqiang Wang, Hongqiang Lin, Alan Chen-Yu Hsu, Jinping Liu, and Fang Wang. 2019. "Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism" International Journal of Molecular Sciences 20, no. 17: 4216. https://doi.org/10.3390/ijms20174216
APA StylePang, Z., Ran, N., Yuan, Y., Wang, C., Wang, G., Lin, H., Hsu, A. C.-Y., Liu, J., & Wang, F. (2019). Phenotype-Specific Therapeutic Effect of Rhodiola wallichiana var. cholaensis Combined with Dexamethasone on Experimental Murine Asthma and Its Comprehensive Pharmacological Mechanism. International Journal of Molecular Sciences, 20(17), 4216. https://doi.org/10.3390/ijms20174216






