Comparative Quantitative Analysis of Porcine Optic Nerve Head and Retina Subproteomes
Abstract
:1. Introduction
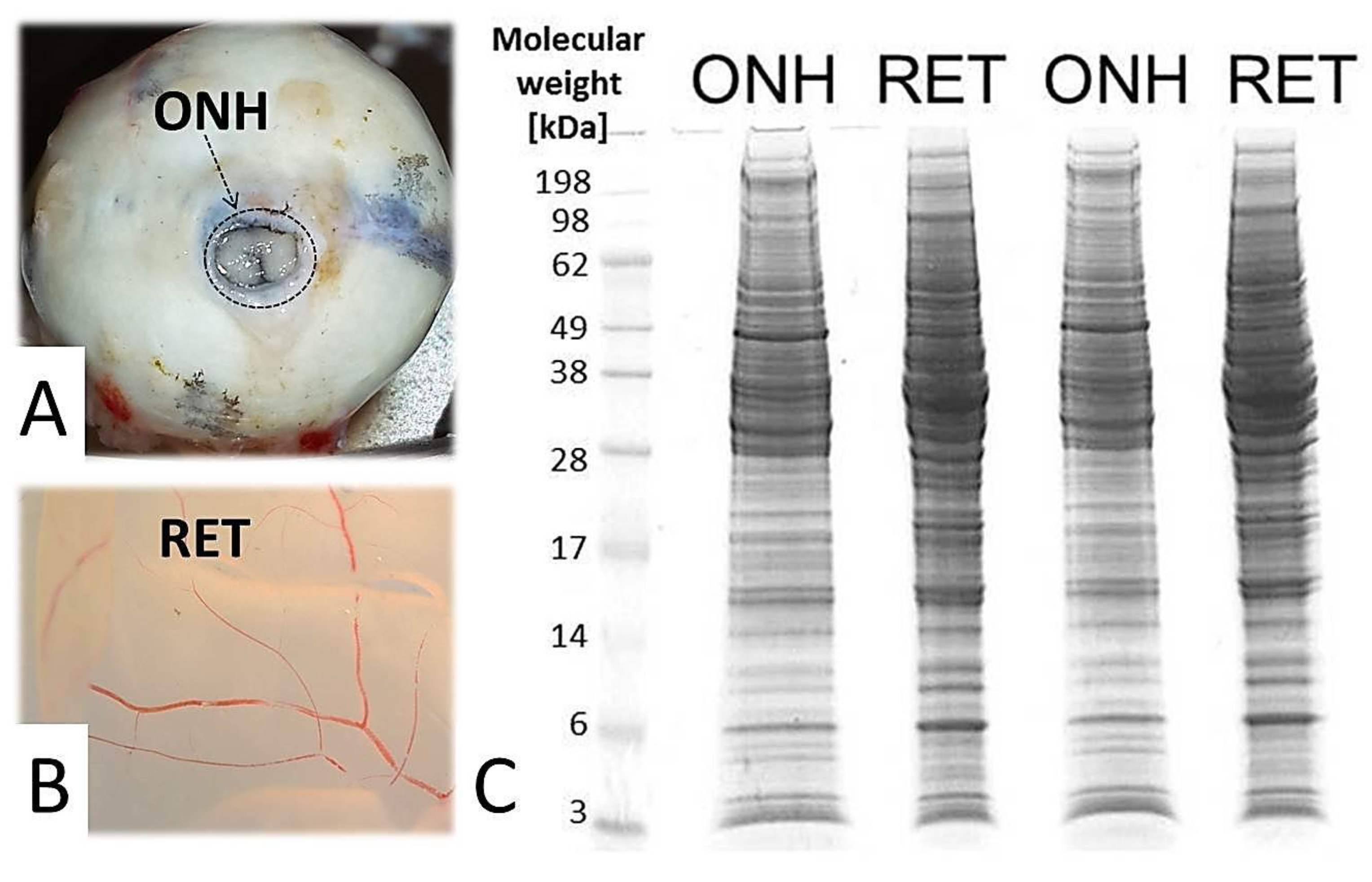
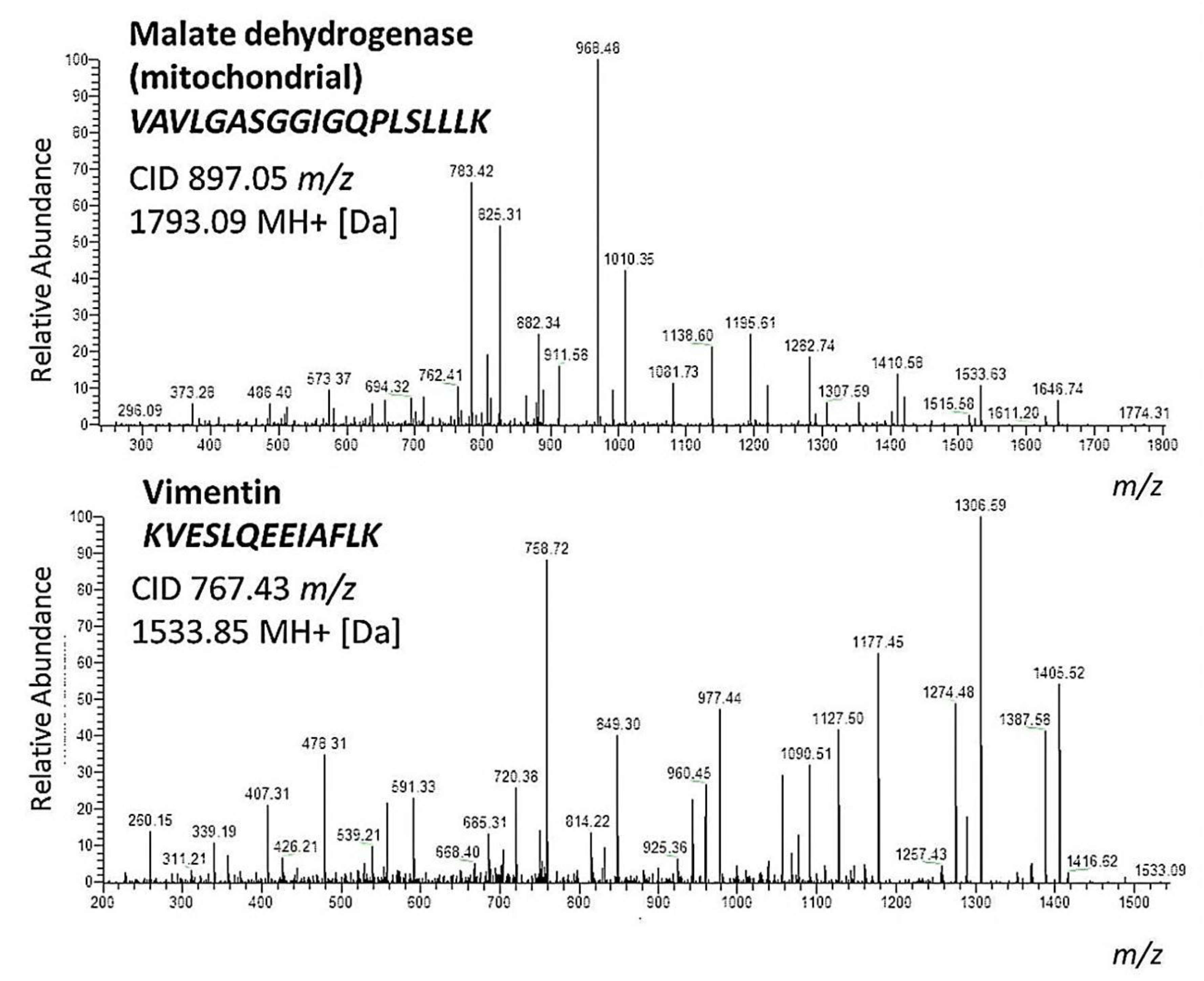
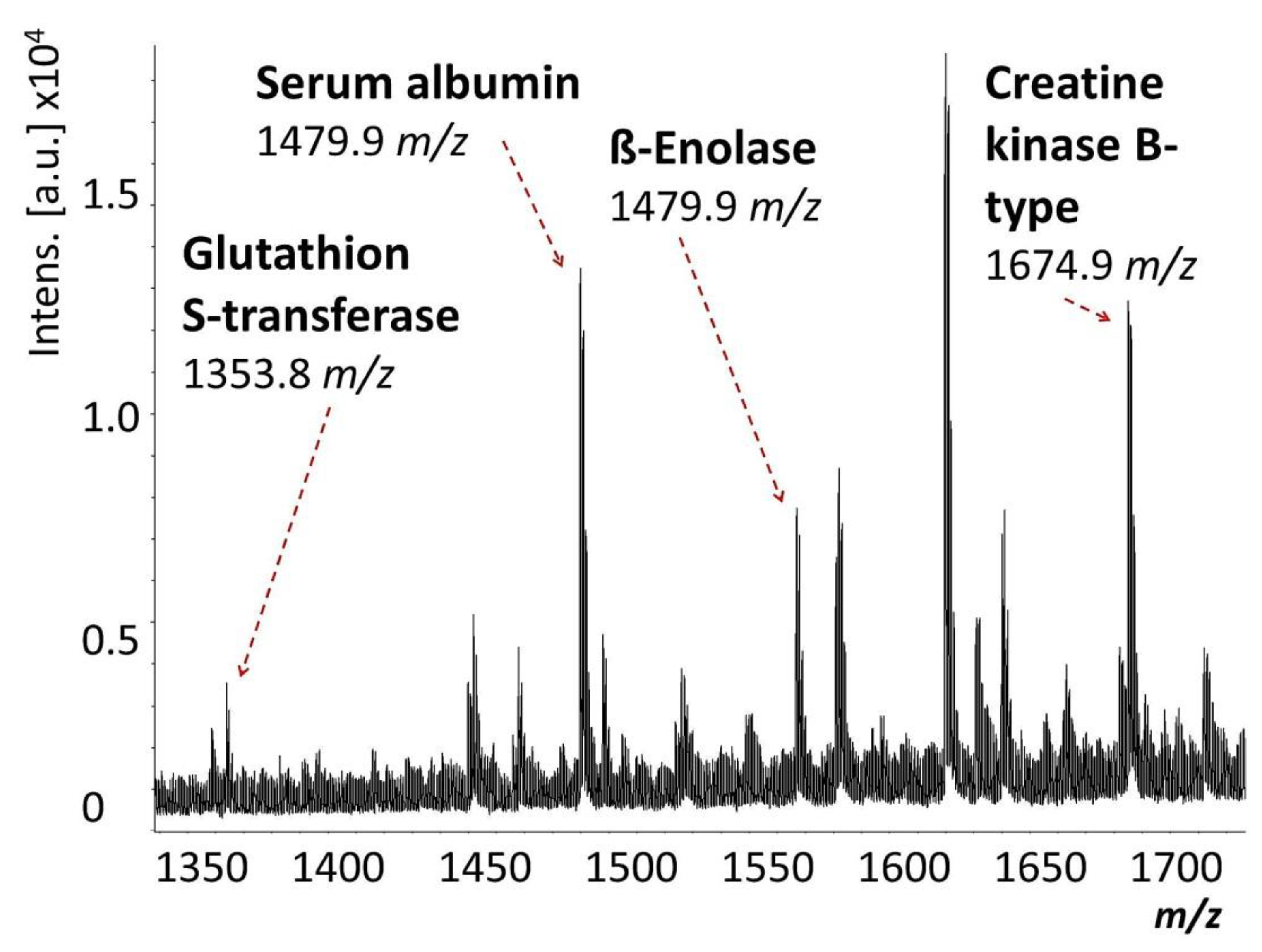
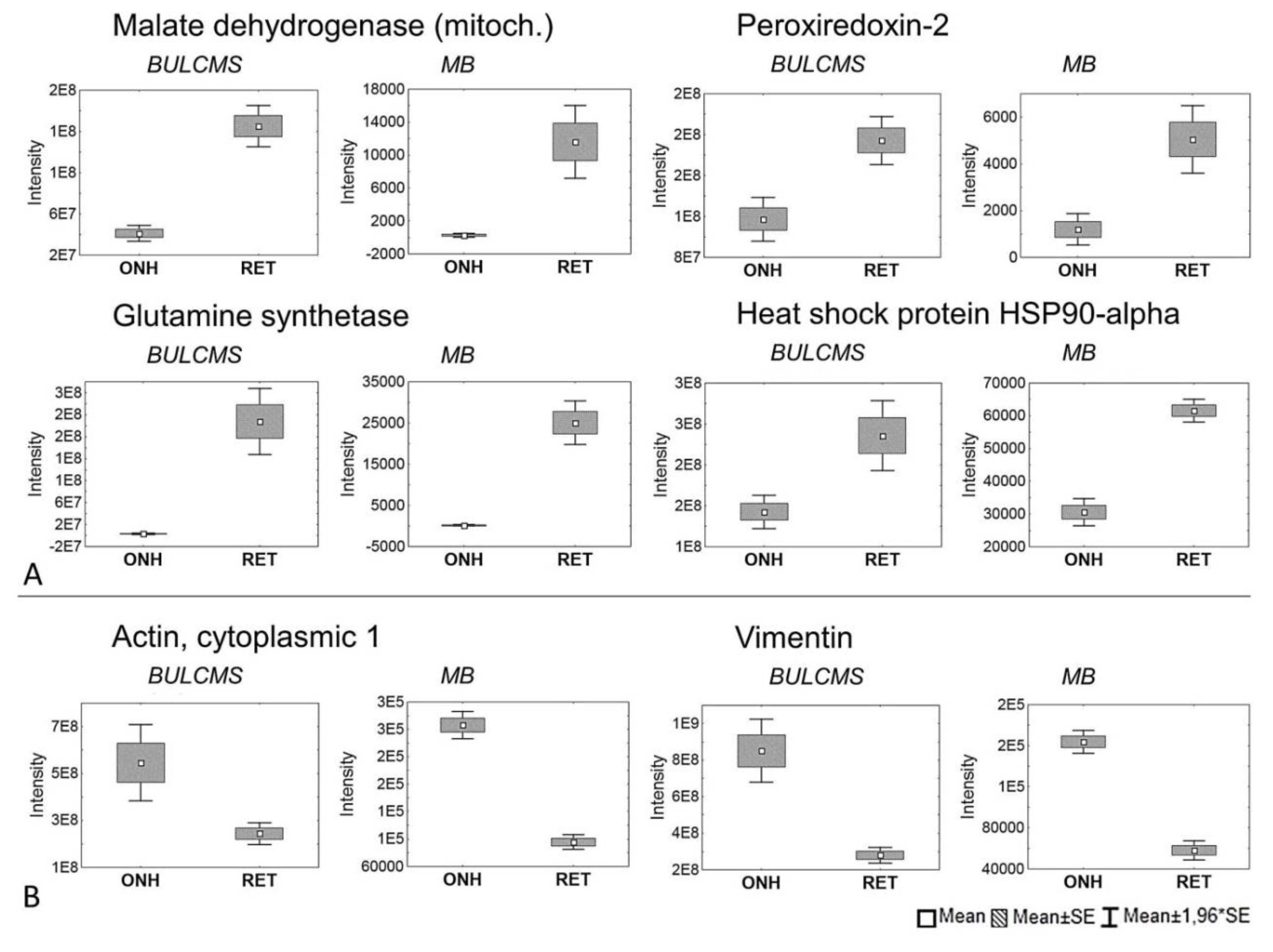
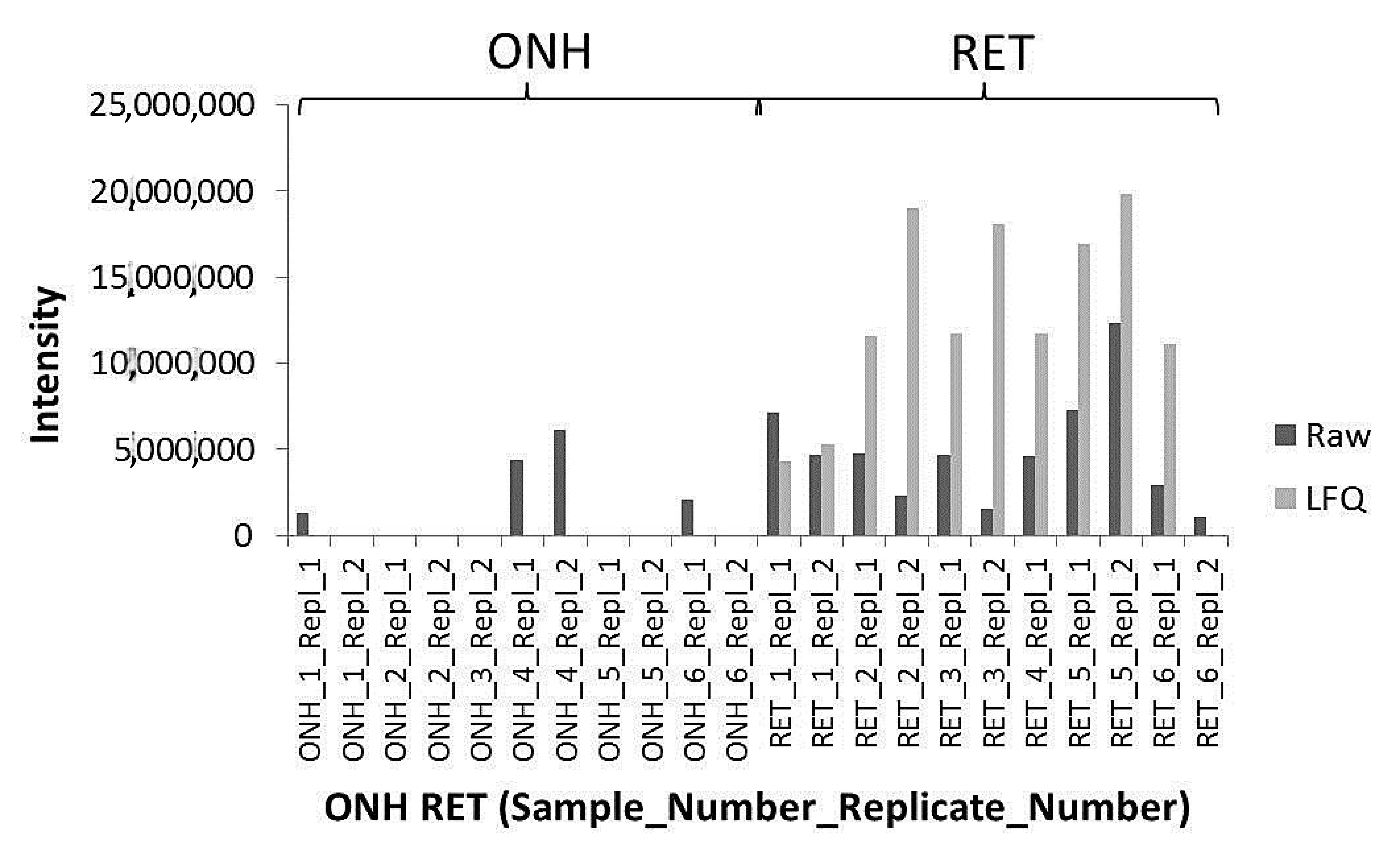
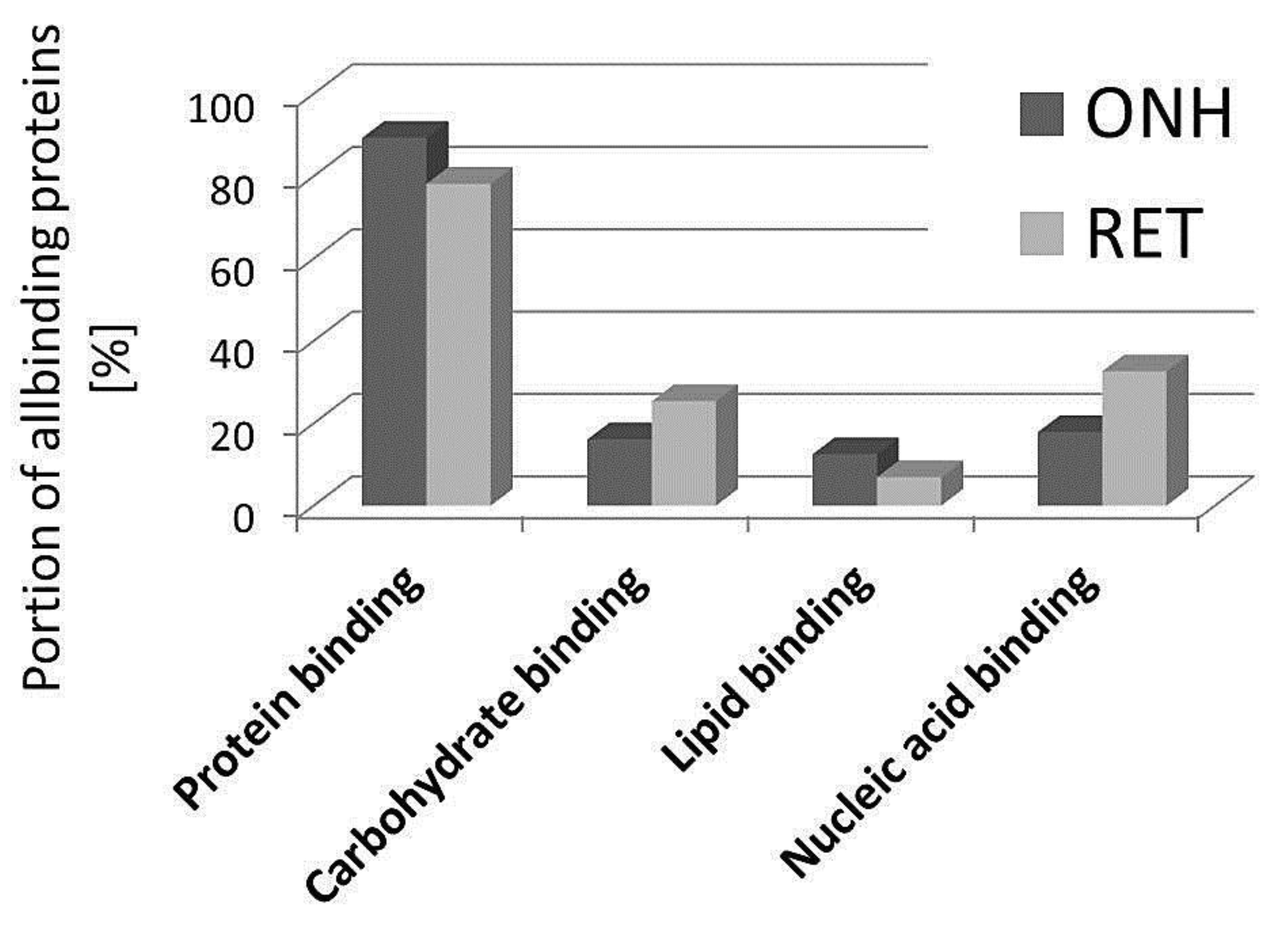
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. BULCMS Analysis
4.3. MECP2 Targeted Detection
4.4. MALDI-TOF MS Analysis
4.5. Functional Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Funke, S.; Perumal, N.; Bell, K.; Pfeiffer, N.; Grus, F.H. The potential impact of recent insights into proteomic changes associated with glaucoma. Expert Rev. Proteom. 2017, 14, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bendixen, E.; Danielsen, M.; Larsen, K.; Bendixen, C. Advances in porcine genomics and proteomics-a toolbox for developing the pig as a model organism for molecular biomedical research. Brief. Funct. Genom. 2010, 9, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Fan, N.; Lai, L.C. Genetically modified pig models for human diseases. J. Genet. Genom. 2013, 40, 67–73. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, A.M.; Bendixen, E. Pig proteomics: A review of a species in the crossroad between biomedical and food sciences. J. Proteom. 2012, 75, 4296–4314. [Google Scholar] [CrossRef] [PubMed]
- Ceciliani, F.; Restelli, L.; Lecchi, C. Proteomics in farm animals models of human diseases. Proteom. Clin. Appl. 2014, 8, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabria, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom. Clin. Appl. 2014, 8, 715–731. [Google Scholar] [CrossRef]
- Ruiz-Ederra, J.; Garcia, M.; Hernandez, M.; Urcola, H.; Hernandez-Barbachano, E.; Araiz, J.; Vecino, E. The pig eye as a model of glaucoma. Exp. Eye Res. 2005, 81, 561–569. [Google Scholar] [CrossRef]
- Marino, L.; Colvin, C.M. Thinking pigs: A comparative review of cognition, emotion, and personality in sus domesticus. Int. J. Comp. Psychol. 2015, 28, 1–22. [Google Scholar]
- Verma, N.; Rettenmeier, A.W.; Schmitz-Spanke, S. Recent advances in the use of sus scrofa (pig) as a model system for proteomic studies. Proteomics 2011, 11, 776–793. [Google Scholar] [CrossRef]
- Funke, S.; Markowitsch, S.; Schmelter, C.; Perumal, N.; Mwiiri, F.K.; Gabel-Scheurich, S.; Pfeiffer, N.; Grus, F.H. In-depth proteomic analysis of the porcine retina by use of a four step differential extraction bottom up lc ms platform. Mol. Neurobiol. 2016, 54, 7262–7275. [Google Scholar] [CrossRef]
- Funke, S.; Beutgen, V.M.; Bechter, L.; Schmelter, C.; Zurawski, V.; Perumal, N.; Pfeiffer, N.; Grus, F.H. An in-depth view of the porcine trabecular meshwork proteome. Int. J. Mol. Sci. 2019, 20, 2526. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, C.; Funke, S.; Treml, J.; Beschnitt, A.; Perumal, N.; Manicam, C.; Pfeiffer, N.; Grus, F.H. Comparison of two solid-phase extraction (spe) methods for the identification and quantification of porcine retinal protein markers by lc-ms/ms. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Funke, S.; Perumal, N.; Beck, S.; Gabel-Scheurich, S.; Schmelter, C.; Teister, J.; Gerbig, C.; Gramlich, O.W.; Pfeiffer, N.; Grus, F.H. Glaucoma related proteomic alterations in human retina samples. Sci. Rep. 2016, 6, 29759. [Google Scholar] [CrossRef] [PubMed]
- Cehofski, L.J.; Kruse, A.; Kjaergaard, B.; Stensballe, A.; Honore, B.; Vorum, H. Dye-free porcine model of experimental branch retinal vein occlusion: A suitable approach for retinal proteomics. J. Ophthalmol. 2015, 2015, 839137. [Google Scholar] [PubMed]
- Hauck, S.M. Proteomic analysis of the porcine interphotoreceptor matrix. Proteomics 2005, 5, 3623–3636. [Google Scholar] [CrossRef] [PubMed]
- Hauck, S.M.; Suppmann, S.; Ueffing, M. Proteomic profiling of primary retinal muller glia cells reveals a shift in expression patterns upon adaptation to in vitro conditions. Glia 2003, 44, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Chahrour, M.; Jung, S.Y.; Shaw, C.; Zhou, X.; Wong, S.T.; Qin, J.; Zoghbi, H.Y. Mecp2, a key contributor to neurological disease, activates and represses transcription. Science 2008, 320, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Minh, V.C.N.; Du, F.; Felice, C.A.; Shan, X.W.; Nigam, A.; Mandel, G.; Robinson, J.K.; Ballas, N. Mecp2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. J. Neurosci. 2012, 32, 10021–10034. [Google Scholar]
- Hesselager, M.O.; Codrea, M.C.; Sun, Z.; Deutsch, E.W.; Bennike, T.B.; Stensballe, A.; Bundgaard, L.; Moritz, R.L.; Bendixen, E. The pigpeptideatlas: A recource for systems biology in animal production and biomedicine. Proteomics 2016, 16, 634–644. [Google Scholar] [CrossRef]
- Brandstätter, J.H.; Wässle, H.; Betz, H.; Morgans, C.W. The plasma membrane protein snap-25, but not syntaxin, is present at photoreceptor and bipolar cell synapses in the rat retina. Eur. J. Neurosci. 1996, 8, 823–828. [Google Scholar] [CrossRef]
- Bhattacharya, S.K.; Crabb, J.S.; Bonilha, V.L.; Gu, X.R.; Takahara, H.; Crabb, J.W. Proteomics implicates peptidyl arginine deiminase 2 and optic nerve citrullination in glaucoma pathogenesis. Investig. Ophthalmol. Vis. Sci. 2006, 47, 2508–2514. [Google Scholar] [CrossRef]
- Cafaro, T.A.; Santo, S.; Robles, L.A.; Crim, N.; Urrets-Zavalia, J.A.; Serra, H.M. Peptidyl arginine deiminase type 2 is over expressed in the glaucomatous optic nerve. Mol. Vis. 2010, 16, 1654–1658. [Google Scholar] [PubMed]
- Nagelhus, E.A.; Veruki, M.L.; Torp, R.; Haug, F.M.; Laake, J.H.; Nielsen, S.; Agre, P.; Ottersen, O.P. Aquaporin-4 water channel protein in the rat retina and optic nerve: Polarized expression in muller cells and fibrous astrocytes. J. Neurosci. 1998, 18, 2506–2519. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.J.; Tezel, G.; Patil, R.V.; Romano, C.; Wax, M.B. Serum autoantibody against glutathione s-transferase in patients with glaucoma. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1273–1276. [Google Scholar]
- Hernandez, M.R.; Agapova, O.A.; Yang, P.; Salvador-Silva, M.; Ricard, C.S.; Aoi, S. Differential gene expression in astrocytes from human normal and glaucomatous optic nerve head analyzed by cdna microarray. Glia 2002, 38, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Tezel, G.; Yang, X.; Cai, J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure-induced rat model of glaucoma. Investig. Ophthalmol. Vis. Sci. 2005, 46, 3177–3187. [Google Scholar] [CrossRef]
- Shen, F.; Chen, B.; Danias, J.; Lee, K.C.; Lee, H.; Su, Y.; Podos, S.M.; Mittag, T.W. Glutamate-induced glutamine synthetase expression in retinal müller cells after short-term ocular hypertension in the rat. Investig. Ophthalmol. Vis. Sci. 2004, 45, 3107–3112. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.C.; Sande, P.; Marcos, H.A.; de Zavalia, N.; Sarmiento, M.I.K.; Rosenstein, R.E. Effect of glaucoma on the retinal glutamate/glutamine cycle activity. Faseb J. 2005, 19, 1161–1162. [Google Scholar] [CrossRef] [Green Version]
- Kulijewicz-Nawrot, M.; Sykova, E.; Chvatal, A.; Verkhratsky, A.; Rodriguez, J.J. Astrocytes and glutamate homoeostasis in alzheimer’s disease: A decrease in glutamine synthetase, but not in glutamate transporter-1, in the prefrontal cortex. Asn Neuro 2013, 5, 273–282. [Google Scholar] [CrossRef]
- Karaca, M.; Maechler, P. Development of mice with brain-specific deletion of floxed glud1 (glutamate dehydrogenase 1) using cre recombinase driven by the nestin promoter. Neurochem. Res. 2014, 39, 456–459. [Google Scholar] [CrossRef]
- Ross, C.D.; Godfrey, D.A. Distributions of asparte aminotransferase and malate dehydrogenase activities in rat retinal layers. J. Histochem. Cytochem. 1985, 30, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.D.; Godfrey, D.A. Distribution of activities of aspartate aminotransferase isoenzymes and malate dehydrogenase in guinea pig retinal layers. J. Histochem. Cytochem. 1987, 35, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Barrera, G.; Gentile, F.; Pizzimenti, S.; Canuto, R.A.; Daga, M.; Arcaro, A.; Cetrangolo, G.P.; Lepore, A.; Ferretti, C.; Dianzani, C.; et al. Mitochondrial dysfunction in cancer and neurodegenerative diseases: Spotlight on fatty acid oxidation and lipoperoxidation products. Antioxidants 2016, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Middeldorp, J.; Hol, E.M. Gfap in health and disease. Prog. Neurobiol. 2011, 93, 421–443. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, K.K.W. Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015, 38, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Arjamaa, O.; Nikinmaa, M. Oxygen-dependent diseases in the retina: Role of hypoxia-inducible factors. Exp. Eye Res. 2006, 83, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Y.; Pedersen, C.; Day, B.J.; Patel, M. Dependence of excitotoxic neurodegeneration on mitochondrial aconitase inactivation. J. Neurochem. 2001, 78, 746–755. [Google Scholar] [CrossRef] [Green Version]
- Vandewalle, E.; Pinto, L.A.; Olafsdottir, O.B.; De Clerck, E.; Stalmans, P.; Van Calster, J.; Zeyen, T.; Stefansson, E.; Stalmans, I. Oximetry in glaucoma: Correlation of metabolic change with structural and functional damage. Acta Ophthalmol. 2014, 92, 105–110. [Google Scholar] [CrossRef]
- Stasi, K.; Nagel, D.; Yang, X.; Ren, L.; Mittag, T.; Danias, J. Ceruloplasmin upregulation in retina of murine and human glaucomatous eyes. Investig. Ophthalmol. Vis. Sci. 2007, 48, 727–732. [Google Scholar] [CrossRef]
- Sarnat-Kucharczyk, M.; Rokicki, W.; Zalejska-Fiolka, J.; Pojda-Wilczek, D.; Mrukwa-Kominek, E. Determination of serum ceruloplasmin concentration in patients with primary open angle glaucoma with cataract and patients with cataract only: A pilot study. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016, 22, 1384–1388. [Google Scholar] [CrossRef]
- Padhy, B.; Nanda, G.G.; Chowdhury, M.; Padhi, D.; Rao, A.A.; Alone, D.P. Role of an extracellular chaperone, clusterin in the pathogenesis of pseudoexfoliation syndrome and pseudoexfoliation glaucoma. Exp. Eye Res. 2014, 127, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Doudevski, I.; Rostagno, A.A.; Cowman, M.; Liebmann, J.; Ritch, R.; Ghiso, J. Clusterin and complement activation in exfoliation glaucoma. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2491–2499. [Google Scholar] [CrossRef] [PubMed]
- Charnay, Y.; Imhof, A.; Vallet, P.G.; Kovari, E.; Bouras, C.; Giannakopoulos, P. Clusterin in neurological disorders: Molecular perspectives and clinical relevance. Brain Res. Bull. 2012, 88, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.R.; Di Paolo, G.; Werner, H.; Shchedrina, V.A.; Pypaert, M.; Pieribone, V.A.; De Camilli, P. A role for talin in presynaptic function. J. Cell Biol. 2004, 167, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, S.; Shattil, S.J.; Eto, K.; Tai, V.; Liddington, R.C.; de Pereda, J.M.; Ginsberg, M.H.; Calderwood, D.A. Talin binds to integrin ß tails: A final common step in integrin activation. Science 2003, 302, 103–106. [Google Scholar] [CrossRef]
- Pereira, A.M.; Tudor, C.; Kanger, J.S.; Subramanianm, V.; Martin-Blanco, E. Integrin-dependent activation of jnk signaling pathway by mechanical stress. PLoS ONE 2011, 6, e26182. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J. Integrins in the optic nerve head: Potential roles in glaucomatous optic neuropathy (an american ophthalmological society thesis). Trans. Am. Ophthalmol. Soc. 2006, 104, 453–477. [Google Scholar]
- Gallego, B.I.; Salazar, J.J.; de Hoz, R.; Rojas, B.; Ramirez, A.I.; Salinas-Navarro, M.; Ortin-Martinez, A.; Valiente-Soriano, F.J.; Aviles-Trigueros, M.; Villegas-Perez, M.P.; et al. Iop induces upregulation of gfap and mhc-ii and microglia reactivity in mice retina contralateral to experimental glaucoma. J. Neuroinflamm. 2012, 9, 92. [Google Scholar] [CrossRef]
- Huang, W.; Fileta, J.B.; Filippopoulos, T.; Ray, A.; Dobberfuhl, A.; Grosskreutz, C.L. Hsp27 phosphorylation in experimental glaucoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 4129–4135. [Google Scholar] [CrossRef]
- Piri, N.; Kwong, J.M.; Caprioli, J. Crystallins in retinal ganglion cell survival and regeneration. Mol. Neurobiol. 2013, 48, 819–828. [Google Scholar] [CrossRef]
- Chader, G.J. Animal models in research on retinal degenerations: Past progress and future hope. Vis. Res. 2002, 42, 393–399. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.; Funke, S.; Pfeiffer, N.; Grus, F.H. Serum and antibodies of glaucoma patients lead to changes in the proteome, especially cell regulatory proteins, in retinal cells. PLoS ONE 2012, 7, e46910. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.; Wilding, C.; Funke, S.; Perumal, N.; Beck, S.; Wolters, D.; Holz-Muller, J.; Pfeiffer, N.; Grus, F.H. Neuroprotective effects of antibodies on retinal ganglion cells in an adolescent retina organ culture. J. Neurochem. 2016, 139, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, C.; Perumal, N.; Funke, S.; Bell, K.; Pfeiffer, N.; Grus, F.H. Peptides of the variable igg domain as potential biomarker candidates in primary open-angle glaucoma (poag). Hum. Mol. Genet. 2017, 26, 4451–4464. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed maxlfq. Mol. Cell. Proteom. MCP 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Ausio, J.; de Paz, A.M.; Esteller, M. Mecp2: The long trip from a chromatin protein to neurological disorders. Trends Mol. Med. 2014, 20, 487–498. [Google Scholar]
- Guy, J.; Cheval, H.; Selfridge, J.; Bird, A. The role of mecp2 in the brain. Annu. Rev. Cell Dev. Biol. 2011, 27, 631–652. [Google Scholar] [CrossRef]
- Jaffe, J.D.; Keshishian, H.; Chang, B.; Addona, T.A.; Gillette, M.A.; Carr, S.A. Accurate inclusion mass screening a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol. Cell. Proteom. 2008, 7, 1952–1962. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Funke, S.; Schmelter, C.; Markowitsch, S.D.; Perumal, N.; Heyne, J.C.; Bell, K.; Pfeiffer, N.; Grus, F.H. Comparative Quantitative Analysis of Porcine Optic Nerve Head and Retina Subproteomes. Int. J. Mol. Sci. 2019, 20, 4229. https://doi.org/10.3390/ijms20174229
Funke S, Schmelter C, Markowitsch SD, Perumal N, Heyne JC, Bell K, Pfeiffer N, Grus FH. Comparative Quantitative Analysis of Porcine Optic Nerve Head and Retina Subproteomes. International Journal of Molecular Sciences. 2019; 20(17):4229. https://doi.org/10.3390/ijms20174229
Chicago/Turabian StyleFunke, Sebastian, Carsten Schmelter, Sascha D. Markowitsch, Natarajan Perumal, Janis C. Heyne, Katharina Bell, Norbert Pfeiffer, and Franz H. Grus. 2019. "Comparative Quantitative Analysis of Porcine Optic Nerve Head and Retina Subproteomes" International Journal of Molecular Sciences 20, no. 17: 4229. https://doi.org/10.3390/ijms20174229





