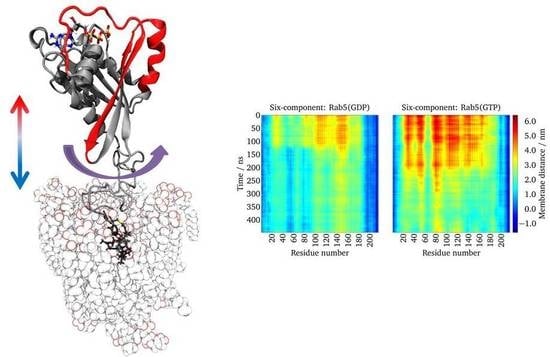
Structure and Dynamics of Mono- vs. Doubly Lipidated Rab5 in Membranes
Abstract
:1. Introduction
2. Results
2.1. Membrane Properties
2.2. Self-Diffusion of Lipid Molecules in the Membrane
2.3. Protein Diffusion in Different Membranes
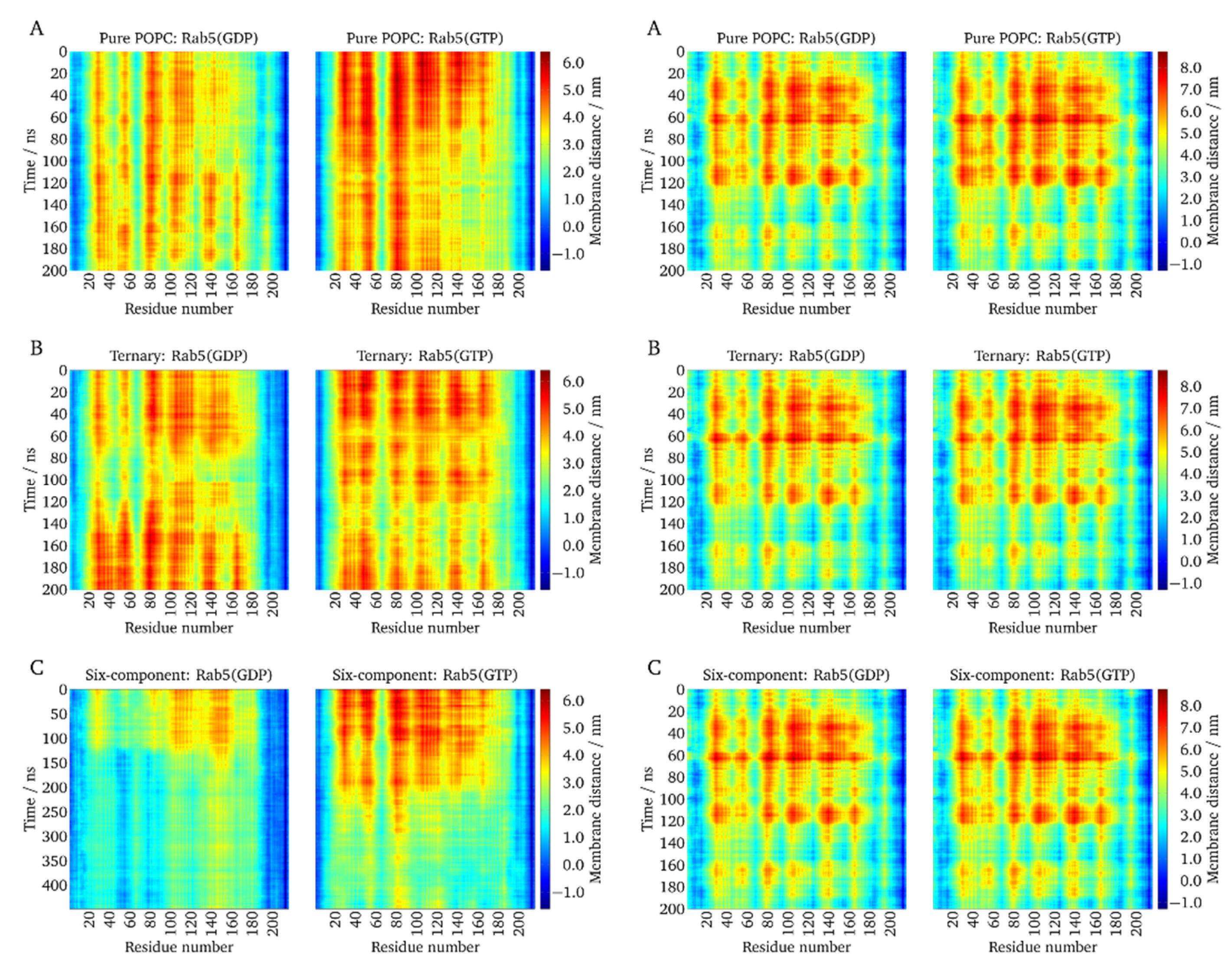
2.4. Local Membrane Perturbations by Geranlygeranyl Anchors
2.5. Lipid Anchor Insertion into Membrane and Protein–Membrane Distance
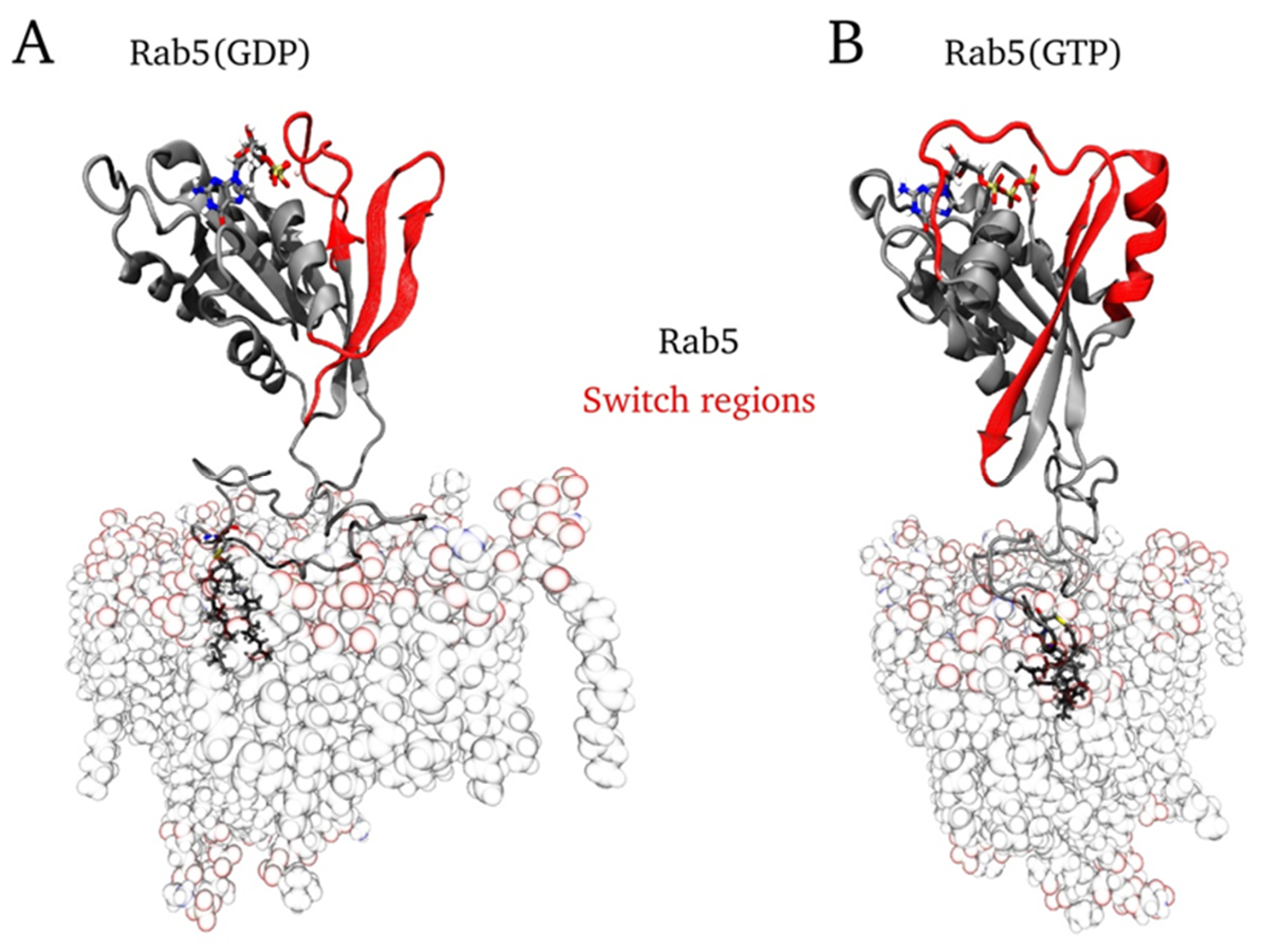
2.6. Protein Orientation at the Membrane Surface
3. Discussion
4. Computational Details
4.1. Protein Model Generation
4.2. Molecular Dynamics- Computational Details
4.3. Parameterization of Geranylgeranyl
4.4. Composition of Different Membranes
4.5. Starting Configuration
4.6. Geometrical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, G.P.; Stahl, P.D. Structure-function relationship of the small GTPase Rab5. J. Biol. Chem. 1993, 268, 24475–24480. [Google Scholar] [PubMed]
- Stenmark, H.; Valencia, A.; Martinez, O.; Ullrich, O.; Goud, B.; Zerial, M. Distinct structural elements ofRrab5 define its functional specificity. EMBO. J. 1994, 13, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.; Pilli, M.; Bernauer, S.; Habermann, B.H.; Zerial, M.; Wade, R.C. The interaction properties of the human Rab GTPase family: A comparative analysis reveals determinants of molecular binding selectivity. PLoS ONE 2012, 7, e34870. [Google Scholar] [CrossRef] [PubMed]
- Edler, E.; Stein, M. Probing the druggability of membrane-bound Rab5 by molecular dynamics simulations. J. Enzym. Inhib. Med. Chem. 2017, 32, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Christoforidis, S.; McBride, H.M.; Burgoyne, R.D.; Zerial, M. The Rab5 effector EEA1 is a core component of endosome docking. Nature 1999, 397, 621–625. [Google Scholar] [CrossRef]
- Pyrzynska, B.; Pilecka, I.; Miaczynska, M. Endocytic proteins in the regulation of nuclear signaling, transcription and tumorigenesis. Mol. Oncol. 2009, 3, 321–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miaczynska, M. Effects of membrane trafficking on signaling by receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, 20. [Google Scholar] [CrossRef]
- Howe, C.L.; Mobley, W.C. Signaling endosome hypothesis: A cellular mechanism for long distance communication. J. Neurobiol. 2004, 58, 207–216. [Google Scholar] [CrossRef]
- Fang, F.; Yang, W.L.; Florio, J.B.; Rockenstein, E.; Spencer, B.; Orain, X.M.; Dong, S.X.; Li, H.Y.; Chen, X.Q.; Sung, K.J. , et al. Synuclein impairs trafficking and signaling of BDNF in a mouse model of parkinson's disease. Sci. Rep. 2017, 7, 3868. [Google Scholar] [CrossRef]
- Veleri, S.; Punnakkal, P.; Dunbar, G.L.; Maiti, P. Molecular insights into the roles of Rab proteins in intracellular dynamics and neurodegenerative diseases. Neuromolecular Med. 2018, 20, 18–36. [Google Scholar] [CrossRef]
- Ravikumar, B.; Imarisio, S.; Sarkar, S.; O'Kane, C.J.; Rubinsztein, D.C. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of huntington disease. J. Cell Sci. 2008, 121, 1649–1660. [Google Scholar] [CrossRef] [PubMed]
- Brunsveld, L.; Waldmann, H.; Huster, D. Membrane binding of lipidated Ras peptides and proteins - the structural point of view. Biochim. Biophys. Acta-Biomembr. 2009, 1788, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Shinde, S.R.; Maddika, S. Post translational modifications of Rab GTPases. Small GTPases 2018, 9, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Baron, R.A.; Seabra, M.C. Rab geranylgeranylation occurs preferentially via the pre-formed Rep-RGGT complex and is regulated by geranylgeranyl pyrophosphate. Biochem. J. 2008, 415, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.H.; Ko, T.P.; Wang, A.H.J. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 2002, 269, 3339–3354. [Google Scholar] [CrossRef]
- Joberty, G.; Tavitian, A.; Zahraoui, A. Isoprenylation of Rab proteins possessing a C-terminalCAAX motif. FEBS Lett. 1993, 330, 323–328. [Google Scholar] [CrossRef]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.Q.; Ali, B.R.; Ramalho, J.S.; Godfrey, R.F.; Barral, D.C.; Hume, A.N.; Seabra, M.C. Membrane targeting of Rab GTPases is influenced by the prenylation motif. Mol. Biol. Cell 2003, 14, 1882–1899. [Google Scholar] [CrossRef]
- Peters, C.; Wagner, M.; Volkert, M.; Waldmann, H. Bridging the gap between cell biology and organic chemistry: Chemical synthesis and biological application of lipidated peptides and proteins. Naturwissenschaften 2002, 89, 381–390. [Google Scholar] [CrossRef]
- Silvius, J.R. Lipidated peptides as tools for understanding the membrane interactions of lipid-modified proteins. Curr. Top. Membranes 2002, 52, 371–395. [Google Scholar]
- Prakash, P.; Gorfe, A.A. Lessons from computer simulations of Ras proteins in solution and in membrane. Biochim. Et Biophys. Acta-Gen. Subj. 2013, 1830, 5211–5218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalli, A.C.; Sansom, M.S.P. Interactions of peripheral proteins with model membranes as viewed by molecular dynamics simulations. Biochem. Soc. Trans. 2014, 42, 1418–1424. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.P.; Jiang, T.; Sun, C.; Lihan, M.; Pant, S.; Mahinthichaichan, P.; Trifan, A.; Tajkhorshid, E. Characterization of lipid–protein interactions and lipid-mediated modulation of membrane protein function through molecular simulation. Chem. Rev. 2019, 119, 6086–6161. [Google Scholar] [CrossRef] [PubMed]
- Gorfe, A.A.; Pellarin, R.; Caflisch, A. Membrane localization and flexibility of a lipidated ras peptide studied by molecular dynamics simulations. J. Am. Chem. Soc. 2004, 126, 15277–15286. [Google Scholar] [CrossRef]
- Leung, K.F.; Baron, R.; Seabra, M.C. Geranylgeranylation of Rab GTPases. J. Lipid Res. 2006, 47, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Edler, E.; Schulze, E.; Stein, M. Membrane localization and dynamics of geranylgeranylated Rab5 hypervariable region. Biochim. Biophys. Acta-Biomembr. 2017, 1859, 1335–1349. [Google Scholar] [CrossRef]
- Siu, S.W.I.; Vacha, R.; Jungwirth, P.; Bockmann, R.A. Biomolecular simulations of membranes: Physical properties from different force fields. J. Chem. Phys. 2008, 128, 12. [Google Scholar] [CrossRef]
- Douliez, J.P.; Ferrarini, A.; Dufourc, E.J. On the relationship between C-C and C-D order parameters and its use for studying the conformation of lipid acyl chains in biomembranes. J. Chem. Phys. 1998, 109, 2513–2518. [Google Scholar] [CrossRef] [Green Version]
- Saxton, M.J. Single-particle tracking: The distribution of diffusion coefficients. Biophys. J. 1997, 72, 1744–1753. [Google Scholar] [CrossRef]
- Siebrasse, J.P.; Veith, R.; Dobay, A.; Leonhardt, H.; Daneholt, B.; Kubitscheck, U. Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc. Natl. Acad. Sci. USA 2008, 105, 20291–20296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunwald, D.; Martin, R.M.; Buschmann, V.; Bazett-Jones, D.P.; Leonhardt, H.; Kubitscheck, U.; Cardoso, M.C. Probing intranuclear environments at the single-molecule level. Biophys. J. 2008, 94, 2847–2858. [Google Scholar] [CrossRef] [PubMed]
- Filippov, A.; Oradd, G.; Lindblom, G. The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys. J. 2003, 84, 3079–3086. [Google Scholar] [CrossRef]
- Jacobson, K.; Ishihara, A.; Inman, R. Lateral diffusion of proteins in membranes. Annu. Rev. Physiol. 1987, 49, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, K.; Liu, P.; Lagerholm, B.C. The lateral organization and mobility of plasma membrane components. Cell 2019, 177, 806–819. [Google Scholar] [CrossRef]
- Edler, E.; Stein, M. Simulation of membrane-bound Rab5: Structure and dynamics at different lipid compositions. Eur. Biophys. J. Biophys. Lett. 2015, 44, 165. [Google Scholar]
- Vogel, A.; Katzka, C.P.; Waldmann, H.; Arnold, K.; Brown, M.F.; Huster, D. Lipid modifications of a Ras peptide exhibit altered packing and mobility versus host membrane as detected by 2h solid-state NMR. J. Am. Chem. Soc. 2005, 127, 12263–12272. [Google Scholar] [CrossRef]
- Edler, E.; Stein, M. Recognition and stabilization of geranylgeranylated human Rab5 by the GDP dissociation inhibitor (GDI). Small GTPases 2019, 10, 227–242. [Google Scholar] [CrossRef]
- BenTal, N.; Honig, B.; Peitzsch, R.M.; Denisov, G.; McLaughlin, S. Binding of small basic peptides to membranes containing acidic lipids: Theoretical models and experimental results. Biophys. J. 1996, 71, 561–575. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [Green Version]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Pastor, R.W.; MacKerell, A.D. Development of the CHARMM force field for lipids. J. Phys. Chem. Lett. 2011, 2, 1526–1532. [Google Scholar] [CrossRef]
- Venable, R.M.; Sodt, A.J.; Rogaski, B.; Rui, H.; Hatcher, E.; MacKerell, A.D.; Pastor, R.W.; Klauda, J.B. CHARMM all-atom additive force field for sphingomyelin: Elucidation of hydrogen bonding and of positive curvature. Biophys. J. 2014, 107, 134–145. [Google Scholar] [CrossRef]
- Lim, J.B.; Rogaski, B.; Klauda, J.B. Update of the cholesterol force field parameters in CHARMM. J. Phys. Chem. B 2012, 116, 203–210. [Google Scholar] [CrossRef]
- Foloppe, N.; MacKerell, A.D. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000, 21, 86–104. [Google Scholar] [CrossRef]
- Pavelites, J.J.; Gao, J.L.; Bash, P.A.; Mackerell, A.D. A molecular mechanics force field for NAD+, NADH, and the pyrophosphate groups of nucleotides. J. Comput. Chem. 1997, 18, 221–239. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.H.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An n(log(n)) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Chiantia, S.; London, E. Sphingolipids and membrane domains: Recent advances. Handb. Exp. Pharmacol. 2013, 33–55. [Google Scholar]
- Douliez, J.P.; Leonard, A.; Dufourc, E.J. Restatement of order parameters in biomembranes - calculation of C-C bond order parameters from c-d quadrupolar splittings. Biophys. J. 1995, 68, 1727–1739. [Google Scholar] [CrossRef]






| Membrane | System | POPC | Chol | PSM | POPE | POPS | PI(3)P |
|---|---|---|---|---|---|---|---|
| Pure POC | HVR206–215 | 1.36 | |||||
| GG-Rab5 | 1.63 ± 0.08 | ||||||
| (GG,GG)-Rab5 | 1.44 ± 0.03 | ||||||
| Ternary Membrane | HVR206–215 | 0.78 | 0.59 | 0.60 | |||
| GG-Rab5 | 0.83 ± 0.03 | 0.65 ± 0.05 | 0.72 ± 0.04 | ||||
| (GG,GG)-Rab5 | 0.75 ± 0.02 | 0.55 ± 0.02 | 0.64 ± 0.02 | ||||
| Six-Component Membrane | HVR206–215 | 0.74 | 0.59 | 0.62 | 0.68 | 0.65 | 0.62 |
| GG-Rab5 | 0.79 ± 0.01 | 0.62 ± 0.02 | 0.66 ± 0.02 | 0.73 ± 0.01 | 0.70 ± 0.03 | 0.67 ± 0.03 | |
| (GG,GG)-Rab5 | 0.74 ± 0.01 | 0.58 ± 0.01 | 0.61 ± 0.01 | 0.67 ± 0.01 | 0.64 ± 0.01 | 0.60 ± 0.01 |
| Diffusion Coefficient/ 10−7 cm2 s−1 | HVR206–215 | Mono-GG Rab5 | Double-GG Rab5 |
|---|---|---|---|
| Pure POPC | 0.78 ± 0.13 | 1.59 ± 0.33 | 0.93 ± 0.09 |
| Ternary Membrane | 0.47 ± 0.10 | 1.31 ± 0.30 | 0.51 ± 0.06 |
| Six-component Membrane | 0.43 ± 0.08 | 1.49 ± 0.08 | 0.43 ± 0.05 |
| Parameter | System | Pure POPC | Ternary Membrane | Six-Component Membrane |
|---|---|---|---|---|
| Membrane thickness/ nm | HVR206–215,a | 3.87 | 4.60 | 4.54 |
| Mono-GG Rab5b | 3.88 ± 0.02 | 4.61 ± 0.01 | 4.57 ± 0.01 | |
| Double-GG Rab5c | 3.88 ± 0.03 | 4.61 ± 0.02 | 4.56 ± 0.02 | |
| Lipid area/ nm2 | HVR206–215,a | 0.662 | 0.437 | 0.466 |
| Mono-GG Rab5b | 0.854 ± 0.006 | 0.534 ± 0.004 | 0.518 ± 0.005 | |
| Double-GG Rab5c | 0.652 ± 0.044 | 0.432 ± 0.002 | 0.460 ± 0.002 | |
| Lipid order parameter | HVR 206–215,a | 0.176 | 0.327 | 0.305 |
| Mono-GG Rab5b | 0.174 ± 0.001 | 0.326 ± 0.002 | 0.309 ± 0.001 | |
| Double-GG Rab5c | 0.176 ± 0.001 | 0.327 ± 0.001 | 0.309 ± 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Münzberg, E.; Stein, M. Structure and Dynamics of Mono- vs. Doubly Lipidated Rab5 in Membranes. Int. J. Mol. Sci. 2019, 20, 4773. https://doi.org/10.3390/ijms20194773
Münzberg E, Stein M. Structure and Dynamics of Mono- vs. Doubly Lipidated Rab5 in Membranes. International Journal of Molecular Sciences. 2019; 20(19):4773. https://doi.org/10.3390/ijms20194773
Chicago/Turabian StyleMünzberg, Eileen, and Matthias Stein. 2019. "Structure and Dynamics of Mono- vs. Doubly Lipidated Rab5 in Membranes" International Journal of Molecular Sciences 20, no. 19: 4773. https://doi.org/10.3390/ijms20194773
APA StyleMünzberg, E., & Stein, M. (2019). Structure and Dynamics of Mono- vs. Doubly Lipidated Rab5 in Membranes. International Journal of Molecular Sciences, 20(19), 4773. https://doi.org/10.3390/ijms20194773